Abstract
RNA molecules perform diverse regulatory functions in natural biological systems, and numerous synthetic RNA-based control devices that integrate sensing and gene-regulatory functions have been demonstrated, predominantly in bacteria and yeast. Despite potential advantages of RNA-based genetic control strategies in clinical applications, there has been limited success in extending engineered RNA devices to mammalian gene-expression control and no example of their application to functional response regulation in mammalian systems. Here we describe a synthetic RNA-based regulatory system and its application in advancing cellular therapies by linking rationally designed, drug-responsive, ribozyme-based regulatory devices to growth cytokine targets to control mouse and primary human T-cell proliferation. We further demonstrate the ability of our synthetic controllers to effectively modulate T-cell growth rate in response to drug input in vivo. Our RNA-based regulatory system exhibits unique properties critical for translation to therapeutic applications, including adaptability to diverse ligand inputs and regulatory targets, tunable regulatory stringency, and rapid response to input availability. By providing tight gene-expression control with customizable ligand inputs, RNA-based regulatory systems can greatly improve cellular therapies and advance broad applications in health and medicine.
Keywords: nucleic acid therapies, RNA controller, synthetic biology, synthetic riboswitch, immunotherapy
The ability to control functional responses in mammalian cells with customizable and compact regulatory systems in vivo addresses a critical need in diverse clinical applications, particularly in cellular therapies (1). As an example, adoptive T-cell therapy seeks to harness the precision and efficacy of the immune system against diseases that escape the body’s natural surveillance. The adoptive transfer of antigen-specific T cells can reconstitute immunity to viruses and mediate tumor regression (2–4). T cells engineered to express tumor-specific T-cell antigen receptors can achieve highly refined target recognition (5), thus minimizing toxic off-target effects associated with conventional chemotherapy. However, considerable research has shown that the persistence of transferred T cells in vivo is both central to therapeutic success and elusive to current technology (6, 7). The efficacy of adoptive immunotherapy in humans is often limited by the failure of transferred T cells to survive in the host (8, 9).
Clonal expansion of T cells is a critical component of T-cell activation mediated by cytokines such as IL-2 and IL-15, which activate JAK-STAT signaling pathways and lead to the expression of genes involved in growth modulation (10) (Fig. 1A). Sustaining the survival and proliferation of T cells following adoptive transfer is challenging because of the limited availability of homeostatic cytokines (IL-15/IL-7) and stimulatory antigen presenting cells. State-of-the-art strategies for improving the persistence of adoptively transferred lymphocytes require that patients be subjected to myeloablative total body irradiation/chemotherapy and toxic levels of IL-2 (11). Alternative strategies with unregulated expression of growth-related genes pose the risk of uncontrolled lymphoproliferation and leukemic transformation (6). Thus, the ability to integrate growth-stimulatory gene expression with tightly controlled regulatory systems has the potential to greatly improve the safety and efficacy of adoptive T-cell therapy.
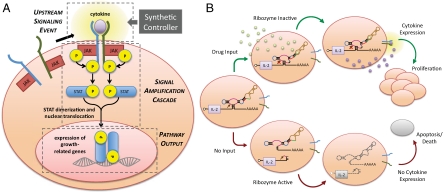
Fig. 1.
An engineered T-cell proliferation regulatory system utilizing a synthetic RNA device to achieve drug-mediated modulation of cell signaling and proliferation. (A) The common γ-chain T-cell proliferation pathway and integration of a synthetic controller targeted to the upstream signaling events. (B) An engineered T-cell proliferation regulatory system on the basis of programmable drug-mediated regulation of cytokine expression from a synthetic ribozyme switch. Ribozyme color scheme is as described in ref. 19.
Inspired by the diverse functional roles exhibited by regulatory RNAs in natural systems (12–14) and the relative ease by which RNA can be modeled and designed (15), researchers have begun developing synthetic RNA-based regulatory systems that integrate discrete gene-regulatory and sensing functions as genetic control strategies (16–18). However, the absence of successful adaptations of these earlier genetic devices to the regulation of functional responses in mammalian cells highlights remaining difficulties in translating designs that regulate reporter gene expression to functional control.
We describe a synthetic, small-molecule-responsive RNA-based gene-regulatory system in mammalian cells and demonstrate its application in advancing cell-based therapies through the control of cell-fate decisions. We develop a genetic strategy for effectively controlling T-cell expansion on the basis of drug-responsive RNA regulators that exert tight control over key upstream signaling molecules in the proliferation pathway. Our work demonstrates a RNA-based regulatory system that exhibits unique properties critical for translation to therapeutic applications, including adaptability to diverse input molecules and genetic targets, tunable regulatory stringency, and rapid input response.
