Abstract
We report the visualization of NO production using fluorescence in tissue slices of the mouse main olfactory bulb. This discovery was possible through the use of a novel, cell-trappable probe for intracellular nitric oxide detection based on a symmetric scaffold with two NO-reactive sites. Ester moieties installed onto the fluorescent probe are cleaved by intracellular esterases to yield the corresponding negatively charged, cell-impermeable acids. The trappable probe Cu2(FL2E) and the membrane-impermeable acid derivative Cu2(FL2A) respond rapidly and selectively to NO in buffers that simulate biological conditions, and application of Cu2(FL2E) leads to detection of endogenously produced NO in cell cultures and olfactory bulb brain slices.
Keywords: fluorescent sensing, NO, olfaction, trappable probe, fluorescence microscopy
Nitric oxide (NO) is important for biological signaling. It activates soluble guanylyl cyclase, initiating a signaling cascade that promotes vascular smooth muscle dilation (1–3). Nitric oxide produced in the nervous system has been implicated in neurotransmission (4), and the immune system generates NO as a defense against pathogens (5). Unregulated nitric oxide production has been associated with pathological conditions such as cancer, ischemia, septic shock, inflammation, and neurodegeneration (6).
Because of its various biological consequences, investigating the generation, translocation, and utilization of NO continues to be an active area of research. A major limitation to advances in the field, however, has been the dearth of selective tools for visualization of biological NO, for example by fluorescence microscopy. Detection of nitric oxide offers many challenges. NO reacts rapidly in vivo with dioxygen, oxygen-generated radicals such as superoxide, amines, thiols, and metal centers (7, 8). It also diffuses readily from its point of origin (9), making rapid detection desirable for uncovering both its production site and function. Moreover, NO is produced at concentrations as low as ∼100 picomolar, so it is essential to have an NO probe with a low detection limit (10). Fluorescent sensors can be designed to accommodate the properties of NO under physiological conditions, making this technique particularly valuable for in vivo nitric oxide imaging.
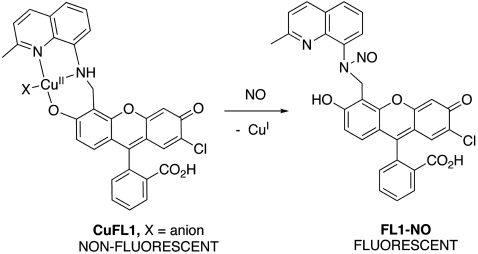
Transition metal complexes have been investigated as platforms for NO detection (11). The strategy is to incorporate a fluorophore into a ligand that is quenched either by intracellular photoinduced electron transfer and/or by coordination to a paramagnetic or heavy metal ion. Fluorescence is restored by reduction of the metal and/or displacement of the ligand upon reaction of the probe with NO. CuFL1 (Fig. 1) is an excellent example of such a metal-based cellular NO imaging agent (12, 13). CuFL1 satisfies many of the requirements of a good sensor. It is nontoxic, cell membrane permeable, has low energy excitation and emission wavelengths, responds directly and selectively to NO, and exhibits dramatic fluorescence enhancement upon reaction with NO. When the nonemissive CuFL1 reacts with NO, Cu(II) is reduced to Cu(I) and the secondary amine of the ligand is N-nitrosated to produce the fluorescent species, FL1-NO (Fig. 1). CuFL1 has been used to monitor NO production in bacterial cell cultures with great success (14, 15), but to improve its applicability a cell-trappable version of the ligand is required. Cell-trappability is a key feature for biological experiments in intact tissues for which continual perfusion of tissue slices with artificial physiological saline solution is required. Under perfusion conditions, CuFL1 diffuses out of cells, making analysis of the results difficult. Incorporating an ester moiety onto the ligand is one way to confer trappability to a probe (16). The probe remains cell membrane permeable until cytosolic esterases cleave the ester to yield a carboxylate, which is negatively charged at physiological pH. This transformation traps the probe within the cell.
Fig. 1.
Structure and NO detection scheme of CuFL1.
