Abstract
Complex psychiatric disorders are resistant to whole-genome analysis due to genetic and etiological heterogeneity. Variation in resting electroencephalogram (EEG) is associated with common, complex psychiatric diseases including alcoholism, schizophrenia, and anxiety disorders, although not diagnostic for any of them. EEG traits for an individual are stable, variable between individuals, and moderately to highly heritable. Such intermediate phenotypes appear to be closer to underlying molecular processes than are clinical symptoms, and represent an alternative approach for the identification of genetic variation that underlies complex psychiatric disorders. We performed a whole-genome association study on alpha (α), beta (β), and theta (θ) EEG power in a Native American cohort of 322 individuals to take advantage of the genetic and environmental homogeneity of this population isolate. We identified three genes (SGIP1, ST6GALNAC3, and UGDH) with nominal association to variability of θ or α power. SGIP1 was estimated to account for 8.8% of variance in θ power, and this association was replicated in US Caucasians, where it accounted for 3.5% of the variance. Bayesian analysis of prior probability of association based upon earlier linkage to chromosome 1 and enrichment for vesicle-related transport proteins indicates that the association of SGIP1 with θ power is genuine. We also found association of SGIP1 with alcoholism, an effect that may be mediated via the same brain mechanisms accessed by θ EEG, and which also provides validation of the use of EEG as an endophenotype for alcoholism.
Keywords: alcoholism, electroencephalogram, endophenotype, genetics, whole-genome association
Genetic studies of behavior and psychiatric disorders are hampered by etiologic heterogeneity of these complex phenotypes. Addiction vulnerability arises from both internalizing (emotional) and externalizing (dyscontrol) behavioral dimensions (1), and both of these broad aspects of behavior are strongly influenced by early life trauma and other gene/environment interactions (2). Etiologic heterogeneity dilutes power to detect genetic effects, and is a reason for failures to detect and replicate genome-wide associations (GWAS) in complex disorders. Increasing sample size does not remove underlying heterogeneity and can introduce additional confounds. In neuropsychiatry, these considerations have led to the use of intermediate phenotypes (or endophenotypes) that are heritable, relevant to disease, and have good measurement properties and assay variation more closely related to gene function (3) as surrogates to probe the underlying biology of complex disorders. Risk genes for schizophrenia have recently been identified using quantitative variables derived from functional magnetic resonance imaging in comparatively small cohorts (4). Therefore, an important strategy to increase power for GWAS of psychiatric disorders might be the use of endophenotypes.
The resting electroencephalogram (EEG) is a safely and inexpensively obtained phenotype relevant to normal behavioral variation and to psychiatric disease. The EEG recorded at the scalp is the sum of postsynaptic currents of synchronously depolarized, radially oriented pyramidal cells in the cerebral cortex, and reflects rhythmic electrical activity of the brain. EEG patterns dynamically and quantitatively index cortical activation, cognitive function, and state of consciousness. EEG traits were among the original intermediate phenotypes in neuropsychiatry, having been first recorded in humans in 1924 by Hans Berger, who documented the α rhythm, seen maximally during states of relaxation with eyes closed, and supplanted by faster β waves during mental activity. EEG can be used clinically for the evaluation and differential diagnosis of epilepsy and sleep disorders, differentiation of encephalopathy from catatonia, assessment of depth of anesthesia, prognosis in coma, and determination of brain death (5, 6). EEG also has moderate predictive value for personality variation and psychiatric disease including depression (7), bipolar disorder (8), attention-deficit/hyperactivity disorder (9), and obsessive-compulsive disorder (10). Increased β power is associated with both alcoholism and family history of alcoholism (11, 12), θ power is altered in alcoholics (13–15), and reduced α power has been associated with a family history of alcoholism and with alcoholism with comorbid anxiety disorders (16, 17). However, the EEG is not clinically useful for diagnosis of any specific psychiatric disorder.
The stability and heritability of the EEG make it suitable for genetic analysis. Under standardized conditions the resting EEG is a stable trait in healthy adults, with high test-retest correlations [e.g., 0.7 even at >10 months (18)]. The marked interindividual variability in the resting EEG spectral band power is largely genetically determined and heritability of EEG spectral power is uniformly high for all wave forms (19, 20).
We have performed GWAS in a sample of Plains American Indians, in which α (8–13 Hz), β (13–30 Hz), and θ (3–8 Hz) EEG spectral power are moderately heritable with high test-retest correlations over 2 years (15). Notably, this sample represents a population isolate evidencing a small but, as it turned out, useful degree of European admixture, and is genetically distinct from other Native American populations. In this dataset, common familial traits such as alcoholism and other psychiatric disorders do not themselves generate statistical signals approaching genome-wide significance. However, we were able to identify a set of genes and possible pathways that affect α and θ EEG wave forms that are relevant to some of these same complex behavioral traits. Two of the gene associations were replicated in a US Caucasian dataset.
Results
Genome-Wide Significant Loci for Resting EEG Power.
