Abstract
TcaR and IcaR are a weak and a strong negative regulator of transcription of the ica locus, respectively, and their presence prevents the poly-N-acetylglucosamine production and biofilm formation in Staphylococcus epidermidis. Although TcaR was shown to interact with the ica promoter, the precise binding region and the mechanism of interaction remained unclear. Here we present the 3D structure of TcaR in its apo form and in complex with salicylate as well as several aminoglycoside and β-lactam antibiotics. A comparison of the native and complex TcaR structures indicates that the mechanism of regulation involves a large conformational change in the DNA-binding lobe. Here, we deduced the consensus binding sequence of two [∼TTNNAA] hexamers embedded in a 16 bp sequence for a TcaR dimer. Six TcaR dimers bind specifically to three approximately 33 bp segments close to the IcaR binding region with varying affinities, and their repressor activity is directly interfered by salicylate and different classes of natural antimicrobial compounds. We also found in this study that the antimicrobial compounds we tested were shown not only to inhibit TcaR–DNA interaction but also to further induce biofilm formation in S. epidermidis in our in vivo assay. The results support a general mechanism for antibiotics in regulating TcaR–DNA interaction and thereby help understand the effect of antibiotic exposure on bacterial antibiotic resistance through biofilm formation.
Keywords: repressor, biofilm, DNA binding, multiple drug resistance, transcription regulation
Staphylococci are among the most common cause of bacterial infections in the community, which have been, and continue to be a major cause of human disease resulting in over than one million infections each year worldwide. Staphylococcus aureus is the best known and by far most studied staphylococcal species that produces hospital- and community-acquired infections, with methicillin-resistant S. aureus posing a serious public health threat (1, 2). S. epidermidis is the sister species of S. aureus, which often causes infection in immunocompromised individuals or those after damage to the epithelium. Similar to S. aureus, it produces biofilm to protect itself from host immune system and enhance their resistance to antibiotic chemotherapy (3). Because biofilm tolerance is of major clinical relevance and > 60% of the bacterial infections currently are involved in the biofilm formation (4), it is important to study those issues.
The key component of the biofilm extracellular matrix in S. epidermidis is polysaccharide intercellular adhesin (PIA) (5), an essential factor in biofilm formation composed of homopolymer of β-1,6-linked N-acetylglucosamine (GlcNAc). The production of PIA depends on the expression of the icaADBC operon, which encodes three membrane proteins (IcaA, IcaD, and IcaC) (6–8). The ica operon is negatively regulated by a repressor encoded by an upstream gene of IcaR (9–11). Even though our knowledge of the regulation of the ica locus is limited, recent studies have shown that TcaR acts as a regulatory factor to affect the transcription of icaADBC (11). The close relation between TcaR and IcaR proteins suggest a synergistic effect and they both regulate IcaA transcription, poly-N-acetylglucosamine production, and biofilm formation (11).
TcaR is a MarR family transcription regulator that is involved in teicoplanin and methicillin resistance (12). Therefore, it was originally described as a putative transcriptional regulator of the teicoplanin-associated locus (tca). As the MarR-type proteins can act as positive, negative, or both positive and negative regulators, TcaR also acts as a multifunctional regulator, not only as a regulatory factor to affect the transcription of icaADBC (11), the first regulator reported for cell wall-anchored proteins (spa and sasF), but also as the regulator of sarS (13, 14). In addition, TcaR upregulates sarS and thus spa expression, and represses the SasF production.
The crystal structures of a number of MarR family proteins have been reported, including MarR from Escherichia coli (15), SlyA from Enterococcus faecalis (16), OhrR from Bacillis subtilis (17), MexR from Pseudomonas aeruginosa (18), and MarR from Xanthomonas campestris (19). These structures revealed that MarR family proteins are all homodimers and that each monomer contains a winged helix-turn-helix (wHTH) DNA-binding domain.
Antibiotic resistance is an increasing problem throughout the developed world. Given the importance of MarR family protein in antibiotic resistance, an understanding of the mechanism of their regulation is urgently needed for efficient treatment of bacterial infections. Here, to study how the ica operon is controlled, and to investigate the role of TcaR in antibiotic resistance, TcaR from S. epidermidis was overexpressed in E. coli with a His tag to facilitate purification. The crystal structure of TcaR was solved by multiwavelength anomalous dispersion (MAD) method using protein containing seleno-methionines. Furthermore, DNA-binding assays, in vivo biofilm formation assay, and computer modeling were used to elucidate the regulation mechanism of the ica operon. Finally, we present the structures of TcaR complexed with salicylate (Sal) and four antibiotics, namely, ampicillin (Amp), kanamycin (Kan), methicillin (Meth), and penicillin G (PnG), to elucidate the regulation mechanism of TcaR. Several mutations of amino acids involved in the antibiotic binding were also made to validate the interactions between the antibiotics and TcaR.
Results and Discussion
Features of the TcaR Structure.