The mammalian olfactory bulb (OB), the first site for synaptic evaluation of olfactory receptor input in the CNS, is an ideal site to elucidate the functions of NO. Immunocytochemistry experiments confirm that the OB is rich in NO-producing cells (17). Direct measurements of NO in OBs, both in vivo and in vitro, using an NO-selective microprobe, reveal significant resting levels of NO and also odor-stimulated increases in NO (18). These results are of interest because of literature linking NO levels in the OB to odor learning and memory storage (19–21). To determine the effects of NO on synaptic interactions in the OB circuit, it would be of great value to localize NO sources and sinks on the seconds time scale with cellular resolution in living brain slices of the OB suitable for electrophysiological analysis (22, 23). The development of the cell-trappable NO probe described here is an important contribution not only to these analyses, but potentially to studies of the cellular and synaptic actions of NO in many other brain regions, e.g., the hippocampus (24–26) and cerebellum (27, 28), where NO has also been implicated in processes of synaptic plasticity, learning, and memory. Ultimately, one might envision application of the trappable NO probe to anaesthetized mice exposed to odorants at the nose while monitoring NO production in the OB by fluorescence optical imaging.
Herein we describe the synthesis and characterization of a set of fluorescein-based symmetrical ligands and their Cu(II) complexes as NO-specific fluorescent probes. These second-generation probes are based on CuFL1 and employ the ester/acid strategy for cell-trappability. The Cu(II) complexes, generated in situ, respond rapidly and selectively to nitric oxide over other biologically relevant reactive oxygen and nitrogen species (RONS). We present data showing fluorescence detection of endogenously produced NO in macrophage and neuroblastoma cells, together with mouse olfactory bulb slice experiments that visually confirm and extend previous electrochemical results.
Results
Ligand Construction and Properties.
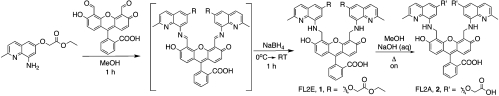
The synthetic procedures that we used to generate the modified fluoresceins are outlined in Fig. 2. Condensation of ethyl[(8-amino-2-methylquinolin-6-yloxy)acetate] with 4′,5′-fluorescein dialdehyde in a 2∶1 ratio in methanol followed by reduction using sodium borohydride afforded FL2E, 1. The acid form of the ligand was prepared by saponification of FL2E in methanol and water to yield FL2A, 2.
Fig. 2.
Syntheses of FL2E and FL2A.
The free ligands emit weakly with maxima at 522 and 516 nm for FL2E and FL2A, respectively, with quantum yields of 0.37 ± 0.05% and 1.8 ± 0.2%. When 1 μM buffered sensor solutions, prepared in situ by combining CuCl2 and FL2E or FL2A in a 2∶1 ratio, were exposed to excess NO (∼1,300 equiv) under anaerobic conditions, the emission maxima shifted to 526 nm for both probes, in accord with formation of the N-nitrosated ligand (12). Accompanying this wavelength shift was a 17 ± 2-fold and 27 ± 3-fold increase in the integrated fluorescence emission relative to the initial copper complex for Cu2(FL2E) and Cu2(FL2A), respectively. The final quantum yields were 40 ± 8% and 36 ± 5%, respectively. A fluorescence response was also observed when the probes were exposed to S-nitrosothiols (RSNO), such as S-nitroso-N-acetyl-DL-penicillamine (SNAP). The observed fluorescence was reduced compared to that generated using gaseous NO, because SNAP must decompose to produce NO before NO can react with the probe, and the decomposition kinetics are relatively slow under the conditions of the experiment. The probes are selective for NO or RSNO over other biologically relevant RONS, exhibiting only modest changes in fluorescence when exposed for 1 h to excess  ,
,  , H2O2, ClO-, ONOO- or HNO (Fig. S1).
, H2O2, ClO-, ONOO- or HNO (Fig. S1).
Fluorescence Microscopic Imaging of Biologically Derived NO Production.