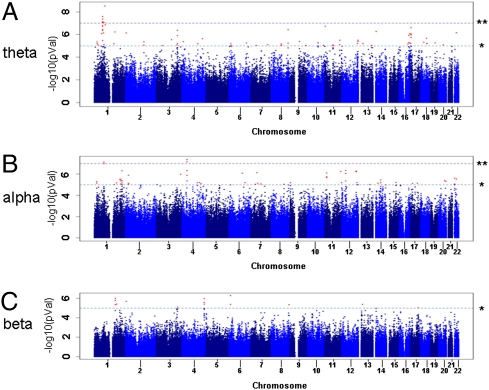
Five separate genomic regions, three for θ power [Fig. 1A, all on chromosome (chr) 1] and two for α power (Fig. 1B, chr 1 and 4), showed genome-wide significant association (P < 1.23 × 10−7) to a single EEG trait (Table 1). No genome-wide significant associations were observed for β power (Fig. 1C) or for the complex psychiatric diagnosis of alcohol use disorder (SI Text), demonstrating that random genome-wide significant signals are not generated for common familial traits in a dataset of this size. Quantile-quantile plots show an excess of low P values for θ power but little or no excess for α power or β power (SI Text). Due to the population structure of the Plains Indians there exist close family relationships between study participants confirmed by λ values of 1.3 (α and θ) and 1.1 (β), and these could lead to spurious association. Empirically derived P values (Table 1) that correct for family structure for nominally significant single-nucleotide polymorphisms (SNPs) (P < 1 × 10−5) remained suggestive of association, although no marker remained significant after Bonferroni correction.
Fig. 1.
Manhattan plots for resting EEG spectral power: θ (A), α (B), and β (C). The three plots show association (−log10P value) (y axis) for individual SNPs (uncorrected for family structure) against chromosome position (x axis). Threshold P values of 1 × 10−7 (**) and 1 × 10−5 (*) are indicated by dotted lines.
Table 1.
Markers with significant association to EEG traits accompanied by subthreshold markers within significant regions
| EEG frequency band | SNP | Location | Position | Gene | HWE P value | Plains Indians minor allele frequency | Plains Indians P value | Plains Indians corrected P value* | Caucasians P value |
| Theta | rs6588207 | 1p31.3 | 66839146 | SGIP1 | 0.5972 | 0.03 | 4.24 × 10−8 | 7.1 × 10−6 | 0.056 |
| Theta | rs10889635 | 1p31.3 | 66848163 | SGIP1 | 0.5526 | 0.03 | 2.52 × 10−8 | 4.9 × 10−6 | 0.013 |
| Theta | rs6656912 | 1p31.3 | 66856259 | SGIP1 | 1 | 0.02 | 4.24 × 10−7 | 1.61 × 10−5 | ND |
| Theta | rs6681460 | 1p31.3 | 66895645 | SGIP1 | 0.4659 | 0.03 | 7.30 × 10−8 | 1.3 × 10−5 | ND |
| Theta | rs10789215 | 1p31.3 | 66923773 | SGIP1 | 1 | 0.03 | 8.51 × 10−8 | 1.2 × 10−5 | NS |
| Theta | rs2146904 | 1p31.3 | 66932924 | SGIP1 | 1 | 0.03 | 4.37 × 10−7 | 3.63 × 10−5 | ND |
| Theta | rs536410 | 1p31.3 | 66963672 | SGIP1 | 0.8271 | 0.04 | 1.69 × 10−7 | 1.43 × 10−5 | ND |
| Theta | rs2483704 | 1p31.3 | 66967941 | SGIP1 | 1 | 0.04 | 3.63 × 10−7 | 2.59 × 10−5 | ND |
| Theta | rs2916 | 1p31.3 | 66989285 | TCTEX1D1 | 0.6424 | 0.03 | 1.87 × 10−7 | 1.92 × 10−5 | ND |
| Theta | rs6696780 | 1p31.1 | 76504495 | ST6GALNAC3 | 0.2083 | 0.02 | 8.47 × 10−8 | 3.89 × 10−5 | ND |
| Alpha | rs410076 | 1p31.1 | 76774219 | ST6GALNAC3 | 0.6564 | 0.35 | 1.03 × 10−7 | 1.1 × 10−6 | NS |
| Alpha | rs172714 | 1p31.1 | 76787701 | ST6GALNAC3 | 0.2563 | 0.35 | 7.34 × 10−8 | 3.0 × 10−7 | NS |
| Theta | rs1245665 | 1p31.1 | 81800834 | LPHN2 | 1 | 0.02 | 3.00 × 10−9 | 7.1 × 10−6 | NS |
| Alpha | rs2608830 | 4p13 | 39135549 | RPL9/LIAS | 0.5768 | 0.11 | 1.29 × 10−6 | 6.8 × 10−6 | ND |
| Alpha | rs2259073 | 4p13 | 39135549 | UGDH | 0.4204 | 0.11 | 4.55 × 10−7 | 2.5 × 10−6 | ND |
| Alpha | rs2687964 | 4p13 | 39151469 | UGDH | 0.6905 | 0.10 | 4.54 × 10−6 | 1.39 × 10−5 | ND |
| Alpha | rs7667766 | 4p13 | 39172667 | UGDH | 0.6632 | 0.11 | 7.04 × 10−8 | 8.0 × 10−7 | NS |
| Alpha | rs6827264 | 4p13 | 39194505 | UGDH | 0.5355 | 0.11 | 4.23 × 10−8 | 7.0 × 10−7 | NS |
HWE, Hardy-Weinberg equilibrium; ND, not determined; NS, not significant.
*Empirical P values correcting for participant relatedness.
Genomic Regions Associated with θ Power.
