Abstract
NK cell-mediated murine cytomegalovirus (MCMV) resistance (Cmvr) is under H-2k control in MA/My mice, but the underlying gene(s) is unclear. Prior genetic analysis mapped Cmvr to the MHC class I (MHC-I) Dk gene interval. Because NK cell receptors are licensed by and responsive to MHC class I molecules, Dk itself is a candidate gene. A 10-kb genomic Dk fragment was subcloned and microinjected into MCMV-susceptible (Cmvs) (MA/My.L-H2b × C57L)F1 or (B6 × DBA/2)F2 embryos. Transgenic founders, which are competent for Dk expression and germline transgene transmission, were identified and further backcrossed to MA/My.L-H2b or C57L mice. Remarkably, Dk expression delivered NK-mediated resistance in either genetic background. Further, NK cells with cognate inhibitory Ly49G receptors for self-MHC-I Dk were licensed and critical in protection against MCMV infection. In radiation bone marrow chimeras, NK resistance was significantly diminished when MHC-I Dk expression was restricted to only hematopoietic or nonhematopoietic cells. Thus, MHC-I Dk is the H-2k-linked Cmvr locus; these findings suggest a role for NK cell interaction with Dk-bearing hematopoietic and nonhematopoietic cells to shape NK-mediated virus immunity.
Keywords: NK cell, virus immunity, Ly49G, MCMV, H-2D transgenic
Natural killer (NK) cells with diverse cell-surface receptors provide front-line innate immunity against tumors and viral pathogens (1, 2). Certain polymorphic human KIR and mouse Ly49 NK receptors recognize MHC class I (MHC-I) or class I-like proteins (3). In recent studies, these NK inhibitory receptors were shown to contribute to NK cell self-tolerance and education and, subsequently, mature NK cell effector functions (4–6). Stimulatory NK receptors recognize host or pathogen ligands, although many ligands for these receptors are still unknown. Recent genetic data from chronic virus-infected patient cohorts have revealed that certain combined KIR and HLA class I genotypes correspond to disease protection (7–10). A greater potential for NK stimulation through KIR3DS1 activation receptor recognition of HLA Bw4 or stronger inhibition via inhibitory receptor KIR3DL1 and Bw4 allotypic binding pairs was implicated in delayed AIDS progression. Relatively weaker inhibitory receptor KIR2DL3-HLA C1 pairings corresponded with resolution of infection in chronic HCV patients. The underlying mechanism(s) of disease resistance influenced by MHC polymorphism and particular NK cell receptors, however, remain elusive.
In an acute virus infection model in mice, MHC (H-2k) loci were found to protect C3H, CBA, and BALB.K mice from lethal murine cytomegalovirus (MCMV) infection (11, 12). Consistent with the previous mortality studies, resistance was also observed in MA/My (H-2k) mice and in H-2k-expressing offspring obtained by crossing MA/My with MCMV-susceptible C57L (H-2b) or BALB/c mice (13–15). Related studies hinted that activated NK cells or NK cytotoxicity contributed to the H-2k resistance effect (16, 17). In support, we found that H-2k was essential to NK cell-mediated MCMV resistance in reciprocal H-2 congenic MA/My and C57L strains (14, 18). Through further positional cloning, we determined that a 0.3-Mb interval, including the MHC-I D gene, was indispensable for the H-2k resistance effect (19).
MHC-I Dk is a known ligand for the inhibitory Ly49G receptor (20, 21). Ly49G2+ NK cells were found to be crucial to resistance in H-2 recombinant congenic R7 (Cmvr) mice (19). MHC-I Dk on MCMV-infected cells is also recognized by the Ly49P activation receptor (15). We hypothesized, therefore, that MHC-I Dk contributes to NK-mediated resistance through Ly49 receptors. Yet it remained to be determined that MHC-I Dk itself is required for in vivo resistance or whether resistance is specifically due to polymorphisms at surrounding genes that could contribute to MCMV phenotypes. To test the hypothesis that MHC-I Dk is required for MCMV resistance, a genomic gene fragment was cloned and used to generate transgenic mice on different MCMV-susceptible genetic backgrounds. We examined transgenic mice for MHC-I Dk expression, virus resistance phenotypes, and the role of NK cells in virus immunity.
Results
Generation and Analysis of MHC Class I Dk Transgenic Mice.
A narrow interval, spanning the MHC-I D locus, is critical in H-2k resistance to MCMV infection (19). Given its inherent polymorphism and potential to interact with membrane-bound receptors at the surface of NK cells, Dk is an important candidate. Nonetheless, Dk itself has not been established as the resistance gene. To study this possibility, we generated MHC-I Dk transgenic mice (Fig. S1, Table S1, and Table 1). Peripheral blood from each of the founder lines was examined for cell surface Dk expression. All or ∼30% of Tg3-Dk or Tg1-Dk leukocytes, respectively, were stained with mAb 15-5-5, specific for MHC-I Dk (Fig. 1A), whereas no specific staining was observed for cells taken from Tg2-Dk (Table 2) or any of the nontransgenic littermate controls. Because Dk surface expression in Tg1-Dk and Tg3-Dk lines was slightly higher than that observed for H-2k/b mice with a single Dk allele (Fig. 1A), it was possible that the founder lines carried more than a single transgene copy, or that the insertion site in the genome affected transgenic expression. This might have contributed to an altered ratio of CD4+ and CD8+ T cells in Tg3-Dk × C57L offspring (Fig. 1B). Nonetheless, splenic T cells were not altered in Tg3-Dk × M.L-H2b offspring, nor were NK cell numbers significantly different in either genetic setting (Fig. 1B).