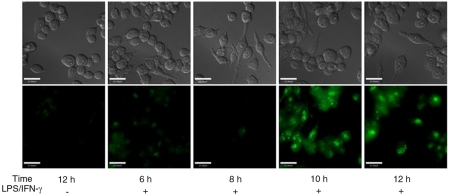
We first evaluated the ability of the trappable probe to detect endogenously produced NO. Macrophage cells of the immune system produce micromolar quantities of NO when inducible nitric oxide synthase (iNOS) is activated in response to a pathogenic attack (5). Endotoxins from pathogens, such as lipopolysaccharide (LPS), and proinflammatory cytokines, such as interferon-γ (IFN-γ), induce expression of the iNOS gene, which results in NO production several hours later. To determine whether Cu2(FL2E) can detect NO under these conditions, RAW 264.7 macrophages were stimulated with LPS (500 ng/mL) and IFN-γ (250–1,000 U/mL) for 4 h, and then incubated with 1 μM Cu2(FL2E) for an additional 2–8 h. In a control experiment, cells were incubated for 12 h with Cu2(FL2E) in the absence of an iNOS inducer. The results of these experiments are shown in Fig. 3. A visible increase in fluorescence was observed 6–8 h following sensor incubation (10–12 h after induction of iNOS) that was not observed in the control cells or in cells visualized at earlier times points (6–8 h postinduction). These results are consistent with previous findings using CuFL1 to image NO production in RAW 264.7 macrophages (13), indicating that Cu2(FL2E) can also detect endogenously produced nitric oxide.
Fig. 3.
RAW 264.7 macrophages coincubated with 1 μM Cu2(FL2E), 500 ng/mL LPS, and 250–1,000 U/mL IFN-γ for 6–12 h. (Top) DIC images; (Bottom) emission from the probe in the green channel. Scale bars, 25 μm.
Brain-derived neuronal nitric oxide synthase (nNOS) is activated following an increase in intracellular Ca2+ levels to produce up to nanomolar concentrations of NO (10). When excess intracellular calcium is present it binds to the regulatory protein calmodulin, which undergoes a conformational change that triggers binding to, and activation of, nNOS (29). Addition of 17-β-estradiol (β-ES) to neuronal cells activates the estrogen receptor, which opens calcium channels to allow an increase in intracellular Ca2+ levels (30). To demonstrate that Cu2(FL2E) can detect low levels of biological NO, human SK-N-SH neuroblastoma cells were coincubated with 5 μM Cu2(FL2E) and 100 nM β-ES for 30 min. As shown in Fig. S2, an increase in fluorescence was observed for plates stimulated with β-ES versus controls with no β-ES, consistent with results obtained with CuFL1 (13).
Probe Localization in SK-N-SH Cells.
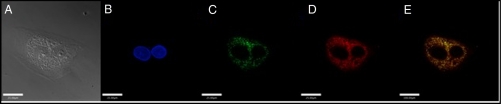
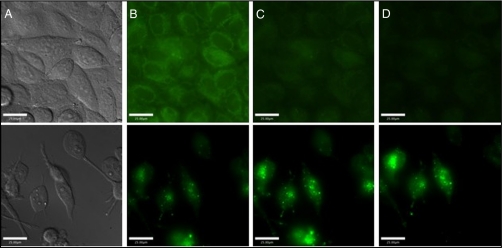
Many fluorescein-based probes localize to specific subcellular regions, such as the mitochondria or Golgi apparatus (31). In the SK-N-SH imaging experiments, punctate staining of the cells was observed, suggesting that Cu2(FL2E) localizes to a subcellular compartment. To investigate this hypothesis, SK-N-SH neuroblastoma cells were coincubated with β-ES (100 nM), Cu2(FL2E) (5 μM), Hoechst 33258 (4.5 μM, nuclear stain), and Mitotracker Red (0.2 μM, mitochondrial stain). Fig. 4 illustrates the findings of this experiment. The probe did not enter the nucleus (Fig. 4B), as expected if the ester is rapidly converted to the acid in the cell, preventing it from traversing the nuclear membrane. Overlay of the green (Fig. 4C) and red (Fig. 4D) detector channels indicates colocalization as demonstrated by the yellow color in the image (Fig. 4E). Fig. S3 presents colocalization scatter plots of the blue vs. green and red vs. green channels as an alternate way to visualize the colocalization. These data clearly reveal that Cu2(FL2E) associates with the mitochondria of SK-N-SH neuroblastoma cells.
Fig. 4.
SK-N-SH neuroblastoma cells coincubated with 5 μM Cu2(FL2E), 100 nM 17-β-estradiol, 0.2 μM Mitotracker Red, and 4.5 μM Hoechst 33258 for 30 min. (A) DIC image; (B) emission from the blue channel (nuclear stain); (C) emission from the green channel (NO probe); (D) emission from the red channel (mitochondrial stain); and (E) overlay of the green and red channels. Scale bars, 25 μm.