The three genomic regions associated with θ power are all located at chr 1p31, each identifying a single candidate gene (Table 1). In Plains Indians, four significant (and two subthreshold) alleles are represented on two closely related low-frequency haplotypes (SI Text) that span almost the whole of the SH3-domain GRB2-like (endophilin) -interacting protein 1 gene (SGIP1), and an additional three subthreshold markers lie within another haplotype block that extends 3′ from SGIP1 into the adjacent Tctex1-containing 1 gene (TCTEX1D1). The association with SGIP1 accounts for 8.8% of observed variation in θ power. Two missense polymorphisms in exon 7 of SGIP1, rs17490057 and rs7526812, can be excluded as the functional locus because of their distributions within haplotypes. Four haplotypes, including a common one (frequency = 0.24) carrying the increased θ power-associated alleles, are present in HapMap Caucasians (SI Text). A single marker significant for θ power was found in both ST6 (α-N-acetylneuraminyl-2,-3-β-galactosyl-1,3)-N-acetylgalactosamine-α-2,6-sialyltransferase 3 (ST-6GALNAC3: rs6696780) and latrophilin 2 (LPHN2: rs12145665). ST6GALNAC3 is an integral Golgi membrane protein which catalyzes the transfer of sialic acids to carbohydrate groups on glycoproteins and glycolipids. LPHN2 is a G-protein-coupled receptor related to the receptor that binds black widow (Latrodectus) spider venom in synaptic membranes (21).
Genomic Regions Associated with α Power.
Two genomic regions show significant association to α power. Although the ST6GALNAC3 gene overlaps with our findings for θ power, the two α power-associated markers (rs410076 and rs172714) both lie within the third intron of ST6GALNAC3 and are in linkage disequilibrium with each other, but not with the θ power-associated rs6696780 located more than 169 kb away. The signals for association to α and θ power are therefore likely to be independent in this gene region. α power was also associated with UDP-glucose dehydrogenase gene (UGDH) through two markers, rs7667766 and rs6817264, that are located in introns 1 and 9, respectively. An additional three SNPs adjacent to this region that show subthreshold association to α power are located in the 5′-regulatory region of UGDH, and in the adjacent lipoic acid synthetase (LIAS) and ribosomal protein-like 9 (RPL9) genes. All five SNPs are within a single haplotype block extending almost 1 Mb (SI Text) and encompassing UGDH, LIAS, and RPL9, along with klotho β (KLB), a component of the receptor for fibroblast growth factor 19 (FGF19). UGDH is an integral Golgi membrane protein involved in posttranslational modification of extrinsic proteins, whose expression is up-regulated in response to TGF-β and hypoxia, a risk factor for schizophrenia.
Validation of SGIP1 Association.
We estimated posterior probabilities of association (PPAs) for the top two hits in SGIP1: rs6588207 and rs10889635. Posterior probability of association is based on Bayesian method and can be interpreted directly as a probability (22). The log10(BF) for the combination of associated markers was 4.11 and for rs10889635 was 4.40 [σa and σd as described by Servin and Stephens (23) for phenotypes normally distributed across genotypes]. We assumed a value of 10−4 for π (prior probability of H1), a choice based on earlier evidence of linkage of EEG variation to this chr 1 region in the Collaborative Study on the Genetics of Alcoholism (COGA) dataset, and nominally significant enrichment for θ power-associated proteins that mediate vesicle transport (P = 0.040) based on gene ontology. A moderate PPA of 0.56 was observed for the region whereas rs10889635 showed a PPA of 0.72, indicating that this region is associated with θ power among Plains Indians, justifying analysis of these markers in an independent sample after consideration of PPAs under defined prior assumptions.
Replication in the United States.
Caucasians.
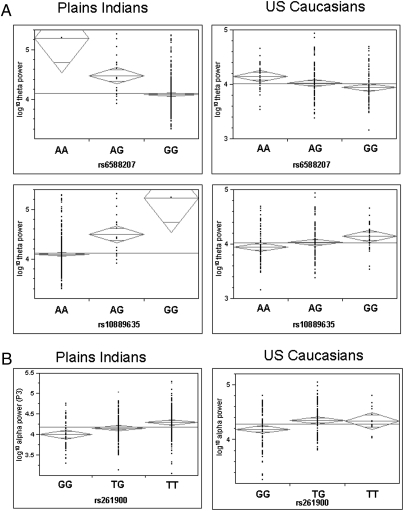
The five genome-wide significant regions were tested in an independent US Caucasian dataset. Association of increased θ power to the G allele of rs10889635 within SGIP1 was replicated (P = 0.013) (Fig. 2A), accounting for 3.5% of the variance in θ power in this population, although the association does not remain significant after correction for multiple testing. Within SGIP1, the A allele of rs6588207, significant in Plains Indians, showed a similar trend (P = 0.056) but did not reach significance. No ST6GALNAC3, LPHN2, or UGDH markers showed association in Caucasians.
Fig. 2.
Replication of EEG/SNP associations in Caucasians. (A) One-way analysis of variance (ANOVA) for rs6588207 and rs10889635 in SGIP1, against (−log10)θ power averaged across posterior electrodes. The G allele of rs10889635 showed association to increased θ power in both Plains Indians (P = 2.52 × 10−8) and Caucasians (P = 0.013). The A allele of rs6588207 showed association to increased θ power in Plains Indians (P = 4.24 × 10−8) with a similar trend in Caucasians (P = 0.056). (B) One-way ANOVA for rs261900 in BICD1, against (−log10) α power in Plains Indians (at the P3 electrode) and Caucasians (−log10)α power averaged across posterior electrodes. The T allele showed association to increased α power in both Plains Indians (P = 9.76 × 10−9) and Caucasians (P = 0.0023).