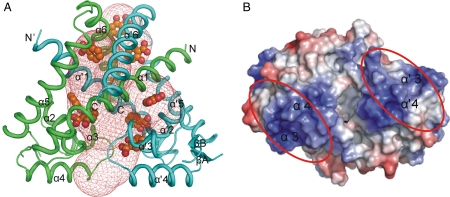
The crystal structures of the native TcaR and SeMet–TcaR derivative in the space group P61 were refined to 2.3 and 2.9 Å, respectively, both yielding low R and Rfree values and stereochemical deviations (Table S1). The refined TcaR structure contains two TcaR molecules (denoted chain A and chain B) in the asymmetric unit, forming a dimer, consistent with the earlier results that MarR family proteins bind to the operator as a dimer (15–18). The sequence alignment of TcaR with other MarR family proteins is shown in Fig. S1. The overall structure of TcaR belongs to the α/β family protein as observed in other MarR family proteins. The TcaR dimer adopts a triangular topology with each monomer consisted of secondary structure α1-α2-α3-α4-βA-βB-α5-α6 (Fig. 1A). The N and the C-terminus α-helices (α1, 5, 6) interdigitate with those of the other monomer to produce dimerization interaction. Strands βA and βB form a β-hairpin, which is slightly twisted and constitutes the typical winged motif. Between βA and βB is the flexible winged region (residues 84–96), which had poor electron density in the initial 2Fo-Fc map. In addition, using the program CAVER, we observed a highly porous structure with several cavities for potential ligand binding in the TcaR dimer. All cavities are located at the dimer interface, surrounded by helices α1, α2, α3, α5, and α6. Furthermore, these intermolecular cavities are large enough to encapsulate the ligands we tested in EMSA assay below.
Fig. 1.
Overall structure of apo TcaR. (A) The overall structure of the TcaR homodimer. The protein structure is shown as a ribbon diagram with chain A in green and chain B in cyan. In addition, we used the program to explore possible cavities in TcaR for ligand binding. The cavities identified by CAVER are shown in mesh representation and using a solvent probe of radius 2 Å. In addition, the binding positions of eight salicylate molecules in the TcaR–salicylate complex are shown in sphere, revealing that the porous structure of TcaR is able to interact with numerous small molecules. (B) The DNA-binding domains. The electrostatic surface of the dimer is viewed after a rotation of approximately 90° from (A), with a horizontal axis in the plane of paper. The electrostatic surfaces are drawn either blue for positive or red for negative. Possible domains involved in binding DNA are labeled as red ovals.
The wHTH DNA-binding domain is composed of α2-α3-α4-βA-W1-βB that adopts the winged-helix fold similar to the winged helix (wH) domain described in other MarR family protein (15–18). The DNA-binding site expected to be facing the DNA, is densely positively charged in a surface patch (Fig. 1B), which includes Lys60, Lys65, Arg70, Arg71, Lys73, Lys74, Lys82, Lys85, Arg93, Lys95, and Lys98, all of which are solvent-exposed and most are contributed by helix α3 and α4. Electrostatic interactions involving those positively charged amino acids must play most important roles for DNA binding.
Noncooperative Binding to the Operator DNA.
As reported earlier, E. coli MarR binds as a dimer at two separate but similar sites in marO (20). Footprinting experiments revealed that MarR protects approximately 21 bp of DNA on both strands at a single site and does not bend its target DNA (21). The crystal structure of OhrR-ohrA operator complex showed that the protein-DNA contact region includes the major groove of the -10 region (TACAAT) and indicated that OhrR, similar to MarR, represses transcription by blocking the access of RNA polymerase to this promoter region (17).
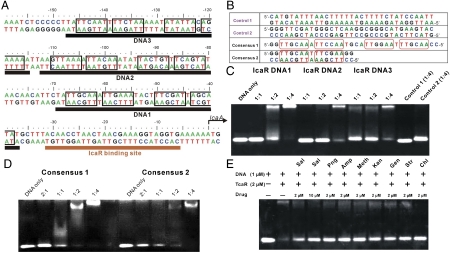
It is known that TcaR binds and regulates ica promoter (11), and the result of band shift assays indicated that there could be multiple TcaR recognition sites within the ica promoter sequence. To investigate the regulation mechanism of TcaR, we inspected the ica promoter sequence and proposed that there are three possible TcaR-binding sites on the ica operon, which are consecutive 33-mer pseudopalindromic sequences each containing consensus sequence TTNNAA (Fig. 2A). Alternatively, another possible arrangement of the consensus sequence TTNNAA is indicated in Fig. S2B. To track down the precise location of the TcaR-binding site, a series of dsDNA segments were designed and tested for TcaR-binding by electrophoretic motility shift assay. To determine whether TcaR binds to each of these three 33-mer sequences, we prepared 33-bp duplex DNAs and analyzed its interaction with TcaR using EMSA, with an increasing concentration of TcaR protein. The negative controls Control 1 and Control 2 are the AT rich and GC rich fragments of IcaA gene, respectively (Fig. 2B). As shown in Fig. 2C, TcaR does not bind to Control 1 and Control 2 DNA sequences, whereas TcaR appears to have a strong interaction with DNA fragments DNA1, DNA2, and DNA3, indicating that each of these DNAs interacts with TcaR. Among the latter segments, DNA1 showed the most significant effect by TcaR on its mobility, suggesting the strongest interaction between them. Consequently, the result indicates that the most favorite TcaR binding site is located immediately adjacent to the IcaR binding site.
