Abstract
Until now, mortality and spreading mechanisms of influenza pandemics have been studied only for the 1918, 1957, and 1968 pandemics; none have concerned the 19th century. Herein, we examined the 1889 “Russian” pandemic. Clinical attack rates were retrieved for 408 geographic entities in 14 European countries and in the United States. Case fatality ratios were estimated from datasets in the French, British and German armies, and morbidity and mortality records of Swiss cities. Weekly all-cause mortality was analyzed in 96 European and American cities. The pandemic spread rapidly, taking only 4 months to circumnavigate the planet, peaking in the United States 70 days after the original peak in St. Petersburg. The median and interquartile range of clinical attack rates was 60% (45–70%). The case fatality ratios ranged from 0.1% to 0.28%, which is comparable to those of 1957 and 1968, and 10-fold lower than in 1918. The median basic reproduction number (R0) was 2.1, which is comparable to the values found for the other pandemics, despite the different viruses and contact networks. R0 values varied widely from one city to another, and only a small minority of those values was within the range in which modelers’ mitigation scenarios predicted effectiveness. The 1889 and 1918 R0 correlated for the subset of cities for which both values were available. Social and geographic factors probably shape the local R0, and they could be identified to design optimal mitigation scenarios tailored to each city.
Keywords: epidemics, history, reproduction number, attack rate, epidemiology
The ongoing A/H1N1 global influenza epidemic was qualified as a “pandemic” on 11 June 2009, when the World Health Organization raised its alert to phase 6. This new episode adds to a series of 11 pandemics that occurred in the 18th, 19th, and 20th century (1). Interpandemic intervals have ranged from 8 (between 1781 and 1789) to a maximum of 42 years (between 1847 and 1889). This maximum value of 42 was just 1 year more than the time elapsed between the last pandemic of the 20th century (1968) and the first pandemic of the 21st century (2009).
During this long period, the threat of the pandemic—and the need for preparedness—increased with the years passed, which fueled the investigation of mitigation scenarios based on social distancing, vaccination, and/or antiviral treatment (2 –7). These studies have relied on mathematical modeling for which “plausible” parameters could be derived only from analysis of past pandemics (2, 6). Most studies focused on 1918, which became de facto the stereotype of the feared pandemic to come. Many researchers extracted quantitative information, especially the value of the basic reproduction number R0 (the mean number of persons infected by a single infectious individual introduced into a totally susceptible population) for that pandemic.
Comparatively, the two other pandemics of the 20th century (1957 and 1968) were much less studied, and no quantitative analysis at all was undertaken for any of the 19th century pandemics, most likely because it was wrongly assumed that not enough data existed to support such analyses. We focused here on the 1889–1890 season when the last pandemic of the 19th century, known as the “Russian flu,” occurred. The 1889 Russian flu, possibly caused by an H3N8 virus (8), was the first pandemic to occur in a highly connected world: at that time, the 19 largest European countries, including Russia, had 202,887 km of railroads (9), which is more than now (10) (Appendix S1, section 1.2 showing the 19th century railway network). Transatlantic travel by boat took less than 6 days at that time (11), instead of less than 1 day now (which is not a substantial difference, given the time scale of the global spread of a pandemic).
We searched all available information on the 1889 pandemic, and analyzed its spread, the sizes and dynamics of the local epidemics, and the associated mortality experience. Our goals were to extend the knowledge on influenza pandemics, and to verify that the study of global pandemics of the 19th century was feasible.
Results
Global Spread.
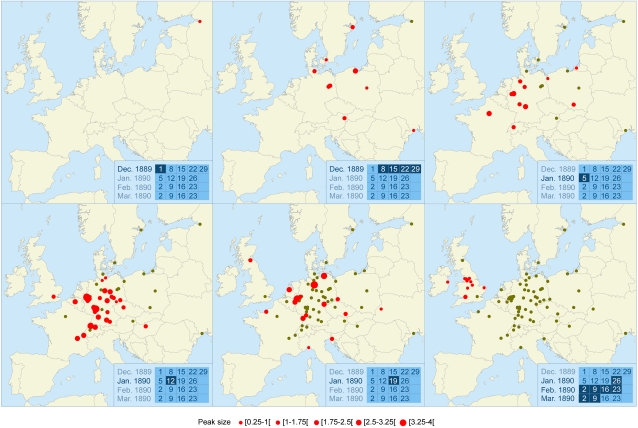
The dissemination of the 1889 pandemic was extremely rapid (Fig. 1 and Movie S1). The mortality peaks occurred during the weeks ending 1 December in St. Petersburg, 22 December in Germany, 5 January in Paris, and 12 January in the United States. The median delay between the notification of the first case and the mortality peak was 5.0 weeks [interquartile range (IQR) = 4.0–6.5] (Appendix S1 , section 1.5). The mean (± SD) speed of the pandemic was estimated at 394 (±255) km/week in continental Europe and 1015 (±727) km/week in the United States.