Subthreshold Associations with θ and α Power.
Other genomic regions showed association in the range of P = 1 × 10−5–1.22 × 10−7. It is likely that several represent chance findings, because either no annotated gene is present in the region or the gene would not appear to affect neuronal reactivity (SI Text). All potential candidate genes identified for each EEG spectral power (at significant or subthreshold level) are listed by function in SI Text.
Golgi Transport Candidates Among Subthreshold Signals.
Analysis of the gene ontology for all candidate genes suggested that a proportion of the genes are involved in similar cellular processes (SI Text). Like SGIP1, four of these genes, BICD1, FAM125B, ANKRD27, and C11orf2, are either involved in retrograde Golgi transport or at least contain a domain similar to those found in vesicular transport proteins. Additionally, RTN1, RPH3AL, and CECR2 are involved in vesicle-mediated transport and neuroendocrine processes. BICD1 (bicaudal D homolog 1) is particularly interesting in that although there was only subthreshold association [rs261900: P = 4.32 × 10−7 (uncorrected)/4.5 × 10−6 (empirical)] to α power across posterior scalp electrodes, the T allele of rs261900 is also associated (P = 0.0023) with increased α power in Caucasians (Fig. 2B).
β EEG Power.
Several candidate genes associated to β power have functions that may affect neuronal electrical activity neurons. These include the α3 glycine receptor gene (GLRA3), a brain-expressed synaptic receptor that inhibits neuroexcitability, with four SNPs in a single haplotype block. rs12027066 is near ALD9A (aldehyde dehydrogenase, 9 family, member A1, an enzyme involved in GABA synthesis). Genetic variation in GABA receptors has been associated with disorders for which EEG has been proposed as an intermediate phenotype, including alcoholism (24) and schizophrenia (25).
SGIP1 Association with θ Power Is Revealed by Admixture Mapping.
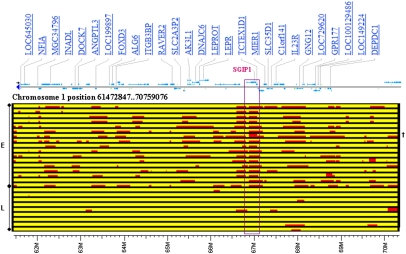
The Plains Indian population was selected because of its characteristics as a population isolate with low admixture (European admixture; mean 5.23% and median 1.6%) and is genetically distinct even from other Indian tribes (SI Text). However, despite the isolate character of Plains Indians, analysis of the θ power association on chr 1 suggests that this signal was detected via admixture mapping, making it feasible to identify the introgressed European-derived chromosomal region and extended haplotypes harboring the putative functional locus. The minor allele frequency (MAF) in Plains Indians for the significant SNP rs10889635 is low (>0.03) compared with Caucasians (MAF = 0.41). When MAF is plotted against European ethnic factor score, a measure of European ancestry, the frequency of the minor G allele increases with increasing European admixture (SI Text), an effect seen also for θ power-associated markers in ST6GALNAC3 and LPHN2, and to a lesser extent in the more distant SEP15 gene. Localized admixture for the region surrounding SGIP1 on chr 1 was assessed for the 17 Plains Indians carrying the G allele of rs10889635 and 9 Plains Indians with low European admixture (mean = 1.3%, median = 1.0%). In all carriers of the G allele, at least one copy of SGIP1 was European-derived (Fig. 3). The relatively small size of the computed blocks of admixture suggests that introduction of European DNA into this population is not recent (26). The association of SGIP1 to θ power is unlikely to be a stratification artifact because there is no correlation between degree of European ancestry and EEG power in any of the frequency bands (SI Text), and because the association at SGIP1 was replicated in Caucasians. Instead, this appears to be a serendipitous example of admixture mapping within a GWAS. The nonreplicating associations of LPHN2 and ST6GALNAC3 to θ power are likely to arise as a consequence of this admixture, and probably do not represent true associations. For α power, European ancestry does not predict the associated minor allele frequencies in ST6GALNAC3 and UGDH (SI Text), indicating these associations may be valid but population-specific.
Fig. 3.
Local admixture of chr 1 region (61.47–70.76 Mb) containing SGIP1 in 17 Plains Indians (E) carrying the θ power-associated G allele of rs10889635 compared with 9 Plains Indians (L) with low European admixture. The analysis shows a common European chromosomal segment at SGIP1 in all Plains Indians carrying the G allele of rs10889635, indicating that the association signal (P = 2.52 × 10−8) is in part due to admixture at the SGIP1 locus. The only Plains Indian homozygous for the G allele of rs10889635 is indicated (†).
EEG as an Endophenotype for Alcoholism.
To evaluate whether SGIP1 is a candidate gene for alcoholism, in addition to EEG variation we analyzed significant SGIP1 markers for association with alcohol use disorders (AUD) in the Plains Indian cohort (Table 2). Alleles significant (but subthreshold) for association with increased θ power were underrepresented in individuals with AUD. Reduced θ power in alcoholics had been reported for this dataset (15). The present data indicate a relationship between SGIP1 genotype and AUD. Significant SNPs at other loci did not show association to AUD except for SNPs at LPHN2 and ST6GALNAC3 that were linked to rs1088935 by their common European ancestry. Moreover, strength of these associations weakened with increasing distance from SGIP1.
