Abstract
Organelle movement is essential for efficient cellular function in eukaryotes. Chloroplast photorelocation movement is important for plant survival as well as for efficient photosynthesis. Chloroplast movement generally is actin dependent and mediated by blue light receptor phototropins. In Arabidopsis thaliana, phototropins mediate chloroplast movement by regulating short actin filaments on chloroplasts (cp-actin filaments), and the chloroplast outer envelope protein CHUP1 is necessary for cp-actin filament accumulation. However, other factors involved in cp-actin filament regulation during chloroplast movement remain to be determined. Here, we report that two kinesin-like proteins, KAC1 and KAC2, are essential for chloroplasts to move and anchor to the plasma membrane. A kac1 mutant showed severely impaired chloroplast accumulation and slow avoidance movement. A kac1kac2 double mutant completely lacked chloroplast photorelocation movement and showed detachment of chloroplasts from the plasma membrane. KAC motor domains are similar to those of the kinesin-14 subfamily (such as Ncd and Kar3) but do not have detectable microtubule-binding activity. The C-terminal domain of KAC1 could interact with F-actin in vitro. Instead of regulating microtubules, KAC proteins mediate chloroplast movement via cp-actin filaments. We conclude that plants have evolved a unique mechanism to regulate actin-based organelle movement using kinesin-like proteins.
Keywords: cp-actin, blue light, organelle movement, phototropin
Organelle movement is pivotal for efficient cellular functions in plants as well as in animals and fungi (1). Chloroplast photorelocation movement is essential for the adaptation of plants under the fluctuating natural light conditions (2). Chloroplasts accumulate toward light under low-light conditions (accumulation response) to maximize light capture for photosynthesis, and they escape from intense light (avoidance response) to avoid chloroplast photodamage (2, 3). These responses are mediated by blue light receptor phototropins in green algae, mosses, and ferns as well as in seed plants (2). Arabidopsis thaliana expresses two phototropins (4–6); phot1 and phot2 are required for the accumulation response whereas phot2 alone is essential for the avoidance response.
Like other plant organelle movements, chloroplast photorelocation movement generally is dependent on actin filaments rather than on microtubules (2). Our recent study suggested that chloroplast photomovement and anchorage to the plasma membranes are mediated by short actin filaments on chloroplasts (cp-actin filaments); formation of cp-actin filaments is associated with phototropin-mediated chloroplast movement (7). The chloroplast outer membrane protein, chloroplast unusual positioning 1 (CHUP1), which can bind to both F-actin and G-actin in vitro (8–10), is implicated in cp-actin filament formation (7). The correlation of cp-actin signal with the velocity of chloroplast movement suggests that cp-actin filaments play a role in generating the motive force (7), but it remains to be determined if cp-actin filaments actually provide the motive force. To identify other components involved in cp-actin filament regulation during chloroplast movement, we screened mutants deficient in chloroplast photorelocation movement using a previously developed band assay (4, 8, 11). Here, we characterized a kinesin-like protein for actin-based chloroplast movement 1 (kac1) mutant.
Results
Positional Cloning of KAC1 Gene and Characterization of kac Mutants.
Two independent kac1 alleles, kac1-1 and kac1-2, were isolated (Fig. 1A). A map-based approach revealed that the KAC1 gene is At5g10470 (Fig. S1 A and B). Analyses of a transfer DNA (T-DNA) insertion line of KAC1 (kac1-3) (Figs. S1B and S2 A and B) and genetic complementation of kac1-1 with an At5g10470 genomic construct confirmed that KAC1 is At5g10470. Moreover, Arabidopsis thaliana has one gene that is highly similar to KAC1; this gene is named “KAC2” (At5g65460) and is located on the lower arm of chromosome 5 (Fig. S1B). Note that KAC1 was identified previously as a protein interacting with various proteins implicated in cell-cycle and microtubule function: CDKA1-interacting protein [designated “KLP2/KCA1” (12, 13)], the geminivirus protein AL1-interacting protein [designated “GRIMP” (14)], and the katanin-interacting protein [designated “KSN1” (15)]. In this paper we refer to KLP2/KCA1/GRIMP/KSN1 as “KAC1” and KCA2 as “KAC2.” Although we cannot exclude completely the possibility that KAC proteins regulate cell-cycle or microtubule-dependent cell growth, under our growth conditions we could not detect obvious growth or developmental defects even in the kac1kac2 double mutant. Moreover, observations of microtubule dynamics using GFP-TUA6 showed that no gross or obvious defect could be observed without a more quantitative analysis (Movie S1 and Movie S2). However, we cannot exclude the possibility that these KAC1 interactive proteins are involved in chloroplast movement.
Fig. 1.