Results
A RNA-Based System Enables Gene Expression and Viability Control in T Cells.
We developed a RNA-based regulatory system for mammalian T-cell proliferation on a platform for assembling RNA devices from modular sensor (aptamer) and gene-regulatory (hammerhead ribozyme) components. The activity of this ribozyme switch platform had been shown in the microorganism Saccharomyces cerevisiae (19, 20); however, the activity of these genetic regulatory elements in mammalian cells had not been previously examined.
Cytokines are potent growth-stimulatory molecules whose effects on cell growth are amplified through the JAK-STAT signaling pathway (Fig. 1A). The ability to regulate upstream pathway molecules and take advantage of signal amplification through an endogenous pathway toward downstream functional responses is an important design strategy supported by our RNA regulatory system. We developed a cell-intrinsic control system for cytokine production utilizing ribozyme ON switches, which are RNA devices that convert a small-molecule input to an increased gene-expression output (Fig. 1B). The system design ensures suppression of cell growth as a default state and induction of cell proliferation only in the presence of an administered small-molecule drug input. In this system, the ribozyme-based device is placed in the 3′ UTR of a target transgene encoding a proliferative cytokine, where self-cleavage by the ribozyme results in rapid degradation of the target transcript and decreased cytokine production. The ribozyme device is designed to adopt at least two conformations (input-unbound and input-bound) associated with either a ribozyme-active or -inactive state. The presence of drug input stabilizes the input-bound, ribozyme-inactive conformation, thereby preserving transcript integrity and up-regulating cytokine production, resulting in autocrine cell growth. The absence of drug input stabilizes the ribozyme-active conformation, resulting in transcript degradation, reduced cytokine production, and diminished cell growth.
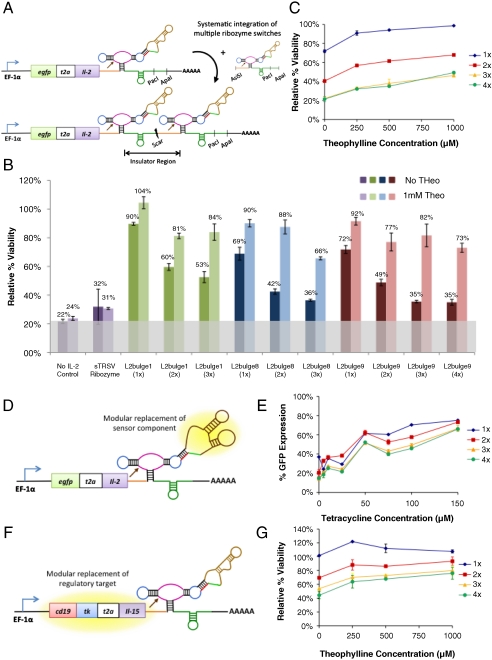
As a model system, a fusion transgene encoding a proliferative cytokine (IL-2) and a quantifiable protein marker (EGFP) served as the regulatory target to permit simultaneous quantification of the regulatory system’s performance at the levels of direct gene expression (fluorescence) and downstream pathway output (viability) (Fig. 2A). The cytokine and reporter proteins were linked through a self-cleaving T2A peptide (SI Text) to ensure that the ribozyme switch activity was equally effective on the linked target genes but that the proteins fold and function as independent molecules. Three theophylline (theo)-responsive ribozyme switches (L2bulge1, 8, and 9; SI Text), which had been tuned through sequence modifications to exhibit different regulatory response properties (19), were inserted into the 3′ UTR of the egfp-t2a-il2 fusion gene. Plasmids incorporating this regulatory system were transiently transfected into the CTLL-2 mouse T-cell line, which, like primary human T cells, is dependent on common γ-chain signaling for survival and proliferation (21). Each of the ribozyme switches resulted in input-responsive regulation over cell viability and fluorescence (Fig. 2B and Fig. S1A; see SI Text for a discussion on controls and normalization methods), confirming the prescribed function of these devices in mammalian cells. The ribozyme-based regulatory systems provided titratable response over a range of input concentrations (Fig. 2C and Fig. S1B), demonstrating the ability to adjust expression levels on the basis of input availability.
Fig. 2.