Table 2.
Comparison of minor allele frequency for seven SGIP1 SNPs between individuals with alcohol use disorder and controls shows significant underrepresentation of alleles associated with increased θ power
| rs6588207 | rs10889635 | rs66881460 | rs10789215 | rs2146904 | rs536410 | rs2483704 | |
| Alcohol use disorder | 0.018 | 0.018 | 0.018 | 0.018 | 0.018 | 0.020 | 0.018 |
| Controls | 0.054 | 0.051 | 0.045 | 0.039 | 0.051 | 0.067 | 0.061 |
| P value, 1 degree of freedom | 0.005 | 0.009 | 0.028 | 0.076 | 0.009 | 0.001 | 0.002 |
| χ2 | 7.843 | 6.78 | 4.824 | 3.139 | 6.87 | 10.903 | 10.024 |
Discussion
This GWAS in Plains American Indians identified two chromosomal regions, both previously implicated in linkage studies, containing multiple candidate genes for variability in resting EEG power. SGIP1 and ST6GALNAC3 lie within a linkage peak on chr 1p (95–115 cM), linked to β2 EEG spectral power (16–20 Hz) in the COGA (27, 28). UGDH, LIAS, RPL9, and KLB lie in a region of chr 4 previously linked to EEG in this Plains Indian dataset (15), together with an additional Native American population (29) and in the COGA dataset (30). Additionally, the study has identified BICD1 located on chr 12p11 as a candidate gene for resting α power variability.
Linkage studies have identified other chromosome regions potentially harboring genes involved in EEG variability. Low-voltage α power mapped to chr 20q (31). Convergence of linkage was found for α, β, and θ resting EEG power to chr 5q13-14 in this Plains Indian dataset, suggesting common loci that regulate EEG across the frequency spectrum (15). This finding was not replicated in this GWAS, most likely because of the relatively small effect of this locus on EEG power. In the present study, failure to replicate associations of ST6GALNAC3 and UGDH in Caucasians may reflect cross-population heterogeneity in loci influencing the EEG. With regard to this point, the haplotype block structure of the chr 4 region containing UGDH is more complex in Caucasians compared with Plains Indians, and it is possible that the association signal arises from variation in other genes in the region including KLB and LIAS. KLB is an interesting candidate because FGF-19 signaling affects cortical development (32) and voltage-gated Na+ channels, thereby altering neuronal reactivity (33).
The EEG, although an intermediate phenotype, is itself a complex trait reflecting the rhythmic electrical activity of the brain, which is probably modulated by many loci. Discovery of a single locus accounting for 8.8% of variance in θ EEG is therefore fortuitous. In US Caucasians this locus accounts for 3.5% of variance, the diminution due to the increased frequency of the informative alleles and relative haplotype frequencies.
The data from the significant and subthreshold genes suggest that many of the genes involved in determining EEG variability are involved in intracellular transport processes rather than cell-surface receptor activity, although two subthreshold genes were involved in glycine ion channel and GABAergic function. SGIP1 is directly involved in vesicle formation at the plasma membrane through interactions with phospholipids and EPS15, an adaptor protein. Knockdown of SGIP1 expression reduces clathrin-mediated endocytosis (34). Alterations in the efficiency of endocytosis might change neuronal activity via receptor turnover or neurotransmitter uptake. The rabphilin 3A-like (without C2 domains) gene (RPHAL) generated only a modest P value [6.2 × 10−6 (uncorrected)/1.2 × 10−5 (empirical)] but is involved in exocytosis and vesicle transport, and is on chr 17 located within a linkage peak identified for the maximum-drinks phenotype in the COGA study (35). Coincidentally, D17S1308, which gave the strongest linkage signal, maps adjacent to the vacuolar protein-sorting 53 homolog gene (VPS53), a gene involved in retrograde vesicle trafficking in the late Golgi. BICD1 is also involved in vesicle trafficking and is essential for retrograde Golgi-to-endoplasmic reticulum transport via interaction with the adaptor protein Rab6a. Disruption of BICD1/Rab6a interaction uncouples vesicles from the dynein-dynactin transport machinery, resulting in the accumulation of vesicles at the cell periphery (36). BICD1 also appears to have an important function in nuclear positioning in neurons during development. The involvement of BICD1 with EEG variability is further suggested because BICD1 interacts directly with LIS1 (37), a protein that also interacts directly with DISC1 (38). Variation at DISC1 has been associated with alteration in the P300-evoked potential, another phenotype of neuronal reactivity (39).
Two other subthreshold genes, cadherin 13 (CDH13) and fibroblast growth factor 14 (FGF14), were identified in a GWAS for nicotine dependence (40) and more recently in a GWAS for alcohol dependence (41). FGF14 potentially alters neuronal reactivity through interaction with voltage-gated Na+ channels (33). Analysis of response to contextual imagery in cigarette smokers suggested that alteration of EEG activity may be a common feature associated with craving (42). The adaptor-related protein complex 2, α subunit2 gene (AP2A2), which encodes another protein involved in endocytosis, is located close to D11S1984, which generated the maximum linkage signal to alcohol dependence in a southwestern Native American population (29). Although it appears that our main findings are for genes that solely influence EEG power, convergences of findings at the subthreshold level to previous results in addictions suggest that by using an intermediate phenotype we potentially identified genes that have a general influence on addiction.