Table 1.
Characterization of H-2k transgenic founders
| Founder | Background | H-2 | Germline transmission | Tg-Dk cell-surface expression |
| Tg1 | (B6 × DBA/2)F2 | b/d | + | Mosaic |
| Tg2 | (B6 × DBA/2)F2 | b | + | − |
| Tg3 | (C57L × M.L-H2b)F2 | b | + | + |
Fig. 1.
Expression of H-2Dk in transgenic mice. PBMCs or splenocytes taken from transgenic founder mice were stained for CD3, NK1.1 or NKp46, CD4, CD8, and cell-surface H-2Dk using mAb 15-5-5. (A Upper) Histograms for H-2Dk expression (solid line) for PBMCs from Tg1-Dk and Tg3-Dk founders. H-2Dk expression for control PBMCs taken from congenic R7 (broken line) and nontransgenic littermate controls (shaded histograms) are also shown. (Lower) Splenocytes from (Tg3-Dk × M.L-H2b)N2 (also called Tg3MN2), (Tg3-Dk × C57L)N2 (also called Tg3LN2), or (Tg1-Dk × M.L-H2b)N1 (also called TgMN1) offspring (solid lines), R7 congenic mice (broken lines), or nontransgenic littermate controls (shaded histograms) are similarly stained. Data are representative of 2–4 mice. (B) Proportions of distinct spleen lymphocyte subsets found in uninfected R7, Tg3LN2, or Tg3MN2 and their nontransgenic littermates. Error bars indicate SD. Statistical analysis was performed for all groups except R7 using a Student's t test. *P < 0.05, **P < 0.01.
Table 2.
Donor cell reconstitution in mixed H-2D bone marrow chimeras
| Percent chimera in PBMCs* | |||||
| Donor | Recipient | Leukocytes | NK cells | CD19+ B | CD3+ T |
| Tg3-Dk | Tg3-Dk | ND | ND | ND | ND |
| Tg3-Dk | Non-Tg | 90.8 ± 3 | 98.8 ± 0.7 | 99.6 ± 0.1 | 64.6 ± 12.9 |
| Non-Tg | Tg3-Dk | 85.2 ± 1.2 | 98 ± 0.9 | 97.5 ± 1.1 | 48.5 ± 8.6 |
| Non-Tg | Non-Tg | ND | ND | ND | ND |
ND, not determined.
*PBMCs stained for Dk, NKp46, CD3, and CD19 in flow cytometry. The proportions of PBMCs displaying H-2Dk were used to assess percent chimerism for lymphoctye subsets.
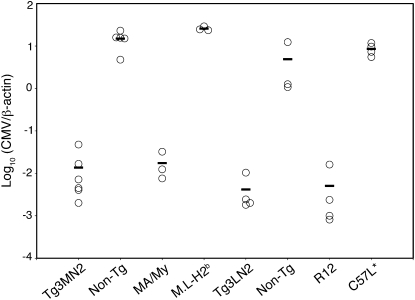
Inasmuch as the MA/My-derived MCMV-resistance allele was dominant in (MA/My × C57L) hybrid offspring (14), we assessed the impact of Tg3-Dk expression in MCMV-susceptible (Fig. S2) F1.H2b mice. As shown in Fig. 2, MHC-I Dk expression in the transgenic mice produced virus resistance just as effective as that observed in MA/My or R12 mice. The effect was evident in animals on either the C57L or M.L-H2b backgrounds; virus levels were ∼3 log10 lower in transgenic mice than in their nontransgenic littermates. Because the magnitude of this difference was comparable to that observed in MA/My and C57L or R12 and M.L-H2b congenic strains derived from them (Fig. 2) (18, 19), these data suggest that Dk corresponds to an H-2k Cmvr locus. In agreement with findings from Tg3-Dk animals, we found that Tg1-Dk expression conferred similar protection in the M.L-H2b background (Fig. S3). MHC-I Dk expression was therefore sufficient to deliver robust MCMV resistance in otherwise susceptible C57L or M.L-H2b genetic backgrounds.
Fig. 2.
H-2Dk expression confers innate MCMV resistance. Tg3MN2, Tg3LN2, nontransgenic littermates, and the designated control strains were infected with 2 × 104 PFU SGV/mouse. Shown are spleen virus levels for individual animals at 90 h postinfection. Average viral titers for each group are also shown. Data are representative of three independent experiments.
NK Cells Are Required in H-2Dk Resistance to MCMV Infection.