Cell-Trappability of Cu2(FL2E).
CuFL1 readily diffuses into cells and responds to NO produced by both cNOS and iNOS in vivo (13). Because CuFL1 is not trappable, it can diffuse back out of cells under many conditions. This is an important shortcoming; experiments in tissue slices require perfusion of media throughout the experiment to maintain tissue health, and experiments in live animals have body fluids continually perfusing the tissues. To demonstrate diffusibility for CuFL1, HeLa cells were incubated with 10 μM of the probe for 30 min and then washed and bathed in phosphate buffered saline (PBS) prior to imaging. An initial wash did not significantly reduce the probe signal. Subsequently, the cells were maintained in a microscope incubator at 37 °C with 5% CO2 and imaged every 10 min over the course of an hour, washing between each image. To wash the cells, the PBS was carefully removed by pipette while the plate remained on the microscope and then fresh PBS was added. The PBS was removed and replaced three times to attempt to mimic conditions where the cells would be perfused with fluids. After 20 min, fluorescence microscopy revealed virtually no CuFL1 remaining in the cells (Fig. 5), consistent with diffusion of CuFL1 out of the cells.
Fig. 5.
Cell imaging experiments demonstrating diffusion of CuFL1 and retention of Cu2(FL2E). (A) DIC image; (B) emission from the green channel at 0 min; (C) 10 min and (D) 20 min. (Top) HeLa control cells incubated with 10 μM CuFL1 for 30 min. (Bottom) RAW 264.7 macrophages incubated with 1 μM Cu2(FL2E), 500 ng/mL LPS, and 250–1,000 U/mL IFN-γ for 12 h.
A similar study was performed using the trappable NO probe. RAW 264.7 macrophages were stimulated with LPS and IFN-γ for 4 h and then incubated with Cu2(FL2E) for 12 h. The cells were washed and imaged while in the microscope incubator under the same conditions as with the HeLa cells. Fig. 5 illustrates that the fluorescence signal is not diminished over time, confirming that the sensor is indeed trappable.
As a second demonstration of trappability, macrophages were coincubated with Cu2(FL2A), the biologically relevant species, and LPS and IFN-γ. Upon washing the cells and imaging, no fluorescent signal was present (Fig. S4). This result is consistent with our expectation that the negative charges of the deprotonated carboxylates prevent the probe from entering cells.
Cytotoxicity Studies.
An ideal probe is minimally cytotoxic. Over the course of experiments in both RAW 264.7 and SK-N-SH cells there was no visible cell death; however, these experiments were performed over relatively short time periods (< 24 h) with low micromolar probe concentrations. To investigate the potential toxicity of Cu2(FL2E) toward SK-N-SH cells, the least robust of the cell lines used for imaging, over a longer time frame, we carried out 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays (32) for a large range of sensor concentrations over 3 d. The survival of SK-N-SH cells incubated with 5 μM Cu2(FL2E) was very good, with 86 ± 1% of cells still alive after 3 d. The 50% lethal concentration (LC50) of the sensor after a 3 d incubation period was ∼50 μM, indicating that the probe concentration can be increased by an order of magnitude before substantial cell death occurs. The survival of SK-N-SH cells incubated with 1 mM Cu2(FL2E) for only 1 d was 75 ± 1%. Thus shorter experiments can tolerate very high probe concentrations, which is sometimes necessary for successful loading into tissue slices.
NO Detection in the Mouse Main Olfactory Bulb.
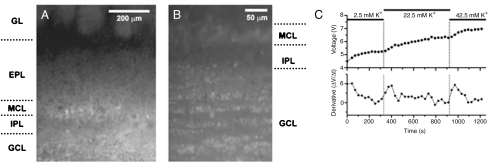
Slices were incubated with probe loading solution and, after washing excess probe from the bathing solution, widespread fluorescence in all cell layers of the OB was observed (Fig. 6). Under low magnification (Fig. 6A), prominently labeled glomeruli and brightly stained cell somata in the mitral and granule cell layers were clearly highlighted against a background of diffuse fluorescence. Under higher magnification, the characteristic laminar arrangements of granule cell somata were revealed (Fig. 6B). The mitral cell layer was separated from granule cells by the inner plexiform layer, which was sparsely labeled. In control slices, only very weak background fluorescence was detected from all layers of the OB, and cell somata were not clearly resolved. Compared to control slices, fluorescence measured after a 1 h recovery period using a FITC filter set and averaged over an area of 0.663 mm2 in OB slices loaded with Cu2(FL2E) for 2 h increased by factors of 4 ± 1 in the glomerular layer, 4 ± 2 in the external plexiform layer, and 14 ± 3 in the granule cell layer.