The Plains Indians are a population isolate, offering advantages of reduced genetic and environmental heterogeneity. As described, this is a relatively nonadmixed tribe, with median European admixture of 1.5%. Although it is a family-based sample (SI Text), we were able to correct our analyses for the relatedness of the participants.
In conclusion, this GWAS suggests that neuronal excitability in the brain is determined in part by the ability to recycle neuronal membrane components. Replication of association of SGIP1 to θ power validates the use of the GWAS approach on intermediate phenotype datasets of modest size. The success of this study in identifying novel genes and cellular processes in determining EEG traits demonstrates that if there are loci of moderate effect size, which is more likely for an intermediate phenotype in a population isolate, it may be unnecessary to have datasets numbering in the thousands, and that the analysis of accurately measured, heritable traits in relatively homogeneous populations is an informative alternative in the genetic analysis of complex psychiatric diseases and behavior.
Methods
Participant Recruitment.
Plains American Indians.
Participants were recruited from a tribe in rural Oklahoma (15) (fully described in SI Text). Written informed consent was obtained according to a human research protocol approved by the Tribal Council and the human research committee of the National Institute on Alcohol Abuse and Alcoholism. EEG was obtained from 359 people. The alcohol use disorder (AUD) cohort comprised 225 cases and 156 controls. AUD was defined as DSM-III alcohol abuse (without dependence) (222 individuals) or DSM-III alcohol dependence (3 individuals).
The replication dataset (described in SI Text) consisted of 185 European Americans (104 women and 81 men: 61 healthy controls and 124 individuals with a variety of DSM-III diagnoses) recruited in Bethesda, MD (43). Written informed consent was obtained via a human research protocol approved by the NIAAA institutional review board.
EEG Acquisition and Analysis.
Details of EEG methods and analysis are in Enoch et al. (15) and SI Text. Resting EEG was recorded using a fitted electrode cap in a field setting while the subject was seated with eyes closed and relaxed in a darkened noise-baffled room. To balance data quality against participant time, data were collected from six scalp electrodes (one frontal, two occipital, three parietal) selected to maximize information for α and θ power. Spectral power was determined for θ (3–8 Hz), α (8–13 Hz), and β (13–30 Hz), and log-transformed to normalize the distribution. For each frequency band, power was averaged for the five posterior electrodes to reduce variability due to individual electrode measurement and was justified by high correlations between α and θ power at the selected electrodes (15), coherence between these signals (44), and the higher θ and α activity in posterior and occipital regions (45). These data were used for association (see SI Text for means and ranges). No outliers were removed.
Genotyping.
Samples (399) were genotyped using the Illumina HapMap550K. Fourteen were discarded due to low genotype call rate (<97%), and four were discarded after gender and allele transmission testing. Genotypes were called using BeadStudio 3.2 (Illumina). Thirty-five replicate pairs showed average genotype reproducibility of 0.99994. Following quality control, association with log10 EEG power was performed on 322 individuals (137 male, 185 female) for whom EEG was available. Replication genotyping was performed by 5′-exonuclease assay.
Statistical Analysis.
Association was tested using the standard linear regression model in PLINK (46). SNPs with MAF < 1% or a call rate < 90% were excluded, leaving 405,281 SNPs. Threshold for association was set at <1.23 × 10−7 using the Bonferroni method. Posthoc analyses using sex and age as covariates both individually and together did not influence associations. P value inflation due to genetic relatedness was estimated using the GC program (47). To correct for family structure, QFAM family-based association tests for SNPs determined to be nominally significant were performed within PLINK with 107 rounds of permutation, yielding empirical P values. Following GWAS, we estimated PPAs for the two top SGIP1 markers, rs6588207 and rs10889635. Bayes factors (BF) were calculated using Bayesian IMputation-Based Association Mapping (http://quartus.uchicago.edu/~yguan/bimbam). PPAs were calculated as described in Stephens and Balding (22). Enrichment in gene ontology terms was detected using GOTM (48).
Local Admixture.
Localized admixture on chr 1 was detected using Local Ancestry in adMixed Populations (LAMP) (49) v2.0 with an r2 cutoff of 0.95 to minimize exclusion of informative SNPs due to variable LD between Europeans and American Indians. Computational runs using generation time estimates of 3, 5, 7, and 9 yielded similar estimates of local admixture. A recombination rate of 1 × 10−8 and seed α values of 0.04 (Europeans) and 0.96 (American Indians) were used.
Population Substructure and Admixture Determination.
Ancestry informative markers (186) (50) were typed in the EEG samples and the HGDP-CEPH Human Genome Diversity Cell Line Panel (http://www.cephb.fr/HGDP-CEPH-Panel). PHASE Structure 2.2 (http://pritch.bsd.uchicago.edu/software.html) was run using ancestry informative marker data from the EEG and CEPH datasets simultaneously to identify population substructure and compute individual ethnic factor scores.
Supplementary Material
Acknowledgments
We thank the study participants for their contributions, and Longina Akhtar and Elisa Moore for technical assistance. This study was funded by the intramural program of National Institute on Alcohol Abuse and Alcoholism.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0908134107/-/DCSupplemental.