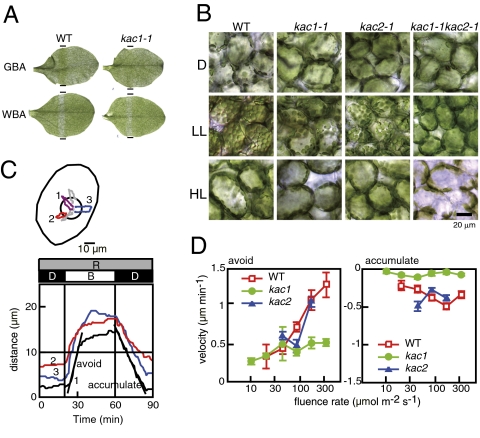
kac mutants are deficient in chloroplast photorelocation movement. (A) Green (GBA) and white band assay (WBA) for detection of chloroplast accumulation and avoidance responses, respectively. The irradiated area is indicated on both sides of the leaves by black bars (ca. 1 mm). (B) Characterization of chloroplast photorelocation movement in mesophyll cells of kac mutants. Plants that were dark-adapted for about 12 h (D) were irradiated with white light at 10 (LL) or 100 μmole m−2 s−1 (HL) for 2 h. All micrographs were taken at the focal plane of the anticlinal wall closest to the epidermis. (C and D) Velocity of chloroplast photorelocation movement in mesophyll cells of wild-type, kac1-1, and kac2-1. (C) Chloroplast movement was induced by irradiation with a blue light microbeam (circle) at various fluence rates (11, 22, 49, 94, 188, and 376 μmole m−2 s−1), and the movement of each chloroplast was traced during the experiments (Upper). Avoidance response was induced during microbeam irradiation, whereas accumulation response was induced after the microbeam turned off. The distances between the center of the microbeam and chloroplasts were plotted as a function of time (black, red, and blue lines), and the velocity was calculated as the slope (indicated by black lines) of the traveled distance during the avoidance or accumulation (Lower). (D) The velocity was plotted as a function of the fluence rate. Bar indicates SE. P values by two-tailed Student's t test were determined for fluence rates of 22, 49, 94, 188, and 376 μmole m−2 s−1: for kac1 versus wild type for avoidance response (P = 0.855, P = 0.706, P = 0.001, P = 0.001, P = 5.7E-05, respectively), for kac1 versus wild type for accumulation response (P = 9.0E-05, P = 0.036, P = 7.8E-06, P = 5.8E-09, P = 2.0E-05, respectively), for kac2 versus wild type for avoidance response (49, 94, and 188 μmole m−2 s−1) (P = 0.062, P = 0.03, P = 0.885, respectively), and for kac2 versus wild type for accumulation response (P = 0.007, P = 0.051, P = 0.058, respectively).
To investigate the function of both KAC1 and KAC2 in chloroplast photorelocation movement, single and double mutants (Figs. S1B and S2A) were examined in detail. If not specified otherwise, kac1-1 and kac2-1 were used in all experiments as representative lines for kac1 and kac2, respectively, because similar results were obtained using other kac1 or kac2 alleles. When dark-adapted wild-type and kac mutant plants were irradiated with a low-fluence (LL) or high-fluence (HL) white light for 2 h, typical chloroplast photorelocation movement was observed in mesophyll cells of both the wild type and kac2. Chloroplasts were positioned on the cell bottom in darkness (dark positioning) and moved to the cell surface under LL conditions in an accumulation response and were at the anticlinal wall under HL conditions in an avoidance response (11) (Fig. 1B). By contrast the dark positioning and avoidance response were normal in kac1 mutants, but fewer chloroplasts accumulated at the cell surface under LL conditions (Fig. 1B), indicating that the kac1 mutant is partially defective in the accumulation response (Fig. 1B). In the kac1kac2 double mutant, the distribution of chloroplasts under both LL and HL conditions was similar to that of wild-type plants under HL conditions (Fig. 1B), indicating that kac1kac2 is defective in the accumulation response. To analyze the avoidance movement in kac1kac2 plants, a small part of the anticlinal wall of a mesophyll cell where chloroplasts localize in kac1kac2 was irradiated with a microbeam of strong blue light, and chloroplast movement was recorded using a video camera. Wild-type chloroplasts escaped from the microbeam-irradiated area (Movie S3), but kac1kac2 chloroplasts did not (Movie S4), indicating that kac1kac2 is deficient in the avoidance response as well as the accumulation response.
KAC1 and KAC2 Genes Are Functionally Equivalent but Have Different Expression Levels.
