Abstract
Bat flight poses intriguing questions about how flight independently developed in mammals. Flight is among the most energy-consuming activities. Thus, we deduced that changes in energy metabolism must be a primary factor in the origin of flight in bats. The respiratory chain of the mitochondrial produces 95% of the adenosine triphosphate (ATP) needed for locomotion. Because the respiratory chain has a dual genetic foundation, with genes encoded by both the mitochondrial and nuclear genomes, we examined both genomes to gain insights into the evolution of flight within mammals. Evidence for positive selection was detected in 23.08% of the mitochondrial-encoded and 4.90% of nuclear-encoded oxidative phosphorylation (OXPHOS) genes, but in only 2.25% of the nuclear-encoded nonrespiratory genes that function in mitochondria or 1.005% of other nuclear genes in bats. To address the caveat that the two available bat genomes are of only draft quality, we resequenced 77 OXPHOS genes from four species of bats. The analysis of the resequenced gene data are in agreement with our conclusion that a significantly higher proportion of genes involved in energy metabolism, compared with background genes, show evidence of adaptive evolution specific on the common ancestral bat lineage. Both mitochondrial and nuclear-encoded OXPHOS genes display evidence of adaptive evolution along the common ancestral branch of bats, supporting our hypothesis that genes involved in energy metabolism were targets of natural selection and allowed adaptation to the huge change in energy demand that were required during the origin of flight.
Keywords: Chiroptera, genetic foundation, mitochondria, OXPHOS
Bats are perhaps the most unusual and specialized of all mammals, as flight is their main mode of locomotion. Although there are several gliding mammals that are able to glide from tree to tree (such as the flying squirrel, gliding possums, and colugos), bats are the only mammal capable of sustaining level flight (1). The evolution of flight in bats was a major factor leading to the success of this amazing group (2, 3). A number of adaptations to flight found in birds are not shared by mammals, thus Darwin in the Origin of Species (Chapter 5) (4) proposed that the evolution of a flying bat from an insectivorous terrestrial mammal was too difficult to imagine.
Bat flight is a highly complex functional system from a morphological, physiological, and aerodynamic perspective (5). As in birds, bat flight requires a metabolic rate that is 3–5 times greater than the maximum observed during exercise in similar-sized terrestrial mammals (2, 6). Hence, a significant metabolic barrier must separate volant from nonvolant vertebrates (6). Therefore, we speculate that energy metabolism is among the primary factors that influenced the development of flight in bats.
The respiratory chain of the mitochondrial produces 95% of the adenosine triphosphate (ATP) needed for locomotion. The enzymes involved in oxidative phosphorylation (OXPHOS) are composed of multisubunit complexes that are encoded by both nuclear and mitochondrial genes (7). Mitochondrial DNA (mtDNA) encodes 13 proteins, all playing vital roles in the electron transport chain. In a previous study, we demonstrated that functional constraints of mtDNA in energy metabolism had influenced the locomotive evolution in birds and mammals (8). In addition to the 13 mitochondrial-encoded proteins, there are more than 70 nuclear genes encoding proteins involved in oxidative phosphorylation (OXPHOS), the metabolic pathway that uses energy released by the oxidation of nutrients to produce adenosine triphosphate (ATP). Genes from the nuclear and mitochondrial genomes must work in concert to generate a functional oxidative phosphorylation (OXPHOS) system (9, 10). The importance of nuclear genes in mitochondrial OXPHOS has been demonstrated by patients with disorders in OXPHOS in which the majority of the gene defects are due to nuclear-encoded OXPHOS genes (11). When mitochondria were introduced into cells with a differing nuclear background, mitochondrial OXPHOS activity was shown to be disrupted (12, 13). These studies demonstrate that mitochondrial and nuclear genes must coevolve in a highly coordinated process, and likely more so as an organism's energetic demands change.
Given the dual (mitochondrial and nuclear) genetic foundation of the respiratory chain, to test whether genes for energy metabolism proteins have evolved adaptively and coevolve during the attainment of flight by bats, in this study we have used both mitochondrial and nuclear genome data from all available species relevant to bat evolution to test for adaptive evolution in OXPHOS genes. In addition to the mitochondrial and nuclear-encoded OXPHOS genes, we also analyzed genes for 888 nuclear-encoded mitochondrial proteins and 7,164 nonmitochondrial proteins to provide a measure of the background of adaptive evolution. Our study demonstrates that a large amount of adaptive evolution occurred specifically on OXPHOS genes and on the common ancestral lineage leading to bats, suggesting that adaptive evolution of OXPHOS genes was necessary for the attainment of flight.
Results and Discussion
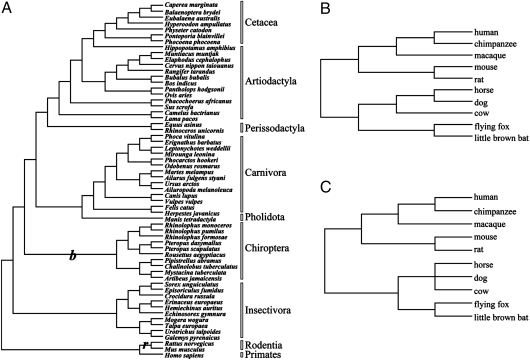
Maximum-likelihood and Bayesian trees were reconstructed with the concatenated 13 protein-coding genes from mitochondrial genomes of 60 mammals (SI Appendix, Table S1) generated a topology the same as that of the traditional phylogeny (Fig. 1A) (14–18). A study (19) based on large amounts of data still failed to resolve the relationships of Laurasiatheria