References
- 1.Kendler KS, et al. The structure of genetic and environmental risk factors for DSM-IV personality disorders: A multivariate twin study. Arch Gen Psychiatry. 2008;65:1438–1446. doi: 10.1001/archpsyc.65.12.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caspi A, et al. Role of genotype in the cycle of violence in maltreated children. Science. 2002;297:851–854. doi: 10.1126/science.1072290. [DOI] [PubMed] [Google Scholar]
- 3.Gottesman II, Gould TD. The endophenotype concept in psychiatry: Etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 4.Potkin SG, et al. FBIRN. A genome-wide association study of schizophrenia using brain activation as a quantitative phenotype. Schizophr Bull. 2009;35:96–108. doi: 10.1093/schbul/sbn155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niedermeyer E. The normal EEG of the waking adult. In: Lopez de Silva F, editor. Electroencephalography: Basic Principles, Clinical Applications, and Related Fields. Baltimore: Williams and Wilkins; 1993. pp. 97–117. [Google Scholar]
- 6.Nunez PL, Srinivasan R. Electric Fields of the Brain. 2nd Ed. New York: Oxford Univ Press; 2005. p. 9. [Google Scholar]
- 7.Knott V, Mahoney C, Kennedy S, Evans K. EEG power, frequency, asymmetry and coherence in male depression. Psychiatry Res. 2001;106:123–140. doi: 10.1016/s0925-4927(00)00080-9. [DOI] [PubMed] [Google Scholar]
- 8.Cook BL, Shukla S, Hoff AL. EEG abnormalities in bipolar affective disorder. J Affect Disord. 1986;11:147–149. doi: 10.1016/0165-0327(86)90021-2. [DOI] [PubMed] [Google Scholar]
- 9.Barry RJ, Clarke AR, Johnstone SJ. A review of electrophysiology in attention-deficit/hyperactivity disorder: I. Qualitative and quantitative electroencephalography. Clin Neurophysiol. 2003;114:171–183. doi: 10.1016/s1388-2457(02)00362-0. [DOI] [PubMed] [Google Scholar]
- 10.Pogarell O, et al. Symptom-specific EEG power correlations in patients with obsessive-compulsive disorder. Int J Psychophysiol. 2006;62:87–92. doi: 10.1016/j.ijpsycho.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Rangaswamy M, et al. Beta power in the EEG of alcoholics. Biol Psychiatry. 2002;52:831–842. doi: 10.1016/s0006-3223(02)01362-8. [DOI] [PubMed] [Google Scholar]
- 12.Pollock VE, Earleywine M, Gabrielli WF. Personality and EEG beta in older adults with alcoholic relatives. Alcohol Clin Exp Res. 1995;19:37–43. doi: 10.1111/j.1530-0277.1995.tb01470.x. [DOI] [PubMed] [Google Scholar]
- 13.Rangaswamy M, et al. Theta power in the EEG of alcoholics. Alcohol Clin Exp Res. 2003;27:607–615. doi: 10.1097/01.ALC.0000060523.95470.8F. [DOI] [PubMed] [Google Scholar]
- 14.Coutin-Churchman P, Moreno R, Añez Y, Vergara F. Clinical correlates of quantitative EEG alterations in alcoholic patients. Clin Neurophysiol. 2006;117:740–751. doi: 10.1016/j.clinph.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 15.Enoch MA, et al. Common genetic origins for EEG, alcoholism and anxiety: The role of CRH-BP. PLoS One. 2008;3:e3620. doi: 10.1371/journal.pone.0003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finn PR, Justus A. Reduced EEG alpha power in the male and female offspring of alcoholics. Alcohol Clin Exp Res. 1999;23:256–262. [PubMed] [Google Scholar]
- 17.Enoch MA, et al. Association of low-voltage alpha EEG with a subtype of alcohol use disorders. Alcohol Clin Exp Res. 1999;23:1312–1319. [PubMed] [Google Scholar]
- 18.Salinsky MC, Oken BS, Morehead L. Test-retest reliability in EEG frequency analysis. Electroencephalogr Clin Neurophysiol. 1991;79:382–392. doi: 10.1016/0013-4694(91)90203-g. [DOI] [PubMed] [Google Scholar]
- 19.Smit DJ, Posthuma D, Boomsma DI, Geus EJ. Heritability of background EEG across the power spectrum. Psychophysiology. 2005;42:691–697. doi: 10.1111/j.1469-8986.2005.00352.x. [DOI] [PubMed] [Google Scholar]
- 20.van Beijsterveldt CE, Molenaar PC, de Geus EJ, Boomsma DI. Heritability of human brain functioning as assessed by electroencephalography. Am J Hum Genet. 1996;58:562–573. [PMC free article] [PubMed] [Google Scholar]
- 21.Lang J, Ushkaryov Y, Grasso A, Wollheim CB. Ca2+-independent insulin exocytosis induced by α-latrotoxin requires latrophilin, a G protein-coupled receptor. EMBO J. 1998;17:648–657. doi: 10.1093/emboj/17.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stephens M, Balding DJ. Bayesian statistical methods for genetic association studies. Nat Rev Genet. 2009;10:681–690. doi: 10.1038/nrg2615. [DOI] [PubMed] [Google Scholar]
- 23.Servin B, Stephens M. Imputation-based analysis of association studies: Candidate regions and quantitative traits. PLoS Genet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Enoch MA. The role of GABA(A) receptors in the development of alcoholism. Pharmacol Biochem Behav. 2008;90:95–104. doi: 10.1016/j.pbb.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lang UE, Puls I, Muller DJ, Strutz-Seebohm N, Gallinat J. Molecular mechanisms of schizophrenia. Cell Physiol Biochem. 2007;20:687–702. doi: 10.1159/000110430. [DOI] [PubMed] [Google Scholar]