Fig. 6.
Static and dynamic fluorescence imaging of Cu2(FL2E) in OB slices. (A) Low magnification fluorescence image of a mouse OB slice after 2 h incubation in loading solution with Cu2(FL2E). (B) High magnification fluorescence image of sensor-loaded mouse OB slice, revealing numerous granule cell somata arrayed in densely packed laminae. (C) Time dependence of cell fluorescence after loading. (Top) Plot of CCD camera readout voltage versus time. Bath KCl was initially 2.5 mM and was incremented by 20 mM at 330 s and 920 s after the start of recording. (Bottom) Plot of the time derivative of the upper trace to reveal the rate of NO binding. Abbreviations: GL, glomerular layer; EPL, external plexiform layer; MCL, mitral cell layer; IPL, internal plexiform layer; GCL, granule cell layer.
After loading, fluorescence from granule layer cell somata was monitored over time, acquiring frames every 30 s using 8 ms of illumination per frame. Under control conditions with 400 μM L-arginine and the basal level of KCl (2.5 mM) in the bath, monotonic increases in fluorescence were measured with a steadily decreasing slope over the time period of 330–760 s (Fig. 6C). The derivatives of the time courses of cell fluorescence signals decayed with a time constant of 263 ± 120 s (n = 4 cells). These observations demonstrate that the probe can be taken up and trapped by cells in brain slice preparations.
The strong initial fluorescence and its persistent increase over time indicate tonic production of NO in the OB slice, consistent with findings obtained by electrochemical microprobe detection (18). To verify the physiological origin of the fluorescence, slices were stimulated by increasing extracellular KCl levels. Applications of 20 mM KCl to the slice were consistently followed by transient elevations in the rate of fluorescence increase, as expected because KCl depolarizes neurons, causing Ca2+ to enter the cells and induce NO synthesis (Fig. 6C). The time derivative of the fluorescence signal increased by a factor of 12 ± 7 (range of increase 3.9–26.2, n = 8 trials from 4 cells, p = 0.0003), from which we estimate that NO levels in the slice were boosted from a basal value of ∼85 nM to ∼1 μM for 1–2 min after KCl addition (18).
Discussion
Efficient synthetic procedures for generating unique ligands for preparing the NO-specific probes FL2E and FL2A have been devised. Cu(II) complexes assembled by combining CuCl2 with either ligand in a 2∶1 ratio display photophysical characteristics that are consistent with the asymmetric probe CuFL1 (12). The resulting complexes are excellent nitric oxide probes and are highly selective for NO and RSNO over other biologically relevant RONS.
The trappable probe detects endogenously produced NO from cNOS and iNOS, consistent with the capabilities of CuFL1 (13). Like CuFL1, the trappable probe is minimally toxic. Unlike CuFL1, however, the trappable probe is maintained within cells despite numerous changes of the extracellular medium, confirming that installing acetoxyethyl esters confers cell trappability. Cu2(FL2E) localizes to the mitochondria, a feature also observed for the Zn(II) sensor ZS5 (31). The structure of ZS5 resembles that of FL2 up to the secondary amine of FL2E. In ZS5 the amine is tertiary and is appended to methyl pyridyl and methyl thiophene units. The fluorescein platform alone is not responsible for the mitochondrial localization, because fluorescein-based probes sometimes localize to other regions of the cell (31). The upper-ring substituents are not similar in ZS5 and FL2E, suggesting that some other, as yet unknown, factor is responsible for directing these sensors to mitochondria.
Our ultimate purpose in devising NO-specific probes is to apply them in experiments pertaining to the time-dependent production of NO in the mammalian brain. Because CuFL1 diffuses out of cells under conditions of tissue perfusion, no conclusive results were obtained from experiments using CuFL1 in mouse OB tissue slices. This shortcoming led to the present design of the cell-trappable probe, and with it OB tissue slices were successfully loaded and fluorescence caused by NO production visualized.