When the velocity of chloroplast avoidance and accumulation were measured and plotted as the function of the fluence rate of the irradiated microbeam, as described previously (16) (see legend of Fig. 1C for details), the velocity in the wild-type chloroplasts increased proportionally to the fluence rate; the relationship was clear for the avoidance movement and slight for the accumulation movement (Fig. 1D). However, in the kac1 mutant, the chloroplast accumulation response was hardly detected (Fig. 1D and Movie S5). Interestingly, the velocity of the avoidance movement was the same as that of the wild type in response to the lower fluence rates of the strong blue light (11, 22, and 49 μmol m−2 s−1) but saturated at around 49 μmol m−2 s−1. Hence, the velocity of avoidance in wild-type chloroplasts was twice or more than that in kac1 at the higher fluence rates (188 and 376 μmol m−2 s−1), indicating that kac1 shows partial impairment of the avoidance response. The kac2 mutant plants showed almost normal chloroplast photorelocation movement (Fig. 1D). RT-PCR analyses revealed that both KAC1 and KAC2 transcripts were expressed in various tissues (Fig. S2C). However, the microarray data showed that KAC1 was more highly expressed than KAC2, especially in chloroplast-containing tissues. The KAC protein level was examined in wild-type, kac1, kac2, and kac1kac2 using KAC antiserum (which can detect KAC1 and KAC2 equally in immunoblotting after SDS/PAGE) (Fig. S3 A and B). A band of approximately 150 kDa, corresponding to the size of full-length KAC proteins, was detected in wild-type and kac2 mutant plants at a similar level, but a severely reduced level was observed in kac1 plants, and the band was not detected in kac1kac2 mutant plants, indicating that KAC1 proteins accumulated at a level several times higher than that of KAC2 (Figs. S3B and S4A). When KAC2 cDNA was expressed under the control of the KAC1 promoter and UTRs in kac1kac2 mutant plants (KAC1pro-KAC2), KAC2 proteins accumulated at a level similar to that of KAC1 proteins in the kac2 mutant and in the KAC1 cDNA-expressing lines in the kac1kac2 background (KAC1pro-KAC1) used as a control line (Fig. S4B). Chloroplast photorelocation movement and positioning were rescued to the same extent in both KAC1pro-KAC1 and KAC1pro-KAC2 lines but not in vector lines (Fig. S4 C and E), indicating that KAC1 and KAC2 are functionally equivalent.
KAC Proteins Were Detected in both Soluble and Membrane Fractions.
Immunoblot analyses of fractionated proteins revealed that KAC proteins were detected in both soluble and microsomal fractions with a slightly larger amount detected in the soluble fraction, in contrast to phototropins and CHUP1, which were detected predominantly in the microsomal fraction (Figs. S3C and S4A). In transgenic lines expressing a GFP-KAC1 or KAC1-GFP fusion gene under the control of the KAC1 promoter and UTRs in the kac1 background, normal chloroplast photorelocation movement was observed, and GFP-KAC1 or KAC1-GFP fluorescence was detected mainly in the cytosol and not in any particular organelles or cytoskeletons (Movie S6). Fractionation profiles and the abundances of KAC proteins were not different from those of the wild type or various mutants deficient in chloroplast movement (Fig. S3C). The fractionation profiles and abundances of phototropins and CHUP1 in kac mutants were also the same as those of the wild type (Fig. S4A), indicating that the defects of these mutants in chloroplast movement are not obviously attributed to impaired accumulation or localization of the assayed proteins regulating chloroplast movement.
KAC Genes Encode Kinesin-Like Proteins.
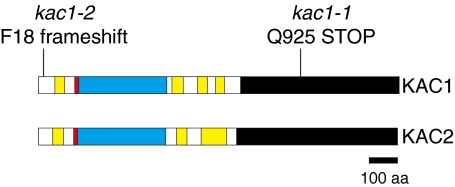
A homology search revealed that KAC genes encode kinesin-like proteins (Fig. 2, Fig. S5). Among the 14 kinesin families (17, 18), KAC proteins belong to the kinesin-14 family. Some are known as minus end-directed motors bearing a motor core domain at their C terminus. The kinesin-14 family is subdivided into two large subfamilies, kinesin-14A and kinesin-14B, and kinesin-14B is further subgrouped into five groups (KIFC2/KIFC3, KatD, KCBP, KIF25, and unclassified) (17, 18). The 18 A. thaliana kinesin-14B members are divided into the KatD (10 members), KCBP (one member), and unclassified (seven members) subgroups. The KAC proteins belong to the unclassified kinesin-14B group (18, 19). The KAC protein is plant specific and exists from moss to seed plants (Fig. S6). Interestingly, KAC proteins have their motor core domain at the N terminus, although it is at the C terminus or middle region in most members of the kinesin-14 family. The KAC proteins are predicted to contain coiled-coil domains at the N-terminal region of the motor core domain and at the middle region of the proteins (Fig. 2). Although the KAC C-terminal domain does not show similarity to any other protein, this region is highly conserved among KAC proteins from various plant species (Fig. 2 and Fig. S6), suggesting that the KAC C-terminal domain is essential for the protein function.
Fig. 2.
KAC1 and KAC2 protein structure. Yellow: coiled-coil domain, red: neck domain, blue: motor core domain, black: conserved C-terminal region. Mutation sites of kac1-1 and kac1-2 mutants are indicated.
Because KAC proteins are members of the kinesin-14 family, ATP-dependent binding to microtubules responsible for microtubule-based motor activity was examined using recombinant KAC motor core domains with or without the N-terminal coiled-coil domain. However, none of the recombinant KAC motor core domains cosedimented with microtubules under any of the nucleotide conditions (Fig. S7). Furthermore, we could not detect intrinsic ATPase activity of KAC proteins. Close inspection of the protein sequences corresponding to the KAC motor domains revealed changes of several amino acids necessary for motor activity and microtubule binding (Discussion).