- 26.Stephens JC, Briscoe D, O'Brien SJ. Mapping by admixture linkage disequilibrium in human populations: Limits and guidelines. Am J Hum Genet. 1994;55:809–824. [PMC free article] [PubMed] [Google Scholar]
- 27.Ghosh S, et al. Linkage mapping of beta 2 EEG waves via non-parametric regression. Am J Med Genet B Neuropsychiatr Genet. 2003;118B:66–71. doi: 10.1002/ajmg.b.10057. [DOI] [PubMed] [Google Scholar]
- 28.Porjesz B, et al. Linkage disequilibrium between the beta frequency of the human EEG and a GABAA receptor gene locus. Proc Natl Acad Sci USA. 2002;99:3729–3733. doi: 10.1073/pnas.052716399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long JC, et al. Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet. 1998;81:216–221. doi: 10.1002/(sici)1096-8628(19980508)81:3<216::aid-ajmg2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 30.Porjesz B, et al. Linkage and linkage disequilibrium mapping of ERP and EEG phenotypes. Biol Psychol. 2002;61:229–248. doi: 10.1016/s0301-0511(02)00060-1. [DOI] [PubMed] [Google Scholar]
- 31.Steinlein O, Anokhin A, Yping M, Schalt E, Vogel F. Localization of a gene for the human low-voltage EEG on 20q and genetic heterogeneity. Genomics. 1992;12:69–73. doi: 10.1016/0888-7543(92)90408-k. [DOI] [PubMed] [Google Scholar]
- 32.Borello U, et al. FGF15 promotes neurogenesis and opposes FGF8 function during neocortical development. Neural Dev. 2008;3:17. doi: 10.1186/1749-8104-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laezza F, et al. The FGF14(F145S) mutation disrupts the interaction of FGF14 with voltage-gated Na+ channels and impairs neuronal excitability. J Neurosci. 2007;27:12033–12044. doi: 10.1523/JNEUROSCI.2282-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uezu A, et al. SGIP1α is an endocytic protein that directly interacts with phospholipids and Eps15. J Biol Chem. 2007;282:26481–26489. doi: 10.1074/jbc.M703815200. [DOI] [PubMed] [Google Scholar]
- 35.Saccone NL, et al. A genome screen of maximum number of drinks as an alcoholism phenotype. Am J Med Genet. 2000;96:632–637. doi: 10.1002/1096-8628(20001009)96:5<632::aid-ajmg8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 36.Matanis T, et al. Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dynein-dynactin motor complex. Nat Cell Biol. 2002;4:986–992. doi: 10.1038/ncb891. [DOI] [PubMed] [Google Scholar]
- 37.Claussen M, Suter B. BicD-dependent localization processes: From Drosophilia development to human cell biology. Ann Anat. 2005;187:539–553. doi: 10.1016/j.aanat.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 38.Brandon NJ, et al. Disrupted in Schizophrenia 1 and Nudel form a neurodevelopmentally regulated protein complex: Implications for schizophrenia and other major neurological disorders. Mol Cell Neurosci. 2004;25:42–55. doi: 10.1016/j.mcn.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 39.Blackwood DH, Muir WJ. Clinical phenotypes associated with DISC1, a candidate gene for schizophrenia. Neurotox Res. 2004;6:35–41. doi: 10.1007/BF03033294. [DOI] [PubMed] [Google Scholar]
- 40.Uhl GR, et al. Molecular genetics of nicotine dependence and abstinence: Whole genome association using 520,000 SNPs. BMC Genet. 2007;8:10. doi: 10.1186/1471-2156-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Treutlein J, et al. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry. 2009;66:773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knott V, et al. EEG correlates of imagery-induced cigarette craving in male and female smokers. Addict Behav. 2008;33:616–621. doi: 10.1016/j.addbeh.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 43.Enoch MA, et al. Association of low-voltage alpha EEG with a subtype of alcohol use disorders. Alcohol Clin Exp Res. 1999;23:1312–1319. [PubMed] [Google Scholar]
- 44.Winterer G, et al. EEG phenotype in alcoholism: Increased coherence in the depressive subtype. Acta Psychiatr Scand. 2003;108:51–60. doi: 10.1034/j.1600-0447.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- 45.Porjesz B, et al. The utility of neurophysiological markers in the study of alcoholism. Clin Neurophysiol. 2005;116:993–1018. doi: 10.1016/j.clinph.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 46.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Devlin B, Bacanu SA, Roeder K. Genomic Control to the extreme. Nat Genet. 2004;36:1129–1130. doi: 10.1038/ng1104-1129. author reply 1131. [DOI] [PubMed] [Google Scholar]
- 48.Zhang B, Schmoyer D, Kirov S, Snoddy J. GOTree Machine (GOTM): A web-based platform for interpreting sets of interesting genes using gene ontology hierarchies. BMC Bioinformatics. 2004;5:16. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sankararaman S, Sridhar S, Kimmel G, Halperin E. Estimating local ancestry in admixed populations. Am J Hum Genet. 2008;82:290–303. doi: 10.1016/j.ajhg.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hodgkinson CA, et al. Addictions biology: Haplotype-based analysis for 130 candidate genes on a single array. Alcohol. 2008;43:505–515. doi: 10.1093/alcalc/agn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.