The detection of NO probe fluorescence in mouse OB slices is consistent with results obtained using an NO-selective microprobe to monitor robust basal NO production in the mitral and granule cell layers of slices (18). The NO microprobe technique provides fast (seconds time scale), sensitive, dynamic detection of extracellular NO signals, but is limited in spatial resolution to a single site ∼10 μm in diameter sampling the extracellular environment of only one or a few neurons. By contrast, the fluorescent probe described herein offers slower (minutes time scale), parallel, time-integrated detection of intracellular NO signals from dozens to hundreds of 10–15 μm sized cells in slices that can trap the fluorophore. Extensive fluorescent labeling of slices suggests that tonic NO production is not confined to the granule cell layer, but is widespread throughout the OB. The most intense fluorescence occurs in the glomerular and granule cell layers, with weaker labeling of the external plexiform layer. This result mirrors the large-scale distribution of nNOS immunoreactive cells and fibers that has been documented in the mouse OB (17, 33–36). The higher density of nNOS expression in the glomerular and granule cell layers might result in elevated binding of NO by locally trapped probe molecules. However, at the cellular level the patterns of fluorescence and NOS immunoreactivity should not be directly correlated, because the probe can be loaded into both NOS-positive and NOS-negative cells. For example, mitral cells in mouse OB are NOS-negative, but their somata displayed strong fluorescence in the present experiments. Probe trapped in mitral cells may be reporting NO that was synthesized in the granule cell layer and subsequently diffused to the mitral cell layer. The basal NO level was previously measured to be ∼85 nM in the vicinity of mitral cell somata (18). One potential caveat of this interpretation is that NO may be continuously generated by cells during the loading period, and therefore some fraction of the sensor could bind NO before entry into cells. FL2E-NO could then reach and load cells beyond the normal spatial signaling range of NO in living tissue. To block this effect, the competitive NOS inhibitor Nω-nitro-L-arginine was added and exogenous L-arginine was omitted during loading. NO production was not completely suppressed, as indicated by persistent probe fluorescence. However, the kinetics of postloading fluorescence should depend on reception of NO signals by target cells because extracellular probe will be removed by perfusion (Fig. 6C), and thus postloading fluorescence signal increases will accurately report cellular patterns of NO synthesis and neurotransmission in the neural circuit.
We expected that our fluorescent probe could provide spatial information about the location of NO sources if extracellular NO were intercepted by an NO scavenger. We tested commonly used scavengers, 2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl 3-oxide (PTIO), its derivative carboxy-PTIO, hemoglobin, and myoglobin, but these were not compatible with the fluorescent probe: PTIO is membrane permeant and competes with the probe for NO binding, carboxy-PTIO reacts with NO to yield NO2 which binds the probe, and both hemoglobin and myoglobin have strong optical absorption at the emission wavelengths of the probe. However, the door is open for synthesis of new scavengers that would be compatible with the probe and enable localization of NO generating cells.
In conclusion, optical recordings of steady state or stimulus-driven increases in fluorescence in slices promise to yield insights into spatiotemporal properties and physiological regulators of NO signaling in the central nervous system. Future improvements in probe design to limit the fluorescence response to intracellular, rather than tonic tissue sources, of NO should move the field toward functional imaging of neuronal signaling at the chemical, in addition to the electrophysiological, level.
Materials and Methods
Details describing the chemical reagents; synthetic materials and methods; synthetic procedures; spectroscopic materials and methods; cell culture; diffusion studies; NO detection experiments; imaging methods; cytotoxicity assays; olfactory bulb imaging; olfactory bulb preparation; nitric oxide probe loading; and animal care can be found in SI Text. Ligand NMR spectra are reported in Figs. S5 and S6.
Supplementary Material
Acknowledgments.
We thank Dr. Mi Hee Lim for preliminary studies and helpful discussions, Drs. Michael Pluth and Zachary Tonzitech for valuable comments, Dr. Wee Han Ang for assistance with the MTT assays, and Dr. Elisa Tomat for help with cell image analysis. This work was supported by the National Science Foundation Grant CHE-061194 (to S.J.L.) and by the National Institute on Deafness and Other Communication Disorders of the National Institutes of Health Grant DC-004208 (to G.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914794107/DCSupplemental.