Fig. 2.
EMSA of TcaR. (A) Part of the DNA sequence of ica operon of S. epidermidis and diagrams of the dsDNA probes with the consensus sequences TTNNAA are shown in black box. (B) The DNA fragments for EMSA experiments. (C) The EMSA of TcaR binding to three 33-mer dsDNA fragments of the ica operator with different DNA ratio. The probe was mixed with 1∶1, 1∶2, and 1∶4 molar ratio of TcaR (dimer), respectively. (D) The EMSA of TcaR binding to consensus DNA probe 1 and 2. The probe was mixed with 2∶1, 1∶1, 1∶2, and 1∶4 molar ratio of TcaR (dimer), respectively. (E) The effects of salicylate and antibiotics to the TcaR binding to DNA1 in EMSA experiment. DNA1 probe duplex of 1 μM was preincubated with 2 μM TcaR (dimer) at room temperature for 15 min before mixing with 2 μM antibiotics, followed by the same procedures as in the other assays.
TcaR is a multifunctional regulator that not only acts as a negative regulator for ica operator but also as a positive regulator for sarS gene and a negative regulator for sasF gene (13). To search for other possible TcaR-binding regions and to further clarify the regulation function of TcaR, we designed two DNA sequences according to the EMSA result in Fig. 2C, designated Consensus 1 and Consensus 2 (Fig. 2B), to test their interaction ability with TcaR. As seen in Fig. 2D, Consensus 1 had virtually the same binding strengths as DNA1 to the TcaR dimer with no cooperativity, whereas Consensus 2 appeared to have weaker interactions with TcaR. Therefore, we can use Consensus 1 DNA sequence to search for more TcaR binding sites on the entire S. epidermidis genome and to further unmask the multifunctional role of TcaR.
The Effects of Salicylate and Antibiotics on TcaR Repressor Activity.
As mentioned earlier, TcaR works as a repressor in the negative regulation of the icaADBC genes, whose expression is required for PIA synthesis and biofilm formation (10, 11). However, the mechanism of how TcaR inactivates icaADBC genes remains unclear. In the previous studies of MarR family protein, diverse anionic lipophilic (usually phenolic) effector molecules were shown to bind to the repressor and altered its conformation (15, 20, 22, 23). Recently, it was reported that biofilm formation in bacteria can be induced by aminoglycoside antibiotics (24). Here, to investigate the possible effect of some drugs on TcaR, eight compounds were tested for their potential inhibition on TcaR–DNA interaction (Fig. S3). These include salicylate, which is known to bind and inactivate MarR in E. coli (20), three beta-lactam antibiotics (penicillin G, ampicillin, and methicillin) that contain a β-lactam nucleus in their molecular structure and act by inhibiting the synthesis of the peptidoglycan layer of bacterial cell walls, three aminoglycoside antibiotics (kanamycin, gentamicin, and streptomycin) that are composed of several sugar groups and amino groups, and bacteriostatic antimicrobial (chloramphenicol), which is considered as a prototypical broad-spectrum antibiotic.
As shown in Fig. 2E, sodium salicylate interfered with the DNA-binding activity of TcaR at a concentration of 2 μM, and this effect was more pronounced at a higher concentration, suggesting that salicylate inhibited the formation of TcaR-ica operon complex. In addition, three beta-lactam antibiotics we identified in TcaR complexes also appeared to antagonize the DNA-binding activity of TcaR. Identical results were obtained with three other aminoglycoside antibiotics in the EMSA experiment. Moreover, another antibiotic, chloromphenicol, was also observed to inhibit the DNA-binding activity of TcaR with a significant greater extent. Thus, we believe that diverse kinds of antibiotics may interact with TcaR to regulate its repressor activity. Because some antibiotics, such as gentamicin and streptomycin, were shown to bind and inactivate IcaR (25), the experimental results suggest that several antibiotics at low concentrations can interact directly with TcaR and IcaR and thus interfere with the repressor activity of TcaR and IcaR to induce the expression of icaADBC and thereby elicit biofilm production in S. epidermidis, as a defense mechanism.
Crystal Structure of TcaR Complexed with Salicylate.