Modularly constructed ribozyme switches exhibit tunable, drug-mediated regulation of gene expression and cell growth in mammalian T cells. (A) A modular insertion strategy allows the implementation of multiple copies of ribozyme switches to tune regulatory stringency. Spacer sequences (Orange and Green) provide structural insulation and maintain the functional independence of each switch. (B) Ribozyme switches provide tunable, small-molecule-mediated regulatory systems. Cell viability levels are reported for constructs encoding theo-responsive switches (L2bulge1, 8, 9) in one (1×), two (2×), three (3×), and four (4×) copies through transient transfections in CTLL-2 cells grown in 0 and 1 mM theo. No IL-2 control, construct not encoding a proliferative cytokine; sTRSV ribozyme, construct encoding a nonswitch hammerhead ribozyme. The gray bar indicates background viability level of cells in the absence of cytokine. (C) Ribozyme switches provide titratable regulatory systems. Cell viability levels are reported for the L2bulge9 regulatory systems at various theo concentrations. (D and E) Tailoring of the input responsiveness of the RNA-based regulatory system through direct replacement of the sensor component generates titratable, tet-responsive switches (L2bulge18tc). (F and G) Tailoring of the output of the RNA-based regulatory system through direct replacement of the target gene (cd19-tk-t2a-il15) achieves enhanced survival response compared to IL-2-based systems. All viability and fluorescence values were normalized to those of controls expressing the appropriate transgene regulated by an inactive ribozyme and cultured at corresponding theo concentrations. Reported values are mean ± SD from at least two replicate samples.
A General Strategy for Tuning the Dynamic Range of Ribozyme Switches Allows for Modulation of T-Cell Proliferation Response.
The potency of the genetic targets in our T-cell regulatory system requires stringent control over basal cytokine expression levels, such that in the absence of input the engineered T cells exhibit proliferation levels similar to cells growing in the absence of cytokine. To engineer a more stringent regulatory system, we implemented a second tuning strategy by linking multiple copies of the ribozyme switches in the 3′ UTR of the transgene. Only one of the switches needs to be in a ribozyme-active state to inactivate the transcript. Therefore, multiple-copy switch devices increase the probability of transcript cleavage and lower basal expression levels (20). We developed a construction strategy for sequentially inserting ribozyme switches in the 3′ UTR of the transgene and insulated the switches through spacer sequences designed to maintain the structural integrity and functional independence of each switch (SI Text and Fig. 2A).
Characterization studies indicated that this tuning strategy effectively decreased basal expression levels (Fig. 2B and Fig. S1A) and that the titratable response of the system was maintained (Fig. 2C and Fig. S1B). Stringent knockdown was achieved with three and four copies of the tightest switch (L2bulge9; Fig. 2B), which resulted in viability levels comparable to cells transfected with no cytokine or with the fully active, nonswitch ribozyme (no IL-2 control and sTRSV ribozyme, respectively). Notably, the regulatory performance of the four-copy switch system was similar to that of the three-copy system (Fig. 2B), indicating that three copies of the tightest switch were sufficient to approach the minimum possible viability levels. The data underscore the nonlinear relationship between direct gene expression and functional pathway outputs from the system. Whereas absolute changes in gene expression (fluorescence) in response to drug input remain similar for constructs with varying copies of a ribozyme switch (Fig. S1A), the absolute changes in pathway output (viability) increased substantially with ribozyme copy number (Fig. 2B).
Ribozyme Switches Can Be Programmed to Respond to Alternative Drug Molecules.
The on-state expression levels of drug-responsive RNA regulatory systems can be limited by the toxicity and cell permeability of the input molecule (22). However, the ribozyme-based regulatory system’s component functions are modular and thus amenable to changes that support customization for diverse applications, such as reprogramming input responsiveness toward clinically usable pharmaceuticals. To verify this critical property of our prototype T-cell proliferation control system, we replaced the theo aptamer (23) with the tetracycline (tet) aptamer (24) to construct a tet-responsive switch (L2bulge18tc; Fig. 2D). In vitro assays demonstrated tet-responsive ON switch activity in CTLL-2 cells (Fig. 2E). The tet-responsive systems demonstrated lower basal expression levels and increased dynamic ranges in response to lower input concentrations relative to the theo-responsive systems. Although tet is not a clinically applicable drug input, the tet switch system demonstrates the ability to improve regulatory stringency and increase the switch dynamic range by using aptamers with higher affinities and input molecules that can be administered to higher intracellular concentrations.
A Clinically Relevant T-Cell Proliferation System with Drug-Modulated Regulation of IL-15 Levels.