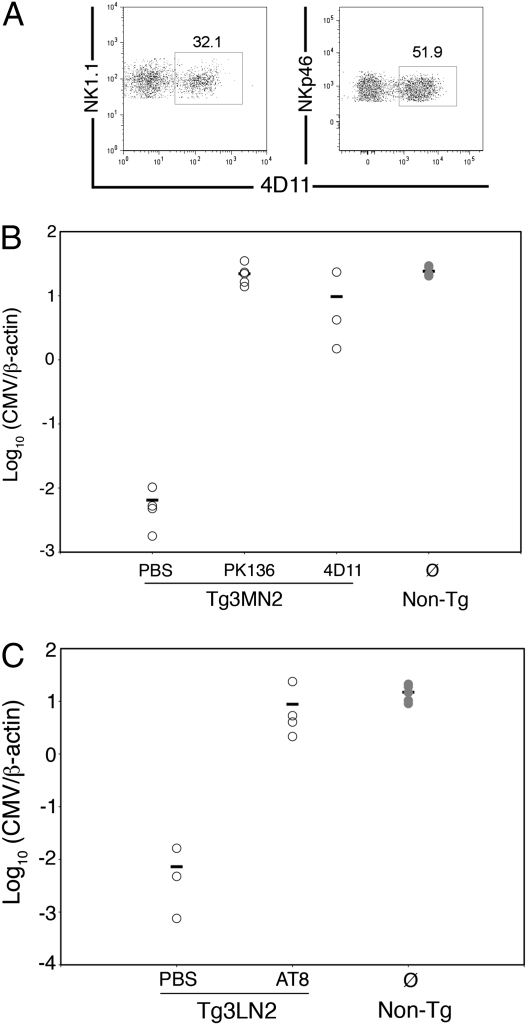
Because a critical role for Ly49G2+ NK cells was previously implicated (19), we next examined their contribution in MHC-I Dk resistance to MCMV. NK cells were depleted from transgenic mice with NK1.1- or Ly49G2-specific mAbs given before MCMV infection. Interestingly, about 30% or 50% of NK cells displayed Ly49G2 receptors in uninfected transgenic mice on MA/My or C57L backgrounds, respectively, indicating that background genes affected the proportion of Ly49G2+ NK cells (Fig. 3A). Moreover, NK cells expressed a lower level of Ly49G2 in the transgenic mice, indicating that MHC-I Dk expression also affected detection of Ly49 receptor alleles, as shown for Ly49A when its cognate MHC ligand is present in cis (22).
Fig. 3.
Ly49G2 expressing NK cells required in MCMV resistance in Tg3-Dk mice. (A) Splenocytes from uninfected Tg3MN2 (Left) or Tg3LN2 (Right) mice were stained for CD3, NK1.1 or NKp46, and Ly49G2 (4D11). The plots are gated on CD3− NK cells, and numbers indicate the percentage of Ly49G2+ splenocytes within the gate. The plots are representative of at least three transgenic mice examined. PBS, 4D11, or PK136-treated Tg3MN2 (B) or AT8-treated Tg3LN2 mice (C) as well as untreated nontransgenic littermates were infected with 2 × 104 PFU (B) or 3 × 104 PFU (C) MCMV. Spleen MCMV genome levels for individual mice at 90 h postinfection are shown. Data in Fig. 4B are representative of two independent experiments.
As expected, NK cell depletions with anti-NK1.1 mAb PK136 effectively abrogated MHC-I Dk resistance (Fig. 3B). Remarkably, mAb 4D11 depletion of Ly49G2+ NK cells had a comparable effect; MCMV resistance was substantially altered by greater than 2 log10 (Fig. 3B). We further found that mAb AT8 depletion of Ly49G2+ NK cells resulted in the complete loss of virus resistance from Tg3LN2 mice (Fig. 3C). These findings suggested that residual Ly49G2− NK cells were unable to limit virus replication. Consistent with this, Ly49G2+ NK cells were selectively expanded in Tg3MN2 spleens in response to MCMV (Fig. S4), as seen before in the R7 congenic strain (19), similar to the expansion of Ly49H+ NK cells in C57BL/6 mice due to cognate interactions between Ly49H and its ligand (23). Because a role for Ly49P had been previously implicated in H-2k MCMV resistance (15, 24), we next flow-sorted NKp46+CD3− splenocytes based on Ly49G expression. As expected, Ly49g expression was readily detected by RT-PCR in the Ly49G+ subset of NK cells (Fig. S5). Ly49p, however, was broadly expressed in Ly49G+, as well as in Ly49G− NK cell subsets. After infection, a similar profile was observed (Fig. S5). These data indicate that Ly49p expression was not restricted to Ly49G+ NK cells; rather, it was abundantly and comparably expressed in both subsets. We conclude that MHC-I Dk virus resistance required NK cells with cognate Ly49G2 receptors ex-pressed; without this inhibitory receptor, Ly49p-expressing NK cells were inadequate to reign in MCMV infection.
We next assessed whether an intrinsic functional defect might explain why residual Ly49G- NK cells were ineffective in MCMV resistance in Ly49G-depleted mice. Splenic NK cells from B6 were sorted into Ly49G+ and Ly49G− NK subsets and assessed for cytotoxicity against several targets. As shown in Fig. 4A, cytokine-activated B6 NK subsets responded with comparable cytotoxicity to YAC-1, YB2/0, or RMA-S target cells, but Ly49G+ NK cell killing was diminished by cognate ligand-bearing YB-Dd target cells. Tg and non-Tg splenocytes were next examined for cytokine production after stimulation with immobilized anti-NK1.1 mAb. Because it has been shown that IL-2 activation can enhance unlicensed NK cell function (4, 5), splenocytes were freshly prepared and tested. As shown in Fig. 4B, NKR-P1 stimulation triggered similar IFN-γ production in both subsets (Fig. 4B). However, there was no difference in the relative frequencies of IFN-γ-producing Ly49G2+ and Ly49G2− NK cells in transgenic mice, or any difference between Ly49G2+ NK cells in Tg and non-Tg mice (Fig. 4C). Nonetheless, the ratio of IFN-γ-productivity of Ly49G2+ to Ly49G2− NK cells in Tg mice was significantly higher than in non-Tg mice (Fig. 4D). Thus, MHC-I Dk effectively licensed transgenic NK cells expressing a cognate Ly49G2 inhibitory receptor. Taken together, these data show that licensed Ly49G+ NK cells efficiently recognize and respond to MCMV infection in mice with self-MHC-I Dk expressed.