Up to now, only two structures of the MarR family protein complexes have been reported, one is complexed with DNA (17, 26), and the other is complexed with salicylate (15, 26, 27). To elucidate the mechanism of how the interaction with the ligands alleviates ica operon binding ability of TcaR, we solved several TcaR complex structures.
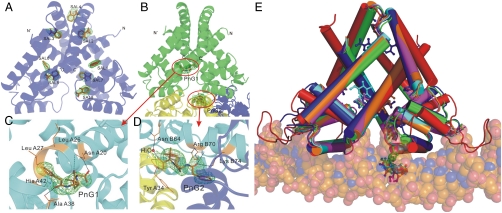
The TcaR–salicylate complex structure was determined to 2.45 Å resolution with eight molecules of salicylates bound to nonequivalent positions on each dimer designated SAL1 to SAL8 (Fig. 3A). Although the TcaR–salicylate structure and apo TcaR are well superimposed with a rmsd of 1.5 Å using 276 corresponding Cα atoms, the binding of salicylate drives a significant conformational change (Fig. S4 A and B). There is a more obvious conformational change in the DNA-binding domain (especially on helices α3 and α4) than the dimerization domain of each monomer. The additional results and discussion of the TcaR–salicylate complex (Fig. S5) could be found in SI Text. This conformational change is asymmetric and may cause the TcaR–DNA interactions to become unfavorable, as discussed below.
Fig. 3.
The structures of TcaR–ligand complex. (A) A ribbon diagram of TcaR in complex with salicylate. Salicylate binds to eight distinct locations in the dimer. (B) A ribbon diagram of TcaR in complex with penicillin G (PnG). PnG binds to two distinct locations in the dimer, with one binding in the interface of three TcaR dimers in the asymmetric unit along the crystallographic c axis. The central TcaR dimer is shown in green, and the right and left dimers are shown in blue and yellow, respectively. (C) One PnG molecule, PnG1 is located in the binding pocket directly at the junction of the DNA-binding domain and the dimerization domain. (D) In the other PnG binding site (PnG2), a PnG molecule is identified at crystal contact between three TcaR molecules related by crystallographic symmetry. (E) Superimposition of the TcaR-DNA model with TcaR–ligand complexes. The TcaR complexes of Sal (blue), PnG (green), Amp (orange), Meth (magenta), and Kan (cyan) revealed a significant conformational change at the WH domain.
Crystal Structure of TcaR Complexed with Antibiotics (Penicillin G).
E. coli MarR has been reported as a multidrug binding protein that can be inhibited from binding its cognate DNA site by several anionic compounds, which include 2,4-dinitrophenicol, plumbagin, menadione, and salicylate, with their apparent binding affinities to MarR being 250, 250, 800, and 500 μM, respectively (28). In addition to these bacterial multidrug resistance regulators, ampicillin at a concentration of 5 mM appeared to antagonize the DNA-binding activity of MarR, and a slight effect was also observed when the chloramphenicol concentration was increased to approximately 10 mM (28). These findings suggest that MarR has a broadly specific, low-affinity substrate binding ability. However, it is still unclear how the binding of these compounds interferes with the DNA-binding ability of MarR.
In the present study, we not only detected several antibiotics that can interfere with the DNA-binding activity of TcaR at a relative low concentration (2 μM, 5,000-fold lower than the determined binding constant for E. coli MarR), but also solved numerous TcaR–antibiotic complex structures to help elucidate the mechanism of drug-induced structural changes in TcaR. The TcaR–penicillin G (PnG) complex structure was determined to 2.55 Å resolution in the space group P61, and the structure was refined to a final Rwork value of 23.3% and an Rfree value of 27.3% (Table S2). There are two distinct PnG binding sites in each dimer, one is located at the interface between the DNA-binding domain and the helical dimerization domain and the other is identified at the interface of three TcaR dimer molecules in the asymmetric unit along the crystallographic c axis (Fig. 3B). As seen in Fig. S4 A and B, significant conformational changes between apo TcaR and the TcaR–PnG complex with a rmsd of 1.4 Å using 274 Cα atoms are also observed, particularly in the DNA-binding domain. These conformational changes shorten the C termini of the α3/α′3 helices from 26.0 Å for the TcaR–DNA model to 18.6 Å for the TcaR–PnG complex (Cα-Cα distance between Asn A61 and Asn B61) and the N termini of the α4/α′4 helices from 31.2 Å for TcaR–DNA model to 26.4 Å for the TcaR–PnG complex (Cα-Cα distance between Lys A65 and Lys B65). The antibiotics induced conformational change further interfering with the DNA binding of TcaR. This observation supports our EMSA assay in which several antibiotics exhibit inhibition effect on the TcaR–DNA formation.