Although IL-2 plays a critical role in the stimulation of activated T cells, it is also involved in activation-induced cell death and the establishment of peripheral tolerance. Several studies have indicated that an alternative γ-chain cytokine, IL-15, provides potent homeostatic T-cell survival/proliferative signals, inhibits IL-2-mediated activation-induced cell death, and may be superior to IL-2 in immunotherapy applications (25, 26). Recently, IL-15 has been shown to function in establishing the long-term persistence of adoptively transferred central memory T (TCM) cells in primates, suggesting significant potential in T-cell therapy for cancer (27). To develop a more clinically relevant regulatory system, we utilized the modularity of the ribozyme switch platform and replaced the egfp-t2a-il2 transgene with a trifunctional fusion transgene (cd19-tk-t2a-il15) encoding IL-15, mutant HSV-1 thymidine kinase (ser39TK, a protein marker that can act as a PET reporter and as a suicide protein in the presence of the drug ganciclovir, providing imaging and safety kill-switch functionalities for downstream clinical implementations), and CD19 (a quantifiable protein marker amenable to FACS- and immunomagnetic-based selections). The alternative transgene was placed directly into the theo-responsive switch systems with L2bulge9 (Fig. 2F). The modified system exhibited ON switch control over cell viability and proliferation in transient transfection experiments (Fig. 2G), confirming modular coupling between the target transgene and the regulatory device. Samples expressing IL-15 showed higher viability levels compared to those expressing IL-2 with the corresponding switch systems (Fig. 2 C and G), suggesting that IL-15 may be a more potent survival/proliferative cytokine and can better amplify the signal response. Therefore, under a low basal expression level, a small increase in IL-15 expression has the potential to significantly enhance T-cell proliferation.
Ribozyme Switches Enable Long-Term, Dynamic Control over Gene Expression.
We generated a CTLL-2 cell line (CffLuc) that stably expressed the firefly luciferase (ffluc) gene to enable biophotonic imaging of cell populations in vivo and subsequently integrated T-cell proliferation regulatory systems with one or three copies of L2bulge9 into CffLuc. Stable integrants were initially sorted on the basis of CD19 expression, and bulk-sorted populations demonstrated theo-reponsive ON switch activity (Fig. S2). We further refined the sorted population by alternating treatment with ganciclovir and IL-2 or with theo and no IL-2 (Fig. S3) to enrich for clones with low basal expression levels and sufficiently high on-state expression levels to sustain cell survival, respectively. We generated clonal cell lines by a final sorting step for CD19-positive cells in the presence of theo.
Growth behavior of individual clones was characterized by culturing under various theo concentrations. Results indicate that the T-cell proliferation regulatory system retained functionality over long time periods when stably integrated and that theo effectively replaced IL-2 as the trigger for cell proliferation (Fig. 3A). Fifteen of the sixteen clones examined showed substantial theo-responsive increase in cell growth (Fig. S4), supporting that the growth modulation effect is specific to the introduced regulatory system.
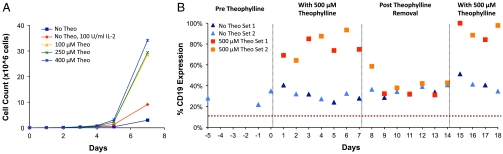
Fig. 3.
T cells stably expressing ribozyme switch systems exhibit drug-mediated regulation of growth over extended time periods in vitro. (A) Clonal cell lines stably expressing the ribozyme switch system exhibit drug-mediated growth. Cell growth was monitored by counting viable cells, and data for a representative clone [1264-48, expressing cd19-tk-t2a-il15-L2bulge9(3×)] are shown. (B) The ribozyme switch system regulates gene expression in response to changes in input concentrations. Duplicate sets of the clonal cell line 1264-48 were cultured in the presence (Red and Orange) and absence (Light Blue and Dark Blue) of theo over 18 days. Expression levels were monitored by staining with PE-CD19 antibody, and values were normalized to those from the inactive ribozyme control. The highest expression level was set to 100%. The red dashed line indicates background staining level of a cell line without a CD19 construct.
Additional assays on a clonal cell line harboring three copies of L2bulge9 (clone 1264-48) were performed to verify the mechanism of growth regulation and examine the dynamic behavior of the regulatory system to variations in theo availability. Cell cultures were grown for one week in the presence of 500 μM theo and continued for a second week in the absence of theo. Theo was reintroduced for another four days at the end of the study. Results indicated rapid (within 24 hours of induction), sustained, and reversible regulation of CD19 protein levels in response to theo (Fig. 3B). IL-15 expression patterns were verified at the transcript level through qRT-PCR (Fig. S5A). In addition, Western blot analysis of phosphorylated STAT5 levels verified activation of the IL-15 receptor-signaling cascade in the presence of theo (Fig. S5B). These results highlight the ability of the ribozyme switch system to quickly, effectively, and robustly switch gene expression on and off in response to drug input.