Fig. 4.
Ly49G+ and Ly49G− NK cells display similar cytotoxicity and cytokine production after stimulation. (A) IL-2-expanded C57BL/6 NK cells were sorted into Ly49G+ and Ly49G− subsets and tested for cytotoxicity against YAC-1, RMA-S, YB2/0, and YB-Dd cell targets using a standard 51Cr release assay. Data are representative of two independent experiments. (B) Freshly prepared Tg3-Dk (TgMN3) and non-Tg splenocytes were incubated with immobilized anti-NK1.1 in the presence of Brefeldin A. PMA and ionomycin (P/I) stimulation was included as a positive control. After 4 h, cells were stained for DX5, CD3, 4D11, and intracellular IFN-γ. Shown are representative dot plots; the numbers represent the percentages of IFN-γ+ cells among the Ly49G2+ or Ly49G2− populations of NK cells. (C) Experimental means measured in triplicate for %IFN-γ+ Tg3-Dk NK cells sorted by Ly49G expression (Left) or %IFN-γ+ Ly49G+ NK cells sorted by Tg3-Dk or non-Tg (Right). Data are representative of four different experiments with 2–3 mice per genotype. Statistical significance was determined by Wilcoxon rank-sum test. (D) Relative IFN-γ productivity is plotted as a ratio of the percentage of IFN-γ+ cells among the Ly49G2+ NK cells to the percentage of IFN-γ+ cells among the Ly49G2− NK cells as described (5).
MHC-I Dk on Both Hematopoietic and Nonhematopoietic Cells Contributes to MCMV Resistance.
MCMV readily infects and replicates within stromal cells of the spleen (25, 26). A possible requirement for MHC-I Dk on nonhematopoetic cells was next examined. Radiation BM chimeras were established by transferring Tg3-Dk BM cells into lethally irradiated nontransgenic recipients, and vice versa. Donor-cell reconstitution in the chimeras was verified by staining for MHC-I Dk on peripheral blood. Donor NK cells and B cells were well established in the chimeras (Table 2). A small proportion of residual recipient CD3+ lymphocytes was also seen in the BM chimeras, but a role for T cells in early H-2k resistance has been excluded (19). Hence, the BM chimeras were directly assessed for resistance to MCMV infection.
As expected, control Tg3-Dk mice reconstituted with Tg3-Dk BM cells displayed full resistance in line with MA/My or Tg3-Dk animals; nontransgenic control animals given nontransgenic BM were unable to limit splenic virus replication (Fig. 5). Unexpectedly however, no greater resistance was observed in nontransgenic recipients of Tg3-Dk BM cells (Fig. 5A), even though donor NK cells in the chimeras were functionally competent and responded with increased IFN-γ and CD69 by 40 h after MCMV infection (Fig. S6). Interestingly, Tg3-Dk recipients of nontransgenic BM were also vulnerable to infection (Fig. 5A), although partial resistance was observed in at least one experiment (Fig. 5B). These data therefore suggest that NK cells recognize and respond to MCMV-induced differences in MHC-I Dk expression on hematopoietic and nonhematopoietic cells to deliver efficient and adequate MCMV resistance.
Fig. 5.
MCMV resistance in radiation bone marrow chimeras. (A) Tg3-Dk (N3) and non-Tg radiation BM chimeras and transfer controls were generated by transfer of donor BM into lethally irradiated hosts. At 6 weeks posttransfer, chimeric and control mice were infected with 1 × 104 PFU MCMV. Shown are spleen virus levels at 3.5 d postinfection. Tg to Tg transfer controls displayed resistance that differed significantly from each of the BM chimeras, non-Tg to non-Tg transfer controls and non-Tg control mice without transfer. No other significant differences were observed. Data are representative of two (non-Tg to Tg) or three (Tg to non-Tg) independent experiments. (B) An independent experiment to test Tg3-Dk (N4) chimeric mice reconstituted with non-Tg BM. Chimeric and control mice were infected with ∼3 × 104 PFU MCMV. Spleen (○) and liver (●) MCMV levels for chimeric, transfer control, and MCMV-resistant control R7 mice are shown. Non-Tg chimeric mice reconstituted with transgenic BM differed significantly from non-Tg transfer control mice, nontransferred R7 control mice, and R7 littermates without MHC-I Dk. *P < 0.05, **P < 0.01, ***P < 0.005.
Discussion
H-2k and non-MHC genetic factors were shown to have a major effect on host survival after MCMV infection (11, 12). Previous genetic studies ruled out the H-2 K/I-A-E region and mapped the MCMV resistance effect to a narrow (0.3-Mb) interval located within the class I D region, with only ∼30 resident gene candidates, including the D gene itself (18, 19). However, because the resistance effect was tightly linked to genes coding for immune-related functions (e.g., Tnf, Lta, and Ltb), the critical gene(s) was still obscure. Importantly, we have shown that a MHC-I Dk transgene alone was sufficient to confer the MCMV resistance effect in strains that were otherwise fully susceptible to infection.