Consistent with the observation in the TcaR–salicylate complex, the binding of PnG produced an asymmetric structural change in the wHTH motif. Superposition of apo TcaR on the PnG–TcaR complex shows that the A chains are superimposed better than the B chains with rmsd values of 0.9 and 1.3, respectively. This nonequivalent conformational change may be due to the nonequivalent position of PnG-binding sites on the dimer molecule. The detailed interactions of PnG1 and PnG2 are shown in Fig. 3 C and D. One PnG molecule, PnG1 is located in the binding pocket between the DNA-binding domain and the dimerization domain (Fig. 3C). This binding site is formed by helices α1 and α2 of chain A. The amino acid composition of this binding site includes Asn A20, Leu A26, Leu A27, Ala A38, and His A42. The amide and the carboxyl groups of Asn A20 are 2.9 and 3.2 Å away from the carboxylic acid of the thiazolidine ring and the carbonyl group on the β-lactam ring, respectively. Moreover, the NH of the backbone amide group of Ala A38 is just 2.9 Å away from the carbonyl group of PnG1. This indicates the interacting residues mentioned above might interact with PnG1 via ionic or hydrogen bond interaction. In the other PnG binding site (PnG2), a PnG molecule is identified at crystal contact between three TcaR molecules related by crystallographic symmetry (Fig. 3D). This binding pocket is formed by the helix α′4 of chain B, helix α1 of chain A on the symmetry related molecule at the bottom left corner, and helix α1 of chain A on the symmetry related molecule at the bottom right corner in the orientation shown in Fig. 3B. In this second binding site, PnG molecule is not only forming interaction with Asn B64, Arg B70, Lys B74, and water but also with Tyr A34 of the symmetry related molecule at the bottom left corner. The carboxylic acid of the thiazolidine ring of PnG2 forms hydrogen bond to water and NH of the backbone amide group of Tyr A43. The carbonyl group of PnG2 interacts with the side chain of residues Arg B70 and Lys B74. We have also solved three other TcaR–antibiotic complexes (two β-lactam antibiotics Amp, Meth, and an aminoglycoside Kan) (Fig. S6) with similar protein–ligand interaction as described above. Please see SI Text for additional results and discussion.
Structure Comparison.
Comparison of apo TcaR with all TcaR–antibiotic complexes we solved in this work reveals that the overall topology is similar (Fig. S7A). Each of those complexes has two antibiotic binding sites, one is located in the cavity at the junction of the helical dimerization domain and the wHTH DNA-binding domain, which we suggest as the first binding site, and the second binding site is found alongside the helix α4 of chain B, which is in the crystal contact interface of three TcaR dimer molecules in the asymmetric unit along the crystallographic c axis. We propose that the first binding site is more important than the second one and is sufficient to modulate the activity of TcaR. This hypothesis is accordance with the observation seen in recently solved structure of MTH313-salicylate complex (27), as discussed below.
When we superimposed all TcaR–ligand complexes, as seen in Fig. S4 A and B, significant conformational changes were observed in the wHTH motifs, especially on chain B. According to the structural differences between these complexes, two kinds of steric movements of the wHTH motif were revealed. One is that the wHTH motif of chain B twisted with the respect to the dimerization domain, making itself apart from the suitable orientation for DNA-binding (i.e., TcaR–Sal complex and TcaR–PnG complex). On the other hand, the wHTH motifs were also twisted to each other to produce a sheared orientation with the most contracted distance between the recognition helices α3/α′3 and α4/α′4 (i.e., TcaR–Amp complex, TcaR–Meth complex, and TcaR–Kan complex). Nevertheless, a displacement of 2.8–4.2 Å was seen along the wHTH motif measured from the Cα of Glu B84 and the N termini of helix α4 showed the displacement of around 3.1–4.5 Å (measured from the Cα of Lys B65). Although the amino acid composition of chain A and chain B is identical in all TcaR–ligand complex structures, the conformational changes are more prominent at chain B in every TcaR–ligand complexes we solved. The asymmetric allosteric mechanism awaits further investigation.
Moreover, superimposition of the TcaR–ligand complexes and the TcaR–DNA model revealed a displacement of α′4 recognition helix especially in the TcaR–Amp, TcaR–Meth, and TcaR–Kan complexes, which would result in severe steric clashes with the DNA backbone (Fig. 3E). Noteworthily, the first binding site in TcaR does not appear to be a direct steric hindrance for the DNA backbone. It is likely that antibiotics could interact with TcaR at the first binding site on TcaR–DNA complex and cause some conformational changes in the wHTH motifs, further weakening the affinity of TcaR to its target DNA sequence and finally induce the departure of TcaR from ica promoter. The hypothesis deduced from our observation clarified that the binding of one antibiotic molecule to the first binding site is sufficient to modulate the DNA-binding activity of TcaR. To confirm this hypothesis, two kinds of TcaR mutant were designed: One is A38W/S41W/H42W for the first binding site, and the other is R70A/K74A for the second binding site. As seen in Fig. S7B, treatment of the antibiotics does not interfere with the DNA-binding activity of the triple mutant A38W/S41W/H42W. However, antibiotics treatment inhibits the formation of R70A/K74A–DNA complex. Consequently, the result shows that the first binding site of TcaR is the critical site for the DNA-binding inhibition.