Fig. 1.
Spread of the 1889 pandemic throughout continental Europe (2006 boundaries) during six successive periods. Each panel refers to a period of one or several weeks shown on the calendar in the Inset. Red dots indicate the places of the mortality peaks that occurred during this period and are proportional to peak size. Green dots indicate cities after the mortality peak has passed. The whole set of European cities studied can be visualized in the last panel (68 in continental Europe and 14 in the United Kingdom and Ireland). Movie S1 shows the week-by-week spread in Europe and in the United States.
Clinical Attack Rate and Mortality Burden.
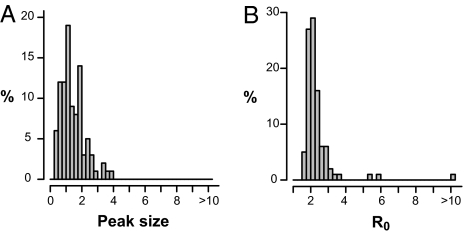
The weighted mean of the 408 clinical attack rates (CAR) found was 50% . The median was 60% and the IQR was 45–70%. The 1888–1889 all-causes mortality time course was studied for 96 studied cities of Europe and the United States. The peak size [defined as (maximum mortality – baseline mortality) / baseline mortality in %] varied widely from city to city (Fig. 2A): the mean peak size was 142%, the median was 124%, and the IQR was 87–186%. The mortality burden was estimated in each of the 96 studied cities. The mean was 0.17%, the median was 0.16% of the population, and the IQR was 0.13–0.25%. The 1889 case fatality ratio (CFR) was directly obtained from data in the French and British armies, and was indirectly estimated from surveys performed in seven Swiss cities and in the German army. The CFR ranged between 0.1% and 0.28% (Appendix S1, section 1.7).
Fig. 2.
Distributions of 1889 pandemic mortality peaks and R0 values. (A) Peak sizes for 96 cities worldwide. The peak size for each city is the relative mortality increase at the peak's highest amplitude compared with baseline mortality; e.g., a peak size of 3 indicates that the excess mortality at the peak was three times the baseline mortality. (B) Estimates of R0 values, assuming CFR = 0.2%, and respective Weibull distributed latent and infectious periods with means of 1.6 and 1.0 days.
Determinants of Local Epidemic Dynamics.
The R0 were estimated in 96 cities studied using a discrete time Susceptible–Exposed (Latent)–Infected–Removed (SEIR) model as described by Mills et al. (12). The median R0 was 2.1 (IQR = 1.9–2.4) (Fig. 2B). In all, 7% of the R0 values were less than 1.8 and 22% were less than 1.9, which are two limits below which some national (4) or global (3) mitigation strategies were found to be effective. These estimates were only weakly (<16% variation) changed by uncertainties in the definition of baseline mortality and assumptions on the CFR (sensitivity analyses in Appendix S1, section 3).
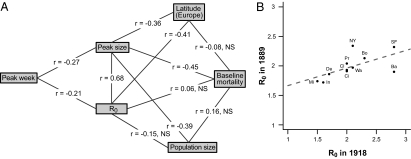
Rank correlations were computed between R0, city population, geographic location (latitude/longitude), peak size, peak time and the average number of inhabitants per dwelling. Peak size and R0, which were strongly correlated as a consequence of the SEIR modeling, were both negatively correlated with latitude in Europe (Fig. 3A), but not in the United States. In the 35 US and European cities with available data, the average number of inhabitants per dwelling (13) was significantly correlated with R0 (r = 0.57, P < 10−3). Finally, the 1889 R0 and those computed by Mills et al. (12) for the 1918 pandemic were positively correlated (Fig. 3B) for the 11 US cities that were common to both studies.
Fig. 3.