We recently reported a critical role for NK cells with Ly49G2 inhibitory receptors in MCMV resistance in MHC-I Dk congenic mice (19). Remarkably, we have shown that Ly49G2+ NK cells were necessary for MCMV resistance in MHC-I Dk transgenic mice. Specific depletion of Ly49G2+ NK cells before MCMV infection fully abrogated MCMV resistance so that virus levels in the spleen 3.5 days after infection were 100- to 1,000-fold higher than in mice with Ly49G2+ NK cells intact. This was substantiated by depletion with two different Ly49G2-specific mAbs. This finding is very different from what has been observed after depletion of Ly49G2+ NK cells in B6 mice where no effect on splenic MCMV titers was observed (27), or where a limited, but significant, effect was recently observed in liver (∼5-fold increase) and salivary gland (∼10-fold increase) by 7 days after infection (28). A profound Ly49G2 effect has so far been restricted to animals displaying H-2k genetic resistance, and it is especially sensitive to MHC polymorphism.
One interpretation—Ly49G2 inhibitory receptors in MHC-I Dk mice may have endowed NK cells with proficient missing-self recognition—is based on several key observations: (i) MCMV immune evasion proteins gp40 and gp48 efficiently down-regulated MHC-I Dk on infected cells (18); (ii) MHC-I Dk is a known ligand of Ly49G2balb/c and Ly49G2129 (20, 21); and (iii) MCMV gp34 associated with MHC-I Dk on infected cells (24). Thus, efficient Ly49G2 missing-self recognition of down-regulated or modified MHC-I Dk could enhance antiviral NK responses through lessening NK inhibition, such that Ly49P, NKG2D, or other stimulatory receptors capable of recognizing ligands expressed on MCMV-infected cells (15, 24, 29–32) might further hone NK responses and cytotoxicity. Interestingly, unlicensed Ly49H+ NK cells were recently shown to contribute greater MCMV resistance after infection than licensed NK cells in B6 mice (28). Indeed, licensed NK cells interacting with self-MHC-I impaired Ly49H-mediated recognition and elimination of m157-bearing infected cells. In striking contrast, here we found that NK cells with a Ly49G2 cognate inhibitory receptor for self-MHC-I Dk were weakly licensed, yet delivered potent MCMV resistance in transgenic mice. Perhaps related to this, Jonsson et al. (33) recently showed that Ly49A was weakly licensed by H-2q, whereas Ly49A-dependent inhibition of NK cell cytotoxicity was much more sensitive to MHC H-2q engagement. Further studies will be needed to precisely define cognate inhibitory Ly49 receptor and self-MHC-I ligand pairings in the MA/My.L-H2b genetic background and the impact on NK cell licensing and effector functions. Despite this, licensed Ly49G2+ cells were critical to MCMV resistance. This finding underscores a major difference in NK cell-mediated immune responses to virus infection controlled mainly through NK cell activation (i.e., in B6 mice) or inhibitory receptor (i.e., in MA/My and other MHC-I Dk mice) recognition of and reactivity with infected cells.
In an alternate model, Ly49P stimulated reporter cells by interacting with Dk-gp34 complexes on infected targets (15, 24). However, without a Ly49P-specific antibody, the in vivo significance of MCMV-infected cell recognition via this receptor is still in question. Together, several findings raise concern with an exclusive Ly49P-based MHC-I Dk MCMV resistance model: (i) Depletion of the Ly49G2 subset fully abrogated virus resistance in MHC-I Dk mice (19) (Fig. 3), but (ii) Ly49G2 subset (∼40% of splenic NK cells) depletion had no impact on MCMV resistance in B6 mice (27). Hence, unless Ly49P and Ly49G2 receptors were strictly coexpressed on NK cells, residual Ly49P+ NK cells in Ly49G2-depleted mice were totally ineffective in MCMV resistance. Preferential coexpression of Ly49D and Ly49H activation receptors has been shown (34), but a similar finding, linking inhibitory and activation receptors, is unprecedented. Indeed, we found that Ly49p was broadly expressed in Ly49G+ and Ly49G− NK cells before or after infection (Fig. S5). Last, (iii) BALB.K mice without Ly49P activation receptors still displayed H-2k protection against lethal MCMV infection (11). Together, these data establish the primacy of Ly49G+ NK cells to deliver efficient MHC-I Dk virus resistance. An intriguing possibility to reconcile potential discrete roles for the Ly49 receptors, Ly49G could give license to Ly49P and/or other stimulatory receptors on the same NK cells to rapidly respond with stimulation and proliferation during MCMV infection. In this scenario, MHC polymorphism may influence NK cell competency for recognition of MCMV-infected cells through inhibitory Ly49G receptors, and consequently the magnitude of the NK cell response toward infected target cells, which also display ligands for NK stimulatory receptors.
The importance of hematopoietic and nonhematopoietic cell types in NK-mediated MCMV resistance is in accord with a proposed model based on missing-self recognition via Ly49G2 inhibitory receptors. This differs from a related study where complete resistance was observed in radiation BM chimeras established by transfer of resistant C57BL/6 BM cells into MCMV-susceptible BALB.B recipients (35). All virus-infected cells in B6→BALB.B BM chimeras were, in principle, competent to display MCMV m157 ligands at the cell surface, irrespective of their cellular origin. Considering the current study revealed a clear requirement for hematopoietic and nonhematopoietic cells; MHC-I Dk should, therefore, represent a major target ligand for NK cell recognition and interaction with infected cells of both lineages.