Model of Antibiotics Binding and its Activation of Transcription.
As discussed above, TcaR in its antibiotic-bound conformation is unlikely to bind its sequence specific DNA-binding site. The inactivation and the regulation of TcaR by antibiotics as deduced from our observation can be summarized into three different mechanisms. First, the binding of antibiotics might reduce the flexibility of the winged DNA-binding domain and stabilize the TcaR dimer conformation that is unable to accommodate the insertion of recognition helices into the major grooves of target DNA. Second, antibiotics probably cause conformational changes on TcaR dimer by shrinking the distance between the wHTH motifs, thereby causing severe steric clashes with the target DNA backbone and making it incompatible with DNA-binding. Third, because the second binding site is close to the DNA-binding helix α4, it would be suitably positioned to influence DNA-binding directly. Taken together, the explanatory hypotheses for the regulatory mechanisms provide a unique explanation of the regulation role for proteins in the MarR transcriptional regulator family.
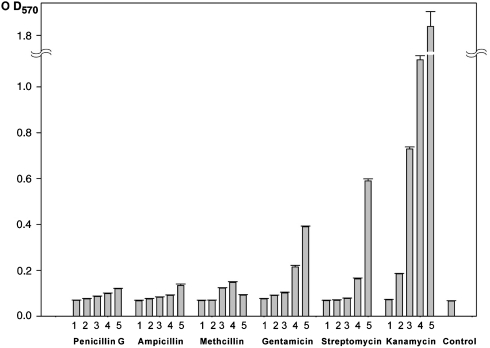
In our present studies, we have noticed that antibiotics would be implicated in eliciting biofilm formation by interfering with the binding of IcaR to DNA (25). Although the exact mechanism is not well understood, we propose a regulatory mechanism for IcaA expression based on the analysis we have done. Schematic representation of the proposed mechanism is shown in Fig. S7C. At first, TcaR repressors bind to the ica promoter in the absence of any inhibitor and prevent transcription of the IcaA gene. There are multiple TcaR recognition sites within the ica promoter sequence and the DNA fragment closest to the IcaR-binding site, which we refer to as “DNA1,” is the most suitable binding site for TcaR. Upon entering of some antibiotics to the cell, significant conformational changes in the DNA-binding domains of TcaR will be exerted, inducing the inactivation and the departure of these transcriptional repressors from the ica promoter, thus increasing the transcription and subsequent expression of IcaA. To investigate the biological effect of antibiotics on the ica operon, biofilm formation was examined in S. epidermidis RP62A in microtiter wells after treatment with several different antibiotics following the protocol of Wakimoto et al. (29). As seen in Fig. 4, the biofilm formation was significantly induced by subinhibitory concentrations of several antibiotics, especially in Kan. The result is consistent with the previous observation that biofilm formation can be stimulated by vancomycin in S. epidermidis clinical strains (30). The regulation of ica promoter transcription activity by TcaR can be further analyzed using β-galactosidase or GFP reporter system, and the analyses are currently under progress.
Fig. 4.
The biofilm formation of the clinical S. epidermidis RP62A supplemented with different concentrations of antibiotics. The biofilm was measured by staining with crystal violet in microtiter wells (A570). The culture was treated with each antibiotic at 5 different concentrations (bar bar 1 = 0.5 μM, bar 2 = 5 μM, bar 3 = 50 μM, bar 4 = 500 μM, bar 5 = 5 mM). Each bar shows the average and standard error of three independent experiments.
Because Staphylococcus is a big threat to the society and the biofilm formation is a fundamental survival mechanism of Staphylococcus, a better understanding of the molecular mechanisms underlying biofilm formation is necessary to achieve a better management of Staphylococcus infection. Our work of the crystal structure of apo TcaR and its complexes with several ligands (including important clinical antibiotics) reveals the allosteric mechanism responsible for derepressing the production of PIA, the major component of biofilm in S. epidermidis. We have also shown that TcaR binds to the DNA fragments in ica operon, especially to DNA1, and its DNA-binding activity is negatively modulated by several antibiotics. Such observations may help us understand the mechanism of the MarR family of transcriptional regulators involved in antimicrobial resistance and virulence.
To combat resistant strains of bacteria, unique molecular targets are being tested for the development of new antibiotics. The golden pigment of staphyloxanthin from S. aures is such an example (31). It is possible the TcaR/MarR system in S. epidermidis and S. aureus may be considered as targets. If ligands that bind more tightly and specifically to the promoter region of IcaA could be discovered or designed, their DNA-binding would shut down the production of PIA and formation of biofilm.
Materials and Methods
Protein Expression, Purification, Crystallization, and Data Collection.
The TcaR gene was amplified directly from the S. epidermidis RP62A genome by PCR and cloned into pET-16b (Novagen). The native TcaR and selenomethionine-labeled protein were produced in E. coli. Additional experimental details can be found in SI Materials and Methods.