Ribozyme Switches Enable Drug-Modulated Control over T-Cell Proliferation in Vivo.
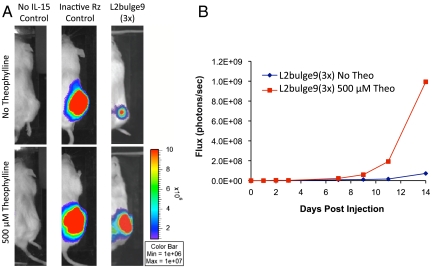
To verify in vivo functionality, we examined several clonal cell lines for theo-dependent growth in mice. Select clones were encased in a hydrogel matrix that contained either 0 or 500 μM theo and injected into the flanks of  mice. Cell lines lacking the transgene regulatory system or stably expressing the inactive ribozyme construct served as negative and positive controls, respectively. In vivo T-cell expansion was not observed from clones lacking cytokine expression (no IL-15 control, Fig. 4A), demonstrating the need for cytokine expression in sustaining cell growth. Uncontrolled T-cell proliferation was observed in the absence of a functional ribozyme-based regulatory device regardless of theo availability (inactive Rz control, Fig. 4A). In contrast, the clonal cell line 1264-48, which harbors three copies of L2bulge9, showed a significantly stronger reporter signal at the conclusion of the 14-day study when injected with 500 μM theo compared to the same clone injected without theo [L2bulge9(3×), Fig. 4A]. Growth rate calculations on the basis of flux measurements over the 14-day period indicated a 32% increase in in vivo growth rate in the presence of 500 μM theo, leading to a 14-fold increase in luciferase signal (Fig. 4B and Fig. S6). The in vivo study was repeated for clone 1264-48 with replicates, with the inactive ribozyme serving as the positive control. Flux measurements over a 9-day period indicated an average of 40% increase in the growth rate of clone 1264-48 in the presence of 500 μM theo (n1 = 4, n2 = 6, P = 0.038 by Mann–Whitney U test). In contrast, the positive control did not show statistically significant changes in growth rate (n1 = 6, n2 = 6, P = 0.394; Figs. S6C and S7), indicating that the input-responsive growth behavior observed in clone 1264-48 was not because of nonspecific effects of theo.
mice. Cell lines lacking the transgene regulatory system or stably expressing the inactive ribozyme construct served as negative and positive controls, respectively. In vivo T-cell expansion was not observed from clones lacking cytokine expression (no IL-15 control, Fig. 4A), demonstrating the need for cytokine expression in sustaining cell growth. Uncontrolled T-cell proliferation was observed in the absence of a functional ribozyme-based regulatory device regardless of theo availability (inactive Rz control, Fig. 4A). In contrast, the clonal cell line 1264-48, which harbors three copies of L2bulge9, showed a significantly stronger reporter signal at the conclusion of the 14-day study when injected with 500 μM theo compared to the same clone injected without theo [L2bulge9(3×), Fig. 4A]. Growth rate calculations on the basis of flux measurements over the 14-day period indicated a 32% increase in in vivo growth rate in the presence of 500 μM theo, leading to a 14-fold increase in luciferase signal (Fig. 4B and Fig. S6). The in vivo study was repeated for clone 1264-48 with replicates, with the inactive ribozyme serving as the positive control. Flux measurements over a 9-day period indicated an average of 40% increase in the growth rate of clone 1264-48 in the presence of 500 μM theo (n1 = 4, n2 = 6, P = 0.038 by Mann–Whitney U test). In contrast, the positive control did not show statistically significant changes in growth rate (n1 = 6, n2 = 6, P = 0.394; Figs. S6C and S7), indicating that the input-responsive growth behavior observed in clone 1264-48 was not because of nonspecific effects of theo.
Fig. 4.
T cells stably expressing the ribozyme switch systems exhibit drug-mediated regulation of growth over extended time periods in vivo. (A) Both cytokine expression and functional ribozyme switches are required for effective regulation of T-cell proliferation. Images are shown for day 14 postinjection of the negative control (no IL-15 Control, CffLuc), positive control (inactive Rz control, stable cell line expressing inactive ribozyme), and stable cell line expressing the ribozyme switch system [L2bulge9(3×), clone 1264-48] in the presence and absence of 500 μM theo. (B) Total luciferase signal flux over 14 days after injection of T cells is reported from the negative control (CffLuc) and clone 1264-48 [L2bulge9(3×)].
Ribozyme Switches Enable Proliferation Control in Primary Human T Cells.