(A) Spearman rank correlations between R0 estimations, population size, peak sizes, peak times (for all cities), and latitudes (for European cities). Significance levels: P < 10−5 for |r| > 0.45; P < 10−4 for |r| > 0.40; P < 10−3 for |r| > 0.35, P < 0.05 for |r| > 0.20. (B) Correlation between 1918 (12) and 1889 R0 values (this study) for the 11 US cities studied in common (Ba, Baltimore; Bo, Boston, Ci, Cincinnati; Cl, Cleveland; De, Detroit; In, Indianapolis; Mi, Minneapolis; NY, New York; Pr, Providence; SF, San Francisco; Wa, Washington). Pearson's correlation = 0.62 (P < 0.05). Dotted line corresponds to linear regression.
Discussion
A first result of the analysis of the 1889 pandemic is that such historical quantitative analyses that extend our knowledge of what is a “typical” influenza pandemic are feasible. The available data were not just aggregated mortality statistics. Huge epidemiologic surveys were already done at that time. For example, the German Sanitary services conducted a survey among the 16,000 physicians of the Empire, with a response rate of 21% (Appendix S2). It would likely be possible to replicate the kind of analysis that we performed of the 1889 pandemic to other previous pandemics.
A second result is that the severity of the 1889 pandemic was mild and similar to the pandemics in 1957 and 1968: the 1889 CFR was ~0.15%, vs. 0.13% in 1957 (14) and less than 0.1% in 1968 (8). This was roughly 10-fold smaller than the 1918 pandemic CFR (12).
The rapid progression of the 1889 pandemic demonstrates that slower surface travel, even with much smaller traveler flows, sufficed to spread the pandemic across all of Europe and the United States in ~4 months. This observation supports mathematical model results, which anticipated that restricting air transportation would have little, if any, effect (15, 16). One possible hypothesis is that the important predictor of the speed of the pandemic is not the absolute numbers of passengers traveling between cities but the “connectedness” of the network of cities: (i.e., the facility to connect any city A with any city B with railway routes).
The negative correlation that we found between the excess mortality (as measured by the peak value) and the R0, on one hand, and the latitude in Europe, on the other hand, is in line with what was observed in 1918, when the mortality burden was higher in the southern region of Europe than in the northern part (17). The correlation analysis showed that there were characteristics of the cities, such as the number of inhabitants per dwelling, that predicted the R0, hence the strength of the local epidemic. The fact that the R0 values were positively correlated almost 30 years apart indicates that there are also intrinsic geographic, weather, and/or sociological characteristics of the cities that could be used to predict which cities are at risk for high R0. The identification of such predictors would be of practical interest, as they could guide local mitigation policies.
In conclusion, adding the 1889 pandemic to the short list of the three pandemics studied previously led to the emergence of some similarities among them: R0 values were consistently ~2 and CAR within the range 30–60% (18 –25). The only striking difference concerned mortality: 1889 was comparable to 1957, 1968, and to the “pseudo-pandemics” of 1947 and 1977–1978 (26), thereby making the 1918 pandemic even more exceptional. Finally, the 2009 A/H1N1, at least so far, seems to be another “mild” pandemic, similar to what our analysis revealed the 1889 Russian flu to be.
Methods
Data.
We retrieved information on the 1889–1890 all-cause mortality time series in government reports and journals for 172 cities in Europe and the United States, from which the 96 cities with populations exceeding 35,000 inhabitants and a unique mortality peak during the period were selected for analysis. The date of notification of the first case was known in 23 of the 96 cities (Appendix S1, section 1, data sources description, and Dataset S1).
The case fatality ratio (CFR), which is the proportion of infected patients who die from the disease, was estimated in young adults based on statistics collected at the time on several hundreds of thousands of soldiers in the French, British, and German armies. We derived CFR estimations from the ratio of mortality to morbidity statistics obtained in a survey made in seven Swiss cities (total population ~170,000 inhabitants) (Appendix S1, section 1.7).
Data on the clinical attack rate (CAR), which is the proportion of the population with clinical signs during the epidemic, were retrieved from 408 administrative units located in 14 European countries and in the United States (Appendix S1, section 1.6, and Dataset S2). Of these administration units, 331 were from Switzerland, 52 from Germany, and the 31 remaining from 13 countries (Appendix S1, section 1.6 clinical attack rate per country). The weighted mean of CAR was computed for each country, with weights proportional to the countries’ population sizes.