Consistent with this, dendritic cells were shown to respond to MCMV infection with cytokines that stimulate NK cell cytotoxicity and proliferation (36–38). In a reciprocal fashion, NK cells produce and release cytokines (e.g., IFN-γ) that augment DC stimulation, maturation, and antigen-presenting functions (39). Indeed, ineffective NK/DC cross-talk has been linked to immune dysfunction characterized by altered NK and DC cytokine responses, altered DC maturation and DC instability, and delayed acquisition of virus-specific CD8+ T cells (37, 40–43). Recent studies to implicate nonhematopoietic cells have shown that MCMV readily infects and replicates in splenic stromal cells, more so than within hematopoietic cells (25, 26). Further, MCMV resistance via Ly49H+ NK cells in C57BL/6 mice corresponded to their redistribution from red to white pulp during infection and preservation of splenic white pulp structure (26). Drawing from the above studies, NK-mediated MHC-I Dk resistance might involve efficient DC priming of Ly49G2+ NK cells responding to missing-self cues. Primed NK cells could then provide further enhancement of virus resistance with efficient NK cell recognition of infected stromal cells, through missing-self and possibly other NK stimulatory receptor signals. It is not known whether Ly49G2 may be more sensitive to MHC-I alteration on MCMV-infected cells than other NK inhibitory receptors, but its effect plainly involves NK cell contacts with MHC-I Dk-expressing hematopoietic and nonhematopoietic cells. Further study using this model will help to elucidate novel mechanisms of NK cell activation and recognition of virus infection, which is causally linked with MHC-I polymorphism.
Materials and Methods
Mice.
MA/My and C57L (Jackson Laboratory) and MHC congenic strains, including MA/My.L-H2b, C57L.M-H2k(R7), and C57L.M-H2k(R12) described previously (18, 19), were maintained in a specific pathogen-free vivarium at the University of Virginia. All animal studies were approved and conducted in accordance with Institutional Animal Care and Use Committee oversight.
Antibodies and Flow Cytometry.
Anti-mouse CD3 (145-2C11) allophycocyanin, NK1.1 (PK136) PE, Ly49G2 (4D11) FITC, and allophycocyanin, CD8 (53-6.7) PE, H-2Dk (15-5-5) PE and biotin, CD19 (1D3) allophycocyanin-Cy7, IFNγ (XMG1.2) FITC, and streptavidin-PerCP were purchased from BD Pharmingen. Anti-mouse CD4 (RM4-5) FITC, CD11c (N418) PE, Ly49G2 (AT8) biotin, and streptavidin-allophycocyanin-Cy7 were purchased from eBioscience. Anti-mouse CD3 (145-2C11) PerCP was purchased from BioLegend. Anti-mouse NKp46 (goat IgG) PE was purchased from R&D Systems. PK136, 4D11 (ATCC), and AT8-purified mAbs were prepared at the University of Virginia Lymphocyte Culture Center. Stained splenocytes were analyzed by flow cytometry on a FACSCanto I or FACSCanto II (BD Biosciences). Data were collected using FACSDiva software (BD Biosciences) and analyzed using FlowJo (version 8.0; Tree Star).
For flow sorting NK cells, uninfected or MCMV-infected splenocytes from transgenic mice were resuspended in MACS buffer and positively selected using CD49b (DX5) microbeads and an autoMACS Separator (Miltenyi Biotec) according to the manufacturer's protocol. DX5-enriched lymphocytes (82–84% NK cells) resuspended in HBSS 1% FBS were stained for surface antigens as described (14). Cells were resuspended in 1 μg/mL DAPI (Sigma) immediately before sorting for live/dead discrimination. NKp46+CD3− cells were sorted into 4D11+ and 4D11− fractions using either a Becton Dickinson FACSVantage SE Turbo Sorter with DIVA Option or an iCyt Reflection.
NK Cytokine and Cytotoxicity Assays.
Stimulation of splenocytes was performed as described previously (5). In brief, splenocytes (8 × 105/well) resuspended in R10 complete media plus low-dose IL-2 (100 U/mL) and Brefeldin A (5 μg/mL) were incubated on immobilized PK136 (32 μg/mL) for 4 h at 37 °C. For control stimulation, splenocytes were incubated with 100 ng/mL PMA (Sigma) plus 0.7 μg/mL ionomycin (Sigma) for 4 h at 37 °C. Afterward, cells were stained for surface antigens followed by permeabilization (BD Cytofix/Cytoperm; BD Biosciences) and staining for intracellular IFN-γ.
For cytotoxicity experiments, C57BL/6 splenic NK cells enriched by negative selection using AutoMACS magnetic separation (Miltenyi Biotech) were stained for DX5, TcRβ, and Ly49G before sorting into Ly49G+ and Ly49G− populations. Both NK cell populations were expanded for 7 days in NK cell medium supplemented with human r-IL2 (1,000 U/mL), and then assessed as effectors in 4 h 51Cr release cytotoxicity assays using standard methods (44).
Virus Assays.
Experimental mice (7-12 weeks) were i.p. infected with SGV stocks (1–3 × 104 PFU). To study the role of NK cells during infection, mice were i.p. injected with 200 μg anti-NK1.1 mAb PK136 or anti-Ly49G2 mAb 4D11 or AT8 48 h before MCMV infection. NK cell depletions were confirmed by staining splenocytes for NKp46 and Ly49G2 (mAb AT8 or 4D11). Spleen virus levels were quantified as described previously (14, 45).
Preparation of Bone Marrow Chimeras.