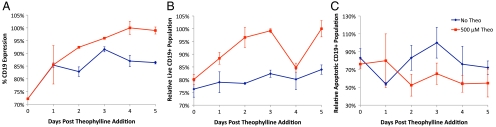
To demonstrate the portability of the regulatory system to human T lymphocytes and translatability to clinical applications, we transduced primary human TCM cells with lentiviral vectors encoding the cd19-tk-t2a-il15 transgene coupled to three copies of the L2bulge9 switch or to an inactive ribozyme. Because a short production timeline and streamlined processing are critical in clinical applications, studies were performed on bulk transduced, unsorted TCM populations to examine the robustness of the regulatory system under no population refinement. Transduced cells were cultured in the presence and absence of 500 μM theo for five days. Cells expressing L2bulge9 showed up to 15% increase in CD19 expression levels (Fig. 5A), 24% increase in the live cell population (Fig. 5B), and 54% reduction in the apoptotic cell population (Fig. 5C) in the presence of theo compared to the inactive ribozyme control, indicating drug-responsive ON switch behavior in gene expression and cell growth. The measured change in CD19 expression is comparable to that observed in CTLL-2 stable cell lines (Fig. S2), and the shift in population distribution between live and apoptotic cells supports that the regulatory system is effective in controlling the fate of primary human TCM cells.
Fig. 5.
The ribozyme switch system effectively regulates gene expression and cell fate in primary human TCM cells. (A) CD19 expression levels are elevated in the presence of theo. The populations of cells that are (B) live and CD19+ or (C) apoptotic and CD19+ indicate an increase in live cells and decrease in apoptotic cells in response to theo. Values for the L2bulge9(3×) sample are normalized to those of the inactive ribozyme control cultured at the same theo concentration. Reported values are mean ± SD from triplicate samples. The highest level is set to 100%.
Discussion
Achieving robust, controlled, long-term persistence of transfused T cells in vivo is a critical objective in adoptive immunotherapy, where efficacy depends on prolonged T-cell survival and safety demands stringent growth regulation. The T-cell proliferation challenge highlights a broader need for programmable regulatory systems that can modulate functional responses in mammalian cells. Whereas numerous gene-regulatory systems have been developed, successful translation to clinical settings has been limited. For example, synthetic inducible promoters require various transcriptional regulatory proteins, thus requiring multiple vectors and transgene expression systems. However, the stable expression of multiple transgenes in primary cell lines presents a significant challenge. In addition, concerns for potential immunogenicity of heterologous proteins limits the use of inducible promoters in clinical settings. Finally, the input specificity and regulatory dynamic range of such protein-based systems are relatively difficult to reprogram, thus limiting the flexibility and broader application of these systems.
An alternative technology that has been pursued with noted success in the context of hematopoietic cell growth regulation is based on chemical inducers of dimerization (CIDs), which induce dimerization of pathway components fused to CID-binding domains (28, 29). However, studies conducted with CID-based systems have required lethally irradiated animal subjects to allow expansion of the transfused T cells with the assistance of homeostatic proliferation. In addition, this system is limited to the regulation of homodimerization events, a mechanism that does not apply to many pathways of interest, including the common γ-chain signaling pathway central to T-cell persistence. The CID-based system is also fixed with respect to the input molecule, which is not available as a pharmaceutical drug.
Our synthetic, drug-responsive RNA-based regulatory system features properties that address critical limitations in existing genetic control strategies for clinical applications, including encapsulation within a compact, RNA-only platform (to avoid the use of immunogenic protein-based components) and allowing direct replacement of sensor and target transgene components (to tailor the technology to diverse applications). Our work describes the successful translation of a RNA device to the regulation of a functional response in mammalian cells. The unique capability to program the regulatory device’s response properties was key to the effective transition from reporter gene-expression regulation to functional control. By developing a rational tuning strategy for the regulatory stringency of our control system and integrating our synthetic controller with upstream signaling components, we were able to substantially enhance control over the functional system response. Furthermore, the observed nonlinear relationship between direct gene-expression levels and downstream pathway outputs highlights the importance of system design strategies. In particular, the ability to link synthetic controllers with relatively moderate gene-regulatory activities to potent upstream pathway components enables dramatic alterations in downstream functional behaviors.