References
- 1.Furchgott RF, Vanhoutte PM. Endothelium-derived relaxing and contracting factors. FASEB J. 1989;3:2007–2018. [PubMed] [Google Scholar]
- 2.Ignarro LJ, Buga GM, Wood KS, Byrns RE, Chaudhuri G. Endothelium-derived relaxing factor produced and released from artery and vein is nitric oxide. Proc Natl Acad Sci USA. 1987;84:9265–9269. doi: 10.1073/pnas.84.24.9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rapoport RM, Draznin MB, Murad F. Endothelium-dependent relaxation in rat aorta may be mediated through cyclic GMP-dependent protein phosphorylation. Nature. 1983;306:174–176. doi: 10.1038/306174a0. [DOI] [PubMed] [Google Scholar]
- 4.Garthwaite J. Concepts of neural nitric oxide-mediated transmission. Eur J Neurosci. 2008;27:2783–2802. doi: 10.1111/j.1460-9568.2008.06285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogdan C. Nitric oxide and the immune response. Nat Immunol. 2001;2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 6.Ignarro LJ. Nitric Oxide Biology and Pathobiology. San Diego: Academic; 2000. [Google Scholar]
- 7.Wink DA, Grisham MB, Mitchell JB, Ford PC. Direct and indirect effects of nitric oxide in chemical reactions relevant to biology. Method Enzymol. 1996;268:12–31. doi: 10.1016/s0076-6879(96)68006-9. [DOI] [PubMed] [Google Scholar]
- 8.Wink DA, Mitchell JB. Chemical biology of nitric oxide: Insights into regulatory, cytotoxic, and cytoprotective mechanisms of nitric oxide. Free Radical Biol Med. 1998;25:434–456. doi: 10.1016/s0891-5849(98)00092-6. [DOI] [PubMed] [Google Scholar]
- 9.Lancaster JR., Jr A tutorial on the diffusibility and reactivity of free nitric oxide. Nitric Oxide. 1997;1:18–30. doi: 10.1006/niox.1996.0112. [DOI] [PubMed] [Google Scholar]
- 10.Hall CN, Garthwaite J. What is the real physiological NO concentration in vivo? Nitric Oxide. 2009;21:92–103. doi: 10.1016/j.niox.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim MH, Lippard SJ. Metal-based turn-on fluorescent probes for sensing nitric oxide. Acc Chem Res. 2007;40:41–51. doi: 10.1021/ar950149t. [DOI] [PubMed] [Google Scholar]
- 12.Lim MH, et al. Direct nitric oxide detection in aqueous solution by copper(II) fluorescein complexes. J Am Chem Soc. 2006;128:14364–14373. doi: 10.1021/ja064955e. [DOI] [PubMed] [Google Scholar]
- 13.Lim MH, Xu D, Lippard SJ. Visualization of nitric oxide in living cells by a copper-based fluorescent probe. Nat Chem Biol. 2006;2:375–380. doi: 10.1038/nchembio794. [DOI] [PubMed] [Google Scholar]
- 14.Patel BA, et al. Endogenous nitric oxide regulates the recovery of the radiation-resistant bacterium Deinococcus radiodurans from exposure to UV light. Proc Natl Acad Sci USA. 2009;106:18183–18188. doi: 10.1073/pnas.0907262106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shatalin K, et al. Bacillus anthracis-derived nitric oxide is essential for pathogen virulence and survival in macrophages. Proc Natl Acad Sci USA. 2008;105:1009–1013. doi: 10.1073/pnas.0710950105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsien RY. A non-disruptive technique for loading calcium buffers and indicators into cells. Nature. 1981;290:527–528. doi: 10.1038/290527a0. [DOI] [PubMed] [Google Scholar]
- 17.Kosaka T, Kosaka K. Heterogeneity of nitric oxide synthase-containing neurons in the mouse main olfactory bulb. Neurosci Res. 2007;57:165–178. doi: 10.1016/j.neures.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Lowe G, Buerk DG, Ma J, Gelperin A. Tonic and stimulus-evoked nitric oxide production in the mouse olfactory bulb. Neuroscience. 2008;153:842–850. doi: 10.1016/j.neuroscience.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kendrick KM, et al. Formation of olfactory memories mediated by nitric oxide. Nature. 1997;388:670–674. doi: 10.1038/41765. [DOI] [PubMed] [Google Scholar]
- 20.Okere CO, Kaba H. Increased expression of neuronal nitric oxide synthase mRNA in the accessory olfactory bulb during the formation of olfactory recognition memory in mice. Eur J Neurosci. 2000;12:4552–4556. doi: 10.1046/j.0953-816x.2000.01325.x. [DOI] [PubMed] [Google Scholar]