Structure Determination, Model Building, and Refinement.
Initial phase angles were calculated by employing the program SOLVE (32) and using the MAD data of SeMet–TcaR in the resolution range of 30–2.9 Å. There are 6 selenium sites located in one molecule, and the phase angles were determined by the single-wavelength anomalous diffraction method using the peak wavelength data. Subsequently, the electron density map was improved using the program RESOLVE (33), and the model was built into density using the program O (34). Manual building of the remaining model and further refinement were carried out with the programs Xtalview (35) and CNS (36) against the 2.9 Å resolution dataset of the SeMet–TcaR crystal. The structure of native TcaR was determined by molecular replacement using CNS, with the refined structure of SeMet–TcaR as a search model. For each structure, iterative cycles of model building with Xtalview and computational refinement with CNS were performed, in which 5% reflections were set aside for Rfree calculation (37). The stereochemical quality was assessed with the program PROCHECK (38). The molecular figures were produced by using PyMOL (DeLano Scientific, http://www.pymol.org) and the possible cavities in TcaR were identified by program CAVER (39).
Other Procedures.
Detailed procedures are available in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We are grateful to Hui-Ling Shr at the Core Facility of X-ray crystallography. We thank the National Synchrotron Radiation Research Center of Taiwan, and SPring-8 of Japan for beam time allocation. This work was supported by grants from the Institute of Biological Chemistry and National Research Program for Genomic Medicine, Academia Sinica.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0913302107/-/DCSupplemental.
Data deposition: The atomic coordinates and structure factors for TcaR (3KP7), TcaR complexed with salicylate (3KP6), penicillin G (3KP2), ampicillin (3KP3), methicillin (3KP4), and kanamycin (3KP5) have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank.
References
- 1.Bancroft EA. Antimicrobial resistance: It’s not just for hospitals. JAMA-J Am Med Assoc. 2007;298(15):1803–1804. doi: 10.1001/jama.298.15.1803. [DOI] [PubMed] [Google Scholar]
- 2.Klevens RM, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA-J Am Med Assoc. 2007;298(15):1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 3.Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358(9276):135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 4.Fux CA, Costerton JW, Stewart PS, Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13(1):34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Vuong C, et al. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol. 2004;6(3):269–275. doi: 10.1046/j.1462-5822.2004.00367.x. [DOI] [PubMed] [Google Scholar]
- 6.Dobinsky S, et al. Glucose-related dissociation between icaADBC transcription and biofilm expression by Staphylococcus epidermidis: Evidence for an additional factor required for polysaccharide intercellular adhesin synthesis. J Bacteriol. 2003;185(9):2879–2886. doi: 10.1128/JB.185.9.2879-2886.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cafiso V, et al. Presence of the ica operon in clinical isolates of Staphylococcus epidermidis and its role in biofilm production. Clin Microbiol Infec. 2004;10(12):1081–1088. doi: 10.1111/j.1469-0691.2004.01024.x. [DOI] [PubMed] [Google Scholar]
- 8.Fluckiger U, et al. Biofilm formation, icaADBC transcription, and polysaccharide intercellular adhesin synthesis by Staphylococci in a device-related infection model. Infect Immun. 2005;73(3):1811–1819. doi: 10.1128/IAI.73.3.1811-1819.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conlon KM, Humphreys H, O’Gara JP. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J Bacteriol. 2002;184(16):4400–4408. doi: 10.1128/JB.184.16.4400-4408.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jefferson KK, Cramton SE, Gotz F, Pier GB. Identification of a 5-nucleotide sequence that controls expression of the ica locus in Staphylococcus aureus and characterization of the DNA-binding properties of IcaR. Mol Microbiol. 2003;48(4):889–899. doi: 10.1046/j.1365-2958.2003.03482.x. [DOI] [PubMed] [Google Scholar]
- 11.Jefferson KK, Pier DB, Goldmann DA, Pier GB. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J Bacteriol. 2004;186(8):2449–2456. doi: 10.1128/JB.186.8.2449-2456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brandenberger M, Tschierske M, Giachino P, Wada A, Berger-Bachi B. Inactivation of a novel three-cistronic operon tcaR-tcaA-tcaB increases teicoplanin resistance in Staphylococcus aureus. Biochim Biophys Acta. 2000;1523(2–3):135–139. doi: 10.1016/s0304-4165(00)00133-1. [DOI] [PubMed] [Google Scholar]
- 13.McCallum N, Bischoff M, Maki H, Wada A, Berger-Bachi B. TcaR, a putative MarR-like regulator of sarS expression. J Bacteriol. 2004;186(10):2966–2972. doi: 10.1128/JB.186.10.2966-2972.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tegmark K, Karlsson A, Arvidson S. Identification and characterization of SarH1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol Microbiol. 2000;37(2):398–409. doi: 10.1046/j.1365-2958.2000.02003.x. [DOI] [PubMed] [Google Scholar]