Descriptive Analysis of the Mortality Patterns in the 96 Cities.
The speed of the epidemic was estimated from the ratios of the distance to the times elapsed between mortality peaks observed in all combinations of two cities.
Baseline mortality was defined by averaging mortality rates over the first 4 weeks of the datasets. This estimation of baseline was found highly correlated to those performed by considering control periods in 1888, or longer, when available (Appendix S1, section 1.4.2). The variation found in the estimates of the baseline guided the sensitivity analyses (Appendix S1, section 3.3). The quality of the excess mortality estimations was assessed via comparison with the mortality rate by respiratory diseases, which was known directly in eight US cities in this study (Appendix S1, section 1.4.2).
The peak size, defined as the relative increase of mortality at the peak's highest amplitude compared with baseline mortality, was chosen as an indicator of the pandemic impact on the mortality in each studied city.
The mortality burden in each city was estimated as the difference between all-cause and baseline mortalities over the period (peak week –4 to peak week +4). In eight cities with sufficiently long datasets, an alternative estimate of the mortality burden was obtained by using a baseline computed with a periodic regression model (27). The two estimates differed by less than 25% on average.
SEIR Model.
We analyzed all-cause–mortality kinetics for the 96 selected cities; their R0 were estimated using a discrete time SEIR model describing the dynamics of local epidemics after introduction of one infectious case (Appendix S1, section 2). The estimation of the generation time and infectious period was based on the data accrued in a comprehensive review of virus-excretion profiles obtained after experimental influenza infection of 1,280 volunteers (28). That review did not find significant differences between the virus-load kinetics of the groups infected with different influenza strains (B, H1N1, and H3N2). The average generation time (i.e., the time for a primary infected individual to transmit the infection to a secondary case) was 2.6 days. The model that we developed (Appendix S1, section 2.1) found latent and infectious periods lasting respectively 1.6 ± 0.3 and 1.0 ±1.0 days (means ± SD). We then fitted the observed excess mortality to the SEIR model, assuming a CFR of 0.2% (Fig. 4 and Appendix S1 , section 2.2).
Fig. 4.
Fit of influenza mortality rates obtained with the SEIR model for the 10 cities with the largest population sizes. Thin lines indicate raw data; bold lines indicate model fits.
Supplementary Material
Acknowledgments
We thank Dr. Mills and Dr. Lipsitch for providing the individual 1918 R0 values that they estimated, and Dr. Lipsitch for critical comments on an earlier version of this paper. We also thank Camille Pelat and Amandine Lacombe for help in database management and programming, and Georges Marion and Virginie Hogtchoko for help in translation of the German reports. This work was supported by European Union contracts InfTrans (6th PCRD) and FluModCont (7th PCRD).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000886107/-/DCSupplemental.
References
- 1.Morens DM, Fauci AS. The 1918 influenza pandemic: Insights for the 21st century. J Infect Dis. 2007;195:1018–1028. doi: 10.1086/511989. [DOI] [PubMed] [Google Scholar]
- 2.Cauchemez S, Valleron AJ, Boëlle PY, Flahault A, Ferguson NM. Estimating the impact of school closure on influenza transmission from Sentinel data. Nature. 2008;452:750–754. doi: 10.1038/nature06732. [DOI] [PubMed] [Google Scholar]
- 3.Colizza V, Barrat A, Barthelemy M, Valleron AJ, Vespignani A. Modeling the worldwide spread of pandemic influenza: Baseline case and containment interventions. PLoS Med. 2007;4:e13. doi: 10.1371/journal.pmed.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferguson NM, et al. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature. 2005;437:209–214. doi: 10.1038/nature04017. [DOI] [PubMed] [Google Scholar]
- 5.Halloran ME, et al. Modeling targeted layered containment of an influenza pandemic in the United States. Proc Natl Acad Sci USA. 2008;105:4639–4644. doi: 10.1073/pnas.0706849105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Longini IM, Jr, Halloran ME, Nizam A, Yang Y. Containing pandemic influenza with antiviral agents. Am J Epidemiol. 2004;159:623–633. doi: 10.1093/aje/kwh092. [DOI] [PubMed] [Google Scholar]
- 7.Longini IM, Jr, et al. Containing pandemic influenza at the source. Science. 2005;309:1083–1087. doi: 10.1126/science.1115717. [DOI] [PubMed] [Google Scholar]
- 8.Taubenberger JK, Morens DM, Fauci AS. The next influenza pandemic: Can it be predicted? JAMA. 2007;297:2025–2027. doi: 10.1001/jama.297.18.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell BR. International Historical Statistics. 6th edition. New York: Palgrave Macmillan; 2007. [Google Scholar]
- 10.Fremdling R. European Railways 1825-2001, An Overview. Groningen: Groningen Growth and Development Centre; 2002. [Google Scholar]
- 11.Newman J. The Blue Riband of the North Atlantic, Westbound and Eastbound Holders. Available at: http://www.greatships.net/riband.html. Accessed December 3, 2009.