KAC1 Neck/Core Interaction Mutations Affected KAC1 Protein Accumulation but Not the Directionality of Chloroplast Movement.
Neck/motor core interactions are essential for the directionality of minus end-directed motor Ncd (20). Mutations of interacting amino acids N340K (neck) and K640N (motor core) of Ncd (Fig. S5) destabilize the neck/motor core interaction, resulting in the neck being positioned unstably toward either the plus or minus end (21); thus the motor moves toward either the plus or minus end of the microtubules (22). These residues are highly conserved among the kinesin-14 family, including within the KAC proteins (Figs. S5 and S6). We constructed transgenic kac1kac2 plants expressing the corresponding mutant KAC1 (N134K[NK] and K426N[KN]) under the control of the native KAC1 promoter and UTRs. Mutant KAC1 proteins accumulated at a drastically reduced level (Fig. S4D). Microbeam experiments revealed that in the NK and KN lines, the direction of chloroplast movement in both the accumulation and avoidance responses was not altered. However, the chloroplast accumulation response was severely impaired, and the velocity of chloroplast avoidance was less than half that in wild type (Fig. S4E). These findings may have resulted from attenuated accumulation of mutant KAC1 proteins. An unstable neck/motor core interaction also may make KAC1 unstable in vivo.
kac Mutants Were Defective in cp-Actin Filament Accumulation.
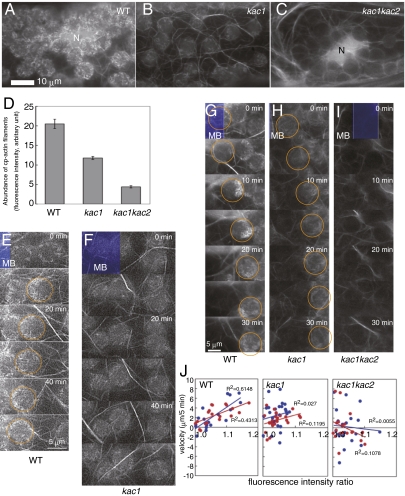
In petiole cells of the lines expressing GFP-mouse talin in the kac1, kac2, and kac1kac2 mutant backgrounds, cp-actin filament dynamics were analyzed (Fig. 3 and Movie S7, Movie S8, Movie S9, Movie S10, Movie S11, and Movie S12). In stationary chloroplasts of wild-type and kac2 mutant plants, cp-actin filaments were found mostly around the chloroplast periphery (Fig. 3A). However, significantly fewer cp-actin filaments were observed in kac1 chloroplasts (Fig. 3 B and D), and none was found in kac1kac2 chloroplasts (Fig. 3 C and D), similar to the chup1 mutant that also lacked cp-actin filaments (7). Biased cp-actin filaments appearing at the front half of moving chloroplasts were found in wild-type and kac2 mutant chloroplasts during the accumulation response toward weak blue light (Fig. 3E). However, in kac1 plants under weak light, the biased cp-actin filaments were not evident, and the accumulation response was not induced (Fig. 3F). In response to a strong blue light microbeam, cp-actin filaments in wild-type and kac2 chloroplasts initially disappeared but then reappeared gradually at the front half of moving chloroplasts before and during the avoidance movement (Fig. 3G and Movie S2) (7). This strong light-dependent disappearance of cp-actin filaments occurred normally in the kac1 mutant, but biased cp-actin filament formation was diminished compared with that of the wild type (Fig. 3H and Movie S8). When the avoidance response was induced by continuous irradiation with blue GFP excitation light, less biased cp-actin filament formation and reduced avoidance were notably detected in kac1 mutant as compared with the wild type (Movie S9 and Movie S10). In kac1kac2, neither cp-actin accumulation nor chloroplast avoidance occurred (Fig. 3I, Movie S11 and Movie S12). In wild-type plants, the difference in GFP fluorescence intensities between the front and rear halves of chloroplasts, indicative of different amounts of cp-actin filaments, showed a positive correlation with the velocity of chloroplast avoidance movement (Fig. 3J) (7). However, this correlation was severely attenuated in the kac1 mutant and could not be detected in the kac1kac2 mutant (Fig. 3J), possibly because of slow avoidance movement and nondirectional spontaneous rapid movements caused by a decrease in or lack of cp-actin filament accumulation, respectively.
Fig. 3.