- 21.Romero-Grimaldi C, Gheusi G, Lledo PM, Estrada C. Chronic inhibition of nitric oxide synthesis enhances both subventricular zone neurogenesis and olfactory learning in adult mice. Eur J Neurosci. 2006;24:2461–2470. doi: 10.1111/j.1460-9568.2006.05127.x. [DOI] [PubMed] [Google Scholar]
- 22.Ma J, Lowe G. Action potential backpropagation and multiglomerular signaling in the rat vomeronasal system. J Neurosci. 2004;24:9341–9352. doi: 10.1523/JNEUROSCI.1782-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma J, Lowe G. Calcium permeable AMPA receptors and autoreceptors in external tufted cells of rat olfactory bulb. Neuroscience. 2007;144:1094–1108. doi: 10.1016/j.neuroscience.2006.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hopper RA, Garthwaite J. Tonic and phasic nitric oxide signals in hippocampal long-term potentiation. J Neurosci. 2006;26:11513–11521. doi: 10.1523/JNEUROSCI.2259-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szabadits E, et al. Hippocampal GABAergic synapses possess the molecular machinery for retrograde nitric oxide signaling. J Neurosci. 2007;27:8101–8111. doi: 10.1523/JNEUROSCI.1912-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taqatqeh F, et al. More than a retrograde messenger: Nitric oxide needs two cGMP pathways to induce hippocampal long-term potentiation. J Neurosci. 2009;29:9344–9350. doi: 10.1523/JNEUROSCI.1902-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Namiki S, Kakizawa S, Hirose K, Iino M. NO signalling decodes frequency of neuronal activity and generates synapse-specific plasticity in mouse cerebellum. J Physiol. 2005;566:849–863. doi: 10.1113/jphysiol.2005.088799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schweighofer N, Ferriol G. Diffusion of nitric oxide can facilitate cerebellar learning: A simulation study. Proc Natl Acad Sci USA. 2000;97:10661–10665. doi: 10.1073/pnas.97.19.10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alderton WK, Cooper CE, Knowles RG. Nitric oxide synthases: Structure, function and inhibition. Biochem J. 2001;357:593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia Y, Krukoff TL. Estrogen induces nitric oxide production via activation of constitutive nitric oxide synthases in human neuroblastoma cells. Endocrinology. 2004;145:4550–4557. doi: 10.1210/en.2004-0327. [DOI] [PubMed] [Google Scholar]
- 31.Nolan EM, et al. Zinspy sensors with enhanced dynamic range for imaging neuronal cell zinc uptake and mobilization. J Am Chem Soc. 2006;128:15517–15528. doi: 10.1021/ja065759a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mosmann T. Rapid colorimetric assay for cellular growth and survival—application to proliferation and cyto-toxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 33.Giuili G, Luzi A, Poyard M, Guellaën G. Expression of mouse brain soluble guanylyl cyclase and NO synthase during ontogeny. Devl Brain Res. 1994;81:269–283. doi: 10.1016/0165-3806(94)90313-1. [DOI] [PubMed] [Google Scholar]
- 34.Gotti S, Sica M, Viglietti-Panzica C, Panzica G. Distribution of nitric oxide synthase immunoreactivity in the mouse brain. Microsc Res Tech. 2005;68:13–35. doi: 10.1002/jemt.20219. [DOI] [PubMed] [Google Scholar]
- 35.Kishimoto J, Keverne EB, Hardwick J, Emson PC. Localization of nitric oxide synthase in the mouse olfactory and vomeronasal system—a histochemical, immunological and in-situ hybridization study. Eur J Neurosci. 1993;5:1684–1694. doi: 10.1111/j.1460-9568.1993.tb00236.x. [DOI] [PubMed] [Google Scholar]
- 36.Weruaga E, et al. NADPH-diaphorase histochemistry reveals heterogeneity in the distribution of nitric oxide synthase-expressing interneurons between olfactory glomeruli in two mouse strains. J Neurosci Res. 1998;53:239–250. doi: 10.1002/(SICI)1097-4547(19980715)53:2<239::AID-JNR13>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.