C57L.Cg-Tg3-Dk (N3 or N4) mice and their nontransgenic littermates were used to establish radiation bone marrow (BM) chimeras as described previously (46). Briefly, donor BM cells (4–8 × 106/200 μL PBS) from 8- to 10-week mice were i.v. injected into the tail vein of lethally irradiated (two 5.75-Gy doses given 3 h apart) recipients (7-16 weeks). Recipients and donors were sex and NKC haplotype matched to enhance donor cell reconstitution. BM transfers were withheld from several transgenic and nontransgenic radiation control animals. The control animals died within 8–13 days. Recipients were given sulfate water for the first 3 weeks and analyzed ∼6 weeks after transplantation.
Supplementary Material
Acknowledgments
We thank Joanne Lannigan (University of Virginia Flow Cytometry Core Facility) and Claudia Rival for assistance with cell sorting. We thank Victor Engelhard and Virginia Carroll for helpful discussion and Jessica Prince for technical support. This work was supported by National Institutes of Health (NIH) Grants AI050072 and AI083024 (to M.G.B.), NIH Biotechnology Training Award T32 GM08715 (to M.D.S.), and NIH Immunology Training Award T32 AI007496 (to E.R.C.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0913126107/-/DCSupplemental.
References
- 1.Trinchieri G. Biology of natural killer cells. Adv Immunol. 1989;47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yokoyama WM, Plougastel BF. Immune functions encoded by the natural killer gene complex. Nat Rev Immunol. 2003;3:304–316. doi: 10.1038/nri1055. [DOI] [PubMed] [Google Scholar]
- 3.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez NC, et al. A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood. 2005;105:4416–4423. doi: 10.1182/blood-2004-08-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim S, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 6.Anfossi N, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–342. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Martin MP, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 8.Khakoo SI, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 9.Martin MP, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahlenstiel G, Martin MP, Gao X, Carrington M, Rehermann B. Distinct KIR/HLA compound genotypes affect the kinetics of human antiviral natural killer cell responses. J Clin Invest. 2008;118:1017–1026. doi: 10.1172/JCI32400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chalmer JE, Mackenzie JS, Stanley NF. Resistance to murine cytomegalovirus linked to the major histocompatibility complex of the mouse. J Gen Virol. 1977;37:107–114. doi: 10.1099/0022-1317-37-1-107. [DOI] [PubMed] [Google Scholar]
- 12.Grundy JE, Mackenzie JS, Stanley NF. Influence of H-2 and non-H-2 genes on resistance to murine cytomegalovirus infection. Infect Immun. 1981;32:277–286. doi: 10.1128/iai.32.1.277-286.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scalzo AA, et al. Genetic mapping of Cmv1 in the region of mouse chromosome 6 encoding the NK gene complex-associated loci Ly49 and musNKR-P1. Genomics. 1995;27:435–441. doi: 10.1006/geno.1995.1074. [DOI] [PubMed] [Google Scholar]
- 14.Dighe A, et al. Requisite H2k role in NK cell-mediated resistance in acute murine cytomegalovirus-infected MA/My mice. J Immunol. 2005;175:6820–6828. doi: 10.4049/jimmunol.175.10.6820. [DOI] [PubMed] [Google Scholar]
- 15.Desrosiers MP, et al. Epistasis between mouse Klra and major histocompatibility complex class I loci is associated with a new mechanism of natural killer cell-mediated innate resistance to cytomegalovirus infection. Nat Genet. 2005;37:593–599. doi: 10.1038/ng1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bancroft GJ, Shellam GR, Chalmer JE. Genetic influences on the augmentation of natural killer (NK) cells during murine cytomegalovirus infection: Correlation with patterns of resistance. J Immunol. 1981;126:988–994. [PubMed] [Google Scholar]
- 17.Shellam GR, Flexman JP, Farrell HE, Papadimitriou JM. The genetic background modulates the effect of the beige gene on susceptibility to cytomegalovirus infection in mice. Scand J Immunol. 1985;22:147–155. doi: 10.1111/j.1365-3083.1985.tb01867.x. [DOI] [PubMed] [Google Scholar]
- 18.Xie X, et al. Deficient major histocompatibility complex-linked innate murine cytomegalovirus immunity in MA/My.L-H2b mice and viral downregulation of H-2k class I proteins. J Virol. 2007;81:229–236. doi: 10.1128/JVI.00997-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie X, Stadnisky MD, Brown MG. MHC class I Dk locus and Ly49G2+ NK cells confer H-2k resistance to murine cytomegalovirus. J Immunol. 2009;182:7163–7171. doi: 10.4049/jimmunol.0803933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silver ET, Lavender KJ, Gong DE, Hazes B, Kane KP. Allelic variation in the ectodomain of the inhibitory Ly-49G2 receptor alters its specificity for allogeneic and xenogeneic ligands. J Immunol. 2002;169:4752–4760. doi: 10.4049/jimmunol.169.9.4752. [DOI] [PubMed] [Google Scholar]
- 21.Makrigiannis AP, et al. Class I MHC-binding characteristics of the 129/J Ly49 repertoire. J Immunol. 2001;166:5034–5043. doi: 10.4049/jimmunol.166.8.5034. [DOI] [PubMed] [Google Scholar]
- 22.Doucey MA, et al. Cis association of Ly49A with MHC class I restricts natural killer cell inhibition. Nat Immunol. 2004;5:328–336. doi: 10.1038/ni1043. [DOI] [PubMed] [Google Scholar]