Role of KAC proteins in cp-actin filament regulation. (A–C) cp-actin filaments on stationary chloroplasts in petiole cells. N, nucleus. (D) Fluorescence intensity was measured in 37–57 chloroplasts derived from three to six cells. Data are shown as mean ± SE. Because no filamentous structure was found on the chloroplasts of kac1kac2 (kac1-1kac2-2), the value of kac1kac2 represents background level of soluble GFP-talin and chlorophyll autofluorescence. P values determined by two-tailed Student's t test are kac1 versus wild type, P = 1.8E-10; kac1kac2 versus wild type, P = 3.1E-18; kac1 versus kac1kac2, P = 2.6E-22. (E–I) Chloroplast movement was induced by continuous irradiation with a blue light microbeam (MB). Chloroplast movement and cp-actin filament dynamics were observed every 10 min under a low-intensity microbeam (3.8 μmole m−2 s−1) (E and F) and every 5 min under a high-intensity microbeam (377 μmole m−2 s−1) (G–I). Orange circles indicate moving chloroplasts. (J) Correlation between biased localization of cp-actin filaments (fluorescence intensity ratio) and the velocity of chloroplast avoidance during 0–5 min (bluedots) and 5–10 min (red dots) after blue microbeam irradiation. R2, coefficient of determination.
Defective cp-Actin Filament Accumulation in kac Mutants Was Correlated with Impaired Chloroplast Attachment to the Plasma Membrane.
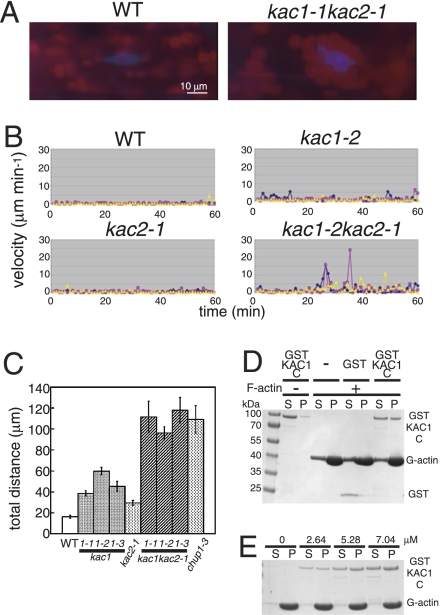
Because cp-actin filaments localize to the plasma membrane side of chloroplasts and help anchor chloroplasts to the plasma membrane (7), kac mutants may be deficient in chloroplast anchoring. Unlike the wild type, chloroplasts in the kac1 single mutant and, especially, in the kac1kac2 double mutant were partially aggregated in petiole cells (Fig. 4A, Movie S13 and Movie S14). Close examination revealed that some chloroplasts aggregated around the nucleus in the kac1kac2 double mutant (Fig. 4A). Without blue light irradiation, chloroplasts were stationary with small Brownian-like movements in the wild type but showed slightly higher motility in kac1 and kac2 plants (Fig. 4 B and C). However, in kac1kac2 plants, chloroplasts translocated long distances and thus were not anchored; this effect was similar to that observed in as the chup1 mutant, which lacks cp-actin filaments (Fig. 4 B and C) (7). Furthermore, a time-lapse movie showed that aggregated chloroplasts in kac1kac2 cells moved rapidly via cytoplasmic streaming (Movie S14). Thus, kac mutants are defective in chloroplast attachment to the plasma membrane, possibly because of their lack of cp-actin filaments.
Fig. 4.
Defective anchorage of chloroplasts to the plasma membrane in kac1kac2. (A) Chloroplast distribution of petiole cells in wild-type and kac1kac2 mutant plants. Nuclei were DAPI-stained (blue). Chloroplasts were identified with red chlorophyll autofluorescence. (B) Chloroplast motility in wild-type and kac mutant lines. Each colored curve represents movement of an independent chloroplast every 1 min during 60-min incubation under red light. (C) Total distance traveled for 60 min in wild-type and kac mutant lines. For the kac1 mutant, three alleles (kac1-1-3) were examined. Data represent the average of nine chloroplasts from three independent experiments. Bar indicates SE. P values determined by two-tailed Student's t test are kac1-1 versus wild type, P = 3.3E-07; kac1-2 versus wild type, P = 3.8E-09; kac1-3 versus wild type, P = 1.8E-05; kac2-1 versus wild type, P = 1.1E-04; kac1-1kac2-1 versus wild type, kac1-1, or kac2-1, P = 1.0E-05, P = 2.0E-04, or P = 6.0E-05, respectively; kac1-2kac2-1 versus wild type, kac1-2, or kac2-1, P = 5.4E-10, P = 8.2E-05, or P = 1.4E-08, respectively; kac1-3kac2-1 versus wild type, kac1-3, or kac2-1, P = 3.4E-07, P = 4.3E-05, or P = 2.3E-06, respectively; chup1-3 versus wild type, P = 2.9E-06. (D and E) Interaction between F-actin and KAC1 C-terminal domain. After incubation of 6 μM GST or GST KAC1 C-terminal domain at 25 °C for 30 min with or without 24 μM F-actin, samples were centrifuged at 228,000 × g for 10 min (D). After incubation of GST KAC1 C-terminal domain (0, 2.64, 5.28, and 7.04 μM) at 25 °C for 60 min with 24 μM F-actin, samples were centrifuged at 228,000 × g for 10 min. Kd = 19.7, 14.7, and 15.6 μM for 2.64, 5.28, and 7.04 μM GST-KAC1 C, respectively (E). Supernatant fraction (S) and precipitant fraction (P) were separated by SDS/PAGE, and the gel was stained with Coomassie brilliant blue.