Another unique property of our RNA-based control system is the modularity of the device framework, which supports rapid and effective tailoring of the regulatory system to respond to clinically applicable inputs. Furthermore, the implementation of higher-order information processing devices (20) on platforms that integrate multiple aptamers to both endogenous and exogenous inputs will enable more sophisticated control strategies with applications in autonomous control systems, in vivo diagnosis, and precise localization of cellular therapeutics to disease targets. The development of RNA aptamers to pharmaceuticals with minimal off-target toxicities and high cell permeability and bioavailability will be critical for the translation of these RNA-based regulatory systems to clinical applications. Therefore, the coupling of our modular device framework with advances in aptamer selection processes (30) will more broadly support the tailoring of RNA-based regulatory systems to diverse applications in health and medicine, including diagnostics, cellular therapeutics, gene therapy, and intelligent molecular therapies.
Materials and Methods
Mammalian Cell Culture Maintenance.
The mouse T-cell line CTLL-2 was obtained from ATCC and maintained in RPMI-1640 media (Lonza) supplemented with 10% heat-inactivated fetal bovine serum (Hyclone), 2 mM sodium pyruvate (Gibco), and 4.5 g/L D-(-)-glucose (Sigma). Cells were fed 100 U/mL IL-2 every 48 hours and maintained between 0.05 × 106 and 0.50 × 106 cells/mL. All cell lines stably expressing the ffLuc:zeocin fusion gene were cultured with 0.20 mg/mL zeocin (Invivogen). Primary human TCM cells were derived from peripheral blood mononuclear cells (SI Text) and maintained in RPMI-1640 media supplemented with 10% heat-inactivated fetal bovine serum. Cells were fed 50 U/mL IL-2 and 0.5 ng/mL IL-15 every 48 hours and maintained between 0.2 × 106 and 1.0 × 106 cells/mL.
Transient Transfection and Fluorescence Quantification.
All transient transfections were performed with an Amaxa Nucleofector II and the Mouse T Cell Nucleofector Kit (Amaxa). Appropriate concentrations of drug input were added immediately after electroporation. Fluorescence and cell viability data were obtained 24 and 48 hours after transfection, respectively, by using a Quanta Cell Lab Flow Cytometer (Beckman Coulter). Plasmid maps are provided in Fig. S8. See SI Text and Fig. S9 for additional details.
In Vivo T-Cell Proliferation Studies in NOD/SCID-IL2(ko) Mice.
Clonal cell lines were expanded under regular culture conditions, washed, and resuspended in PBS. A 100-μL solution containing 0.1 × 106 cells, 50% vol/vol Matrigel (BD Biosciences), and either 0 or 500 μM theo was injected subcutaneously into the flank of NOD/scid-IL2(ko) mice. In vivo growth of the injected cells was monitored by biophotonic imaging using the IVIS Imaging System 100 Series (Xenogen). See SI Text for additional details.
Supplementary Material
Acknowledgments.
We thank members of the Smolke Lab, Y.A. Chen, and D. Endy, for critical reading of the manuscript; M.N. Win for contributing expertise on ribozyme switch design; and B. Aguilar, C. Bautista, C. Brown, L. Brown, W. Chang, R. Diamond, A. Hamlett, M. Hunter, D. Perez, G. Raval, J. Wagner, W. Wong, and C. Wright for technical assistance. This work was supported by the City of Hope’s National Cancer Institute-Cancer Center Support Grant, the National Science Foundation (fellowship to Y.Y.C.), and the Alfred P. Sloan Foundation (fellowship to C.D.S.).
Footnotes
Conflict of interest statement: We declare competing financial interests in the form of a pending patent application whose value may be affected by the publication of this manuscript.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001721107/-/DCSupplemental.