- 12.Mills CE, Robins JM, Lipsitch M. Transmissibility of 1918 pandemic influenza. Nature. 2004;432:904–906. doi: 10.1038/nature03063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anonymous . Census of population and housing. Washington: Department of the interior; 1895. Report 1890 Census. [Google Scholar]
- 14.McDonald JC. Asian influenza in Great Britain 1957-58. Proc R Soc Med. 1958;51:1016–1018. [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper BS, Pitman RJ, Edmunds WJ, Gay NJ. Delaying the international spread of pandemic influenza. PLoS Med. 2006;3:e212. doi: 10.1371/journal.pmed.0030212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferguson NM, et al. Strategies for mitigating an influenza pandemic. Nature. 2006;442:448–452. doi: 10.1038/nature04795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ansart S, et al. Mortality burden of the 1918-1919 influenza pandemic in Europe. Influenza Other Respir Viruses. 2009;3:99–106. doi: 10.1111/j.1750-2659.2009.00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jordan WS, Jr, et al. A study of illness in a group of Cleveland families. XVII. The occurrence of Asian influenza. Am J Hyg. 1958;68:190–212. doi: 10.1093/oxfordjournals.aje.a119962. [DOI] [PubMed] [Google Scholar]
- 19.Hall CE, Cooney MK, Fox JP. The Seattle virus watch. IV. Comparative epidemiologic observations of infections with influenza A and B viruses, 1965-1969, in families with young children. Am J Epidemiol. 1973;98:365–380. doi: 10.1093/oxfordjournals.aje.a121566. [DOI] [PubMed] [Google Scholar]
- 20.Monto AS, Kioumehr F. The Tecumseh Study of Respiratory Illness. IX. Occurence of influenza in the community, 1966–1971. Am J Epidemiol. 1975;102:553–563. doi: 10.1093/oxfordjournals.aje.a112193. [DOI] [PubMed] [Google Scholar]
- 21.Hope-Simpson RE. First outbreak of Hong Kong influenza in a general practice population in Great Britain. A field and laboratory study. BMJ. 1970;3:74–77. doi: 10.1136/bmj.3.5714.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chin TD, Foley JF, Doto IL, Gravelle CR, Weston J. Morbidity and mortality characteristics of Asian strain influenza. Public Health Rep. 1960;75:149–158. [PMC free article] [PubMed] [Google Scholar]
- 23.Dunn FL, Carey DE, Cohen A, Martin JD. Epidemiologic studies of Asian influenza in a Louisiana parish. Am J Hyg. 1959;70:351–371. doi: 10.1093/oxfordjournals.aje.a120083. [DOI] [PubMed] [Google Scholar]
- 24.Woodall J, Rowson KE, McDonald JC. Age and Asian influenza, 1957. BMJ. 1958;2:1316–1318. doi: 10.1136/bmj.2.5108.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis LE, Caldwell GG, Lynch RE, Bailey RE, Chin TD. Hong Kong influenza: the epidemiologic features of a high school family study analyzed and compared with a similar study during the 1957 Asian influenza epidemic. Am J Epidemiol. 1970;92:240–247. doi: 10.1093/oxfordjournals.aje.a121203. [DOI] [PubMed] [Google Scholar]
- 26.Kilbourne ED. Influenza pandemics of the 20th century. Emerg Infect Dis. 2006;12:9–14. doi: 10.3201/eid1201.051254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelat C, et al. Online detection and quantification of epidemics. BMC Med Inform Decis Mak. 2007;7:29. doi: 10.1186/1472-6947-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carrat F, et al. Time lines of infection and disease in human influenza: A review of volunteer challenge studies. Am J Epidemiol. 2008;167:775–785. doi: 10.1093/aje/kwm375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.