Discussion
In this study, we identified kinesin-like KAC proteins as important regulators for cp-actin filament–mediated chloroplast movement and attachment to the plasma membrane.
The crystal structures for the motor domains of Ncd and Kar3 have already been solved (23, 24). Four conserved motifs form a nucleotide-binding pocket with topological positioning similar to that of myosins and GTPase (23, 25): N-1 (P-loop: GQTxxGKS/T), N-2 (switch I: NxxSSR), N-3 (switch II: DxxGxE), and N-4 (RxRP) (23). For a conformational change between the ATP and ADP states, kinesins likely use a mechanism similar to that of myosins and GTPases via ATP hydrolysis in switch regions (23, 26). KAC proteins of various plant species retain the highly conserved P-loop and N-4, but their switch motifs, particularly switch I, are less conserved (Figs. S5 and S6). The glycine in kinesin switch II corresponds to the highly conserved glycine in the myosin and GTPase active site, which is necessary for hydrogen bonding with γ-phosphate and for the conformational change between the nucleoside triphosphate and nucleoside diphosphate states (23, 26). The arginine in switch I and the glutamic acid in switch II form a salt bridge in kinesins similar to that of myosins (25). Mutations of these residues impaired microtubule-activated ATPase activity (26, 27). Furthermore, Kar3 switch I R598A mutant proteins showed reduced microtubule binding compared with the wild-type Kar3 (27). Notably, in seed plant KAC proteins the equivalent switch I arginine is valine, suggesting that seed plant KAC proteins cannot form the salt bridge between switch I and II motifs (Fig. S6). In rice KAC1, switch II glycine is replaced by alanine, the same substitution as the aforementioned switch II glycine mutation (26) (Fig. S6). Therefore, it is likely that vascular plant KAC proteins lack microtubule motor activity. The L12/a5 region forms a microtubule-interacting surface. Some residues for microtubule binding of human kinesin heavy chain were identified by alanine scanning (R623 and K626 in Ncd) (28). In dicot KAC proteins, the corresponding arginine and lysine are changed into acidic and nonpolar amino acids, respectively (the arginine also is different in moss KAC proteins) (Figs. S5 and S6). This amino acid changes also may be a reason why we cannot detect in vitro microtubule-binding activity of recombinant KAC motor domains. Although microtubule motor activity of KAC proteins cannot be excluded totally, the absence of microtubule-motor activity of KAC proteins is consistent with the general view that actin filaments but not microtubules are responsible for chloroplast movement in land plants.
KAC proteins were detected in both the soluble and microsomal fractions by immunoblotting. Although clear plasma membrane or organelle localization of GFP fusion proteins of KAC1 was not detected in the present study, another study revealed localization of KAC1 to the plasma membrane using immunolocalization analysis of A. thaliana root tips and confocal microscopy of overexpressed GFP-KCA1 (i.e., GFP-KAC1) in tobacco BY-2 cells (29). Note that GFP-KAC1 was targeted to the plasma membrane and cell plate during cell division, implicating the involvement of KAC1 in cell division as well as in chloroplast movement (29). Importantly, cp-actin filaments were localized only at the interface between the chloroplast and the plasma membrane and not at the vacuole side (7), and their accumulation was dependent on KAC proteins. Thus, it is plausible that KAC proteins localized on the plasma membrane but not in the cytoplasm may play an important role in cp-actin filament regulation and thus in chloroplast movement and attachment to the plasma membrane. Another cp-actin filament regulator, CHUP1, is localized on the chloroplast outer envelope (8–10). However, we could not detect KAC proteins in CHUP1-enriched chloroplast fractions. Although the relationship between KAC and CHUP1 in cp-actin filament regulation remains to be determined, it is obvious that further analysis of these proteins is essential for full understanding of cp-actin filament–mediated chloroplast movement.
It is interesting that kinesin-like KAC proteins mediate actin-based chloroplast movement independent of microtubules. However, there are two previously reported mechanisms in which kinesins use actin filaments for their cellular functions. First, some kinesins such as mouse KIF5B (30) and budding yeast Smy1p (31) can interact with actin-based motor myosins to use actin filaments. Notably, the function and localization of Smy1p were not perturbed by a microtubule inhibitor or by an Smy1p P-loop mutation that disturbs motor activity (32), indicating that motor activity and microtubule binding of Smy1p are dispensable for Smy1p function, similar to effects observed for KAC in this study. Although it is possible that KAC proteins may mediate chloroplast movement via direct interactions with plant myosins, recent comprehensive analyses of myosin mutant lines in A. thaliana and tobacco strongly suggest that myosins are not involved in chloroplast movement (33, 34). Second, some kinesins can interact directly with actin filaments (35–37). We performed a cosedimentation assay between actin filaments and several KAC recombinant proteins. We could detect an interaction between actin filaments and the KAC1 C-terminal domain (Fig. 4D). Both GST alone and GST-KAC1 C-terminal fusion proteins were detected in the soluble fraction without F-actin (Fig. 4D). However, a significant amount of the GST-KAC1 C terminus, but not GST alone, was precipitated with F-actin (Kd = ∼15 μM) (Fig. 4 D and E), indicating that the KAC1 C terminus can interact with F-actin. This interaction may be necessary for cp-actin generation or maintenance in vivo. Although the precise mechanism by which KAC proteins mediate actin-based chloroplast movement is unknown, our study indicates that plants have evolved a unique kinesin-like protein-dependent mechanism for organelle movement involving specific short actin filaments but not microtubules.