- 23.Dokun AO, et al. Specific and nonspecific NK cell activation during virus infection. Nat Immunol. 2001;2:951–956. doi: 10.1038/ni714. [DOI] [PubMed] [Google Scholar]
- 24.Kielczewska A, et al. Ly49P recognition of cytomegalovirus-infected cells expressing H2-Dk and CMV-encoded m04 correlates with the NK cell antiviral response. J Exp Med. 2009;206:515–523. doi: 10.1084/jem.20080954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benedict CA, et al. Specific remodeling of splenic architecture by cytomegalovirus. PLoS Pathog. 2006;2:e16. doi: 10.1371/journal.ppat.0020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bekiaris V, et al. Ly49H+ NK cells migrate to and protect splenic white pulp stroma from murine cytomegalovirus infection. J Immunol. 2008;180:6768–6776. doi: 10.4049/jimmunol.180.10.6768. [DOI] [PubMed] [Google Scholar]
- 27.Tay CH, et al. The role of LY49 NK cell subsets in the regulation of murine cytomegalovirus infections. J Immunol. 1999;162:718–726. [PubMed] [Google Scholar]
- 28.Orr MT, Murphy WJ, Lanier LL. ‘Unlicensed’ natural killer cells dominate the response to cytomegalovirus infection. Nat Immunol. 2010;11:321–327. doi: 10.1038/ni.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lodoen M, et al. NKG2D-mediated natural killer cell protection against cytomegalovirus is impaired by viral gp40 modulation of retinoic acid early inducible 1 gene molecules. J Exp Med. 2003;197:1245–1253. doi: 10.1084/jem.20021973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lodoen MB, et al. The cytomegalovirus m155 gene product subverts natural killer cell antiviral protection by disruption of H60-NKG2D interactions. J Exp Med. 2004;200:1075–1081. doi: 10.1084/jem.20040583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krmpotic A, et al. NK cell activation through the NKG2D ligand MULT-1 is selectively prevented by the glycoprotein encoded by mouse cytomegalovirus gene m145. J Exp Med. 2005;201:211–220. doi: 10.1084/jem.20041617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenac T, et al. The herpesviral Fc receptor fcr-1 down-regulates the NKG2D ligands MULT-1 and H60. J Exp Med. 2006;203:1843–1850. doi: 10.1084/jem.20060514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jonsson AH, Yang L, Kim S, Taffner SM, Yokoyama WM. Effects of MHC class I alleles on licensing of Ly49A+ NK cells. J Immunol. 2010;184:3424–3432. doi: 10.4049/jimmunol.0904057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith HR, et al. Nonstochastic coexpression of activation receptors on murine natural killer cells. J Exp Med. 2000;191:1341–1354. doi: 10.1084/jem.191.8.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scalzo AA, et al. The effect of the Cmv-1 resistance gene, which is linked to the natural killer cell gene complex, is mediated by natural killer cells. J Immunol. 1992;149:581–589. [PubMed] [Google Scholar]
- 36.Dalod M, et al. Interferon alpha/beta and interleukin 12 responses to viral infections: Pathways regulating dendritic cell cytokine expression in vivo. J Exp Med. 2002;195:517–528. doi: 10.1084/jem.20011672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andoniou CE, et al. Interaction between conventional dendritic cells and natural killer cells is integral to the activation of effective antiviral immunity. Nat Immunol. 2005;6:1011–1019. doi: 10.1038/ni1244. [DOI] [PubMed] [Google Scholar]
- 38.Lucas M, Schachterle W, Oberle K, Aichele P, Diefenbach A. Dendritic cells prime natural killer cells by trans-presenting interleukin 15. Immunity. 2007;26:503–517. doi: 10.1016/j.immuni.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Degli-Esposti MA, Smyth MJ. Close encounters of different kinds: Dendritic cells and NK cells take centre stage. Nat Rev Immunol. 2005;5:112–124. doi: 10.1038/nri1549. [DOI] [PubMed] [Google Scholar]
- 40.Gerosa F, et al. Reciprocal activating interaction between natural killer cells and dendritic cells. J Exp Med. 2002;195:327–333. doi: 10.1084/jem.20010938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dalod M, et al. Dendritic cell responses to early murine cytomegalovirus infection: Subset functional specialization and differential regulation by interferon alpha/beta. J Exp Med. 2003;197:885–898. doi: 10.1084/jem.20021522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robbins SH, et al. Natural killer cells promote early CD8 T cell responses against cytomegalovirus. PLoS Pathog. 2007;3:e123. doi: 10.1371/journal.ppat.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mavilio D, et al. Characterization of the defective interaction between a subset of natural killer cells and dendritic cells in HIV-1 infection. J Exp Med. 2006;203:2339–2350. doi: 10.1084/jem.20060894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karlhofer FM, Yokoyama WM. Stimulation of murine natural killer (NK) cells by a monoclonal antibody specific for the NK1.1 antigen. IL-2-activated NK cells possess additional specific stimulation pathways. J Immunol. 1991;146:3662–3673. [PubMed] [Google Scholar]
- 45.Wheat RL, Clark PY, Brown MG. Quantitative measurement of infectious murine cytomegalovirus genomes in real-time PCR. J Virol Methods. 2003;112:107–113. doi: 10.1016/s0166-0934(03)00197-6. [DOI] [PubMed] [Google Scholar]
- 46.Day YJ, et al. Renal protection from ischemia mediated by A2A adenosine receptors on bone marrow-derived cells. J Clin Invest. 2003;112:883–891. doi: 10.1172/JCI15483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.