- 15.Alekshun MN, Levy SB, Mealy TR, Seaton BA, Head JF. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 A resolution. Nat Struct Biol. 2001;8(8):710–714. doi: 10.1038/90429. [DOI] [PubMed] [Google Scholar]
- 16.Wu RY, et al. Crystal structure of Enterococcus faecalis SlyA-like transcriptional factor. J Biol Chem. 2003;278(22):20240–20244. doi: 10.1074/jbc.M300292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong M, Fuangthong M, Helmann JD, Brennan RG. Structure of an OhrR–ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol Cell. 2005;20(1):131–141. doi: 10.1016/j.molcel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 18.Lim D, Poole K, Strynadka NC. Crystal structure of the MexR repressor of the mexRAB-oprM multidrug efflux operon of Pseudomonas aeruginosa. J Biol Chem. 2002;277(32):29253–29259. doi: 10.1074/jbc.M111381200. [DOI] [PubMed] [Google Scholar]
- 19.Chin KH, et al. The crystal structure of XC1739: a putative multiple antibiotic-resistance repressor (MarR) from Xanthomonas campestrisat 1.8 A resolution. Proteins. 2006;65(1):239–242. doi: 10.1002/prot.21017. [DOI] [PubMed] [Google Scholar]
- 20.Martin RG, Rosner JL. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc Natl Acad Sci USA. 1995;92(12):5456–5460. doi: 10.1073/pnas.92.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin RG, Jair KW, Wolf RE, Jr, Rosner JL. Autoactivation of the marRAB multiple antibiotic resistance operon by the MarA transcriptional activator in Escherichia coli. J Bacteriol. 1996;178(8):2216–2223. doi: 10.1128/jb.178.8.2216-2223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkinson SP, Grove A. Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr Issues Mol Biol. 2006;8(1):51–62. [PubMed] [Google Scholar]
- 23.Saito K, Akama H, Yoshihara E, Nakae T. Mutations affecting DNA-binding activity of the MexR repressor of mexR-mexA-mexB-oprM operon expression. J Bacteriol. 2003;185(20):6195–6198. doi: 10.1128/JB.185.20.6195-6198.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffman LR, et al. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature. 2005;436(7054):1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 25.Jeng WY, et al. Crystal structure of IcaR, a repressor of the TetR family implicated in biofilm formation in Staphylococcus epidermidis. Nucleic Acids Res. 2008;36(5):1567–1577. doi: 10.1093/nar/gkm1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumarevel T, Tanaka T, Umehara T, Yokoyama S. ST1710–DNA complex crystal structure reveals the DNA binding mechanism of the MarR family of regulators. Nucleic Acids Res. 2009;37(14):4723–4735. doi: 10.1093/nar/gkp496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saridakis V, Shahinas D, Xu X, Christendat D. Structural insight on the mechanism of regulation of the MarR family of proteins: High-resolution crystal structure of a transcriptional repressor from Methanobacterium thermoautotrophicum. J Mol Biol. 2008;377(3):655–667. doi: 10.1016/j.jmb.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Alekshun MN, Levy SB. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J Bacteriol. 1999;181(15):4669–4672. doi: 10.1128/jb.181.15.4669-4672.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wakimoto N, et al. Quantitative biofilm assay using a microtiter plate to screen for enteroaggregative Escherichia coli. Am J Trop Med Hyg. 2004;71(5):687–690. [PubMed] [Google Scholar]
- 30.Cargill JS, Upton M. Low concentrations of vancomycin stimulate biofilm formation in some clinical isolates of Staphylococcus epidermidis. J Clin Pathol. 2009;62(12):1112–1116. doi: 10.1136/jcp.2009.069021. [DOI] [PubMed] [Google Scholar]
- 31.Liu CI, et al. A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science. 2008;319(5868):1391–1394. doi: 10.1126/science.1153018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D. 1999;55(Pt 4):849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terwilliger T. SOLVE and RESOLVE: Automated structure solution, density modification, and model building. J Synchrotron Radiat. 2004;11(Pt 1):49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 34.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 35.McRee DE. XtalView/Xfit—A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125(2–3):156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 36.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.Brunger AT. Assessment of phase accuracy by cross validation: the free R value. Methods and applications. Acta Crystallogr D. 1993;49(Pt 1):24–36. doi: 10.1107/S0907444992007352. [DOI] [PubMed] [Google Scholar]
- 38.Morris AL, MacArthur MW, Hutchinson EG, Thornton JM. Stereochemical quality of protein structure coordinates. Proteins. 1992;12(4):345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
- 39.Petrek M, et al. CAVER: A new tool to explore routes from protein clefts, pockets, and cavities. BMC Bioinformatics. 2006;7:316. doi: 10.1186/1471-2105-7-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.