Materials and Methods
Plant culture, mutant screening, and map-based cloning were performed as described previously (11). For Agrobacterium-mediated transformation, the T-DNA vector pBI-HI-BSKR (11) was used (Fig. S8). Chloroplast photorelocation movement was analyzed by observation of chloroplast distribution after 2 h of light irradiation (11) or by microbeam irradiation (16). Observation of actin and microtubule dynamics was performed as described previously (7).
RT-PCR, immunoblotting, and the cytoskeleton binding assay are described in SI Text. For GST-tagged KAC proteins, various KAC cDNAs were cloned into pGEX (Amersham Pharmacia), and recombinant proteins were purified according to the manufacturer's instructions.
Full methods and associated references are available in SI Text. Primers used are listed in Table S1.
Supplementary Material
Acknowledgments
We thank Keiko Hirose and K. Yan (National Institute for Advanced Industrial Science and Technology, Japan) for information on KAC ATPase activity. We thank Takashi Hashimoto (Nara Institute of Science and Technology, Japan) for the gift of GFP-TUA6 lines. We also thank Mineko Simizu for assistance in mutant screening. The BAC clones and SALK T-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center. This work was supported in part by the Japanese Ministry of Education, Sports, Science, and Technology (MEXT 13139203 and 17084006 to M.W.; 19039027 to A.K.) and the Japan Society of Promotion of Science (JSPS 13304061, 16107002, and 20227001 to M.W.; 19570045 to A.K.; 20870030 to N.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0912773107/-/DCSupplemental.
References
- 1.Williamson RE. Organelle movements. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:181–202. [Google Scholar]
- 2.Suetsugu N, Wada M. Chloroplast photorelocation movement mediated by phototropin family proteins in green plants. Biol Chem. 2007;388:927–935. doi: 10.1515/BC.2007.118. [DOI] [PubMed] [Google Scholar]
- 3.Kasahara M, et al. Chloroplast avoidance movement reduces photodamage in plants. Nature. 2002;420:829–832. doi: 10.1038/nature01213. [DOI] [PubMed] [Google Scholar]
- 4.Kagawa T, et al. Arabidopsis NPL1: A phototropin homolog controlling the chloroplast high-light avoidance response. Science. 2001;291:2138–2141. doi: 10.1126/science.291.5511.2138. [DOI] [PubMed] [Google Scholar]
- 5.Jarillo JA, et al. Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature. 2001;410:952–954. doi: 10.1038/35073622. [DOI] [PubMed] [Google Scholar]
- 6.Sakai T, et al. Arabidopsis nph1 and npl1: Blue light receptors that mediate both phototropism and chloroplast relocation. Proc Natl Acad Sci USA. 2001;98:6969–6974. doi: 10.1073/pnas.101137598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kadota A, et al. Short actin-based mechanism for light-directed chloroplast movement in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:13106–13111. doi: 10.1073/pnas.0906250106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oikawa K, et al. Chloroplast unusual positioning1 is essential for proper chloroplast positioning. Plant Cell. 2003;15:2805–2815. doi: 10.1105/tpc.016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oikawa K, et al. Chloroplast outer envelope protein CHUP1 is essential for chloroplast anchorage to the plasma membrane and chloroplast movement. Plant Physiol. 2008;148:829–842. doi: 10.1104/pp.108.123075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt von Braun S, Schleiff E. The chloroplast outer membrane protein CHUP1 interacts with actin and profilin. Planta. 2008;227:1151–1159. doi: 10.1007/s00425-007-0688-7. [DOI] [PubMed] [Google Scholar]
- 11.Suetsugu N, Kagawa T, Wada M. An auxilin-like J-domain protein, JAC1, regulates phototropin-mediated chloroplast movement in Arabidopsis. Plant Physiol. 2005;139:151–162. doi: 10.1104/pp.105.067371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanstraelen M, et al. A plant-specific subclass of C-terminal kinesins contains a conserved A-type cyclin-dependent kinase site implicated in folding and dimerization. Plant Physiol. 2004;135:1417–1429. doi: 10.1104/pp.104.044818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geelen DNV, Inze DG. A bright future for the Bright Yellow-2 cell culture. Plant Physiol. 2001;127:1375–1379. [PMC free article] [PubMed] [Google Scholar]
- 14.Kong L-J, Hanley-Bowdoin L. A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression patterns during plant development and infection. Plant Cell. 2002;14:1817–1832. doi: 10.1105/tpc.003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouquin T, Mattsson O, Næsted H, Foster R, Mundy J. The Arabidopsis lue1 mutant defines a katanin p60 ortholog involved in hormonal control of microtubule orientation during cell growth. J Cell Sci. 2003;116:791–801. doi: 10.1242/jcs.00274. [DOI] [PubMed] [Google Scholar]