References
- 1.June CH. Principles of adoptive T cell cancer therapy. J Clin Invest. 2007;117:1204–1212. doi: 10.1172/JCI31446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falkenburg JH, Smit WM, Willemze R. Cytotoxic T-lymphocyte (CTL) responses against acute or chronic myeloid leukemia. Immunol Rev. 1997;157:223–230. doi: 10.1111/j.1600-065x.1997.tb00985.x. [DOI] [PubMed] [Google Scholar]
- 3.Walter EA, et al. Reconstitution of cellular immunity against cytomegalovirus in recipients of allogeneic bone marrow by transfer of T-cell clones from the donor. N Engl J Med. 1995;333:1038–1044. doi: 10.1056/NEJM199510193331603. [DOI] [PubMed] [Google Scholar]
- 4.Morgan RA, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kahlon KS, et al. Specific recognition and killing of glioblastoma multiforme by interleukin 13-zetakine redirected cytolytic T cells. Cancer Res. 2004;64:9160–9166. doi: 10.1158/0008-5472.CAN-04-0454. [DOI] [PubMed] [Google Scholar]
- 6.Leen AM, Rooney CM, Foster AE. Improving T cell therapy for cancer. Annu Rev Immunol. 2007;25:243–265. doi: 10.1146/annurev.immunol.25.022106.141527. [DOI] [PubMed] [Google Scholar]
- 7.Robbins PF, et al. Cutting edge: Persistence of transferred lymphocyte clonotypes correlates with cancer regression in patients receiving cell transfer therapy. J Immunol. 2004;173:7125–7130. doi: 10.4049/jimmunol.173.12.7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yee C, et al. Adoptive T cell therapy using antigen-specific CD8+ T cell clones for the treatment of patients with metastatic melanoma: in vivo persistence, migration, and antitumor effect of transferred T cells. Proc Natl Acad Sci USA. 2002;99:16168–16173. doi: 10.1073/pnas.242600099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mackensen A, et al. Phase I study of adoptive T-cell therapy using antigen-specific CD8+ T cells for the treatment of patients with metastatic melanoma. J Clin Oncol. 2006;24:5060–5069. doi: 10.1200/JCO.2006.07.1100. [DOI] [PubMed] [Google Scholar]
- 10.Johnston JA, et al. Tyrosine phosphorylation and activation of STAT5, STAT3, and Janus kinases by interleukins 2 and 15. Proc Natl Acad Sci USA. 1995;92:8705–8709. doi: 10.1073/pnas.92.19.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gattinoni L, Powell DJ, Jr, Rosenberg SA, Restifo NP. Adoptive immunotherapy for cancer: Building on success. Nat Rev Immunol. 2006;6:383–393. doi: 10.1038/nri1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novina CD, Sharp PA. The RNAi revolution. Nature. 2004;430:161–164. doi: 10.1038/430161a. [DOI] [PubMed] [Google Scholar]
- 13.Fedor MJ, Williamson JR. The catalytic diversity of RNAs. Nat Rev Mol Cell Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- 14.Breaker RR. Complex riboswitches. Science. 2008;319:1795–1797. doi: 10.1126/science.1152621. [DOI] [PubMed] [Google Scholar]
- 15.Mathews DH, Turner DH. Prediction of RNA secondary structure by free energy minimization. Curr Opin Struct Biol. 2006;16:270–278. doi: 10.1016/j.sbi.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 16.Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nat Biotechnol. 2006;24:545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- 17.Win MN, Liang JC, Smolke CD. Frameworks for programming biological function through RNA parts and devices. Chem Biol. 2009;16:298–310. doi: 10.1016/j.chembiol.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isaacs FJ, et al. Engineered riboregulators enable post-transcriptional control of gene expression. Nat Biotechnol. 2004;22:841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- 19.Win MN, Smolke CD. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc Natl Acad Sci USA. 2007;104:14283–14288. doi: 10.1073/pnas.0703961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–460. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gillis S, Smith KA. Long term culture of tumour-specific cytotoxic T cells. Nature. 1977;268:154–156. doi: 10.1038/268154a0. [DOI] [PubMed] [Google Scholar]
- 22.Beisel CL, Smolke CD. Design principles for riboswitch function. PLoS Comput Biol. 2009;5:e1000363. doi: 10.1371/journal.pcbi.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 24.Berens C, Thain A, Schroeder R. A tetracycline-binding RNA aptamer. Bioorg Med Chem. 2001;9:2549–2556. doi: 10.1016/s0968-0896(01)00063-3. [DOI] [PubMed] [Google Scholar]
- 25.Hsu C, et al. Primary human T lymphocytes engineered with a codon-optimized IL-15 gene resist cytokine withdrawal-induced apoptosis and persist long-term in the absence of exogenous cytokine. J Immunol. 2005;175:7226–7234. doi: 10.4049/jimmunol.175.11.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waldmann TA, Dubois S, Tagaya Y. Contrasting roles of IL-2 and IL-15 in the life and death of lymphocytes: implications for immunotherapy. Immunity. 2001;14:105–110. [PubMed] [Google Scholar]
- 27.Berger C, et al. Adoptive transfer of effector CD8+ T cells derived from central memory cells establishes persistent T cell memory in primates. J Clin Invest. 2008;118:294–305. doi: 10.1172/JCI32103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blau CA, Peterson KR, Drachman JG, Spencer DM. A proliferation switch for genetically modified cells. Proc Natl Acad Sci USA. 1997;94:3076–3081. doi: 10.1073/pnas.94.7.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neff T, et al. Pharmacologically regulated in vivo selection in a large animal. Blood. 2002;100:2026–2031. doi: 10.1182/blood-2002-03-0792. [DOI] [PubMed] [Google Scholar]
- 30.Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu Rev Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.