- 16.Kagawa T, Wada M. Velocity of chloroplast avoidance movement is fluence rate dependent. Photochem Photobiol Sci. 2004;3:592–595. doi: 10.1039/b316285k. [DOI] [PubMed] [Google Scholar]
- 17.Laurence CJ, et al. A standardized kinesin nomenclature. J Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: Insights into structure and function. Trends Cell Biol. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 19.Reddy ASN, Day IS. Kinesins in the Arabidopsis genome: A comparative analysis among eukaryotes. BMC Genomics. 2001;2:2. doi: 10.1186/1471-2164-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Endow SA. Determinants of molecular motor directionality. Nat Cell Biol. 1999;1:163–167. doi: 10.1038/14113. [DOI] [PubMed] [Google Scholar]
- 21.Endres NF, Yoshioka C, Milligan RA, Vale RD. A lever-arm rotation drives motility of the minus-end-directed kinesin Ncd. Nature. 2006;439:875–878. doi: 10.1038/nature04320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Endow SA, Higuchi H. A mutant of the motor protein kinesin that moves in both directions on microtubules. Nature. 2000;406:913–916. doi: 10.1038/35022617. [DOI] [PubMed] [Google Scholar]
- 23.Sablin EP, Kull FJ, Cooke R, Vale RD, Fletterick RJ. Crystal structure of the motor domain of the kinesin-related motor ncd. Nature. 1996;380:555–559. doi: 10.1038/380555a0. [DOI] [PubMed] [Google Scholar]
- 24.Gulick AM, Song H, Endow SA, Rayment I. X-ray crystal structure of the yeast Kar3 motor domain complexed with Mg·ADP to 2.3 Å resolution. Biochemistry. 1998;37:1769–1776. doi: 10.1021/bi972504o. [DOI] [PubMed] [Google Scholar]
- 25.Sack S, Kull FJ, Mandelkow E. Motor proteins of kinesin family. Structures, variations, and nucleotide binding sites. Eur J Biochem. 1999;262:1–11. doi: 10.1046/j.1432-1327.1999.00341.x. [DOI] [PubMed] [Google Scholar]
- 26.Rice S, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 27.Yun M, Zhang X, Park C-G, Park H-W, Endow SA. A structural pathway for activation of the kinesin motor ATPase. EMBO J. 2001;20:2611–2618. doi: 10.1093/emboj/20.11.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woehlke G, et al. Microtubule interaction site of the kinesin motor. Cell. 1997;90:207–216. doi: 10.1016/s0092-8674(00)80329-3. [DOI] [PubMed] [Google Scholar]
- 29.Vanstraelen M, et al. Cell cycle-dependent targeting of a kinesin at the plasma membrane demarcates the division site in plant cells. Curr Biol. 2006;16:308–314. doi: 10.1016/j.cub.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 30.Huang J-D, et al. Direct interaction of microtubule- and actin-based transport motor. Nature. 1999;397:267–270. doi: 10.1038/16722. [DOI] [PubMed] [Google Scholar]
- 31.Beningo KA, Lillie SH, Brown SS. The yeast kinesin-related protein Smy1p exerts its effects on the class V myosin Myo2p via a physical interaction. Mol Biol Cell. 2000;11:691–702. doi: 10.1091/mbc.11.2.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lillie SH, Brown SS. Smy1p, a kinesin-related protein that does not require microtubules. Mol Biol Cell. 1998;140:873–883. doi: 10.1083/jcb.140.4.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aviser D, Prokhnevsky AI, Makarova KS, Koonin EV, Dolja VV. Myosin XI-K is required for rapid trafficking of Golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiol. 2008;146:1098–1108. doi: 10.1104/pp.107.113647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peremyslov VV, Prokhnevsky AI, Aviser D, Dolja VV. Two class XI myosins function in organelle trafficking and root hair development in Arabidopsis. Plant Physiol. 2008;146:1109–1116. doi: 10.1104/pp.107.113654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuriyama R, Gustus C, Terada Y, Uetake Y, Matuliene J. CHO1, a mammalian kinesin-like protein, interacts with F-actin and is involved in the terminal phase of cytokinesis. J Cell Biol. 2002;156:783–790. doi: 10.1083/jcb.200109090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iwai S, Ishiji A, Mabuchi I, Sutoh K. A novel actin-bundling kinesin-related protein from Dictyostelium discoideum. J Biol Chem. 2004;279:4696–4704. doi: 10.1074/jbc.M308022200. [DOI] [PubMed] [Google Scholar]
- 37.Preuss ML, et al. A plant-specific kinesin binds to actin microfilaments and interacts with cortical microtubules in cotton fibers. Plant Physiol. 2004;136:3945–3955. doi: 10.1104/pp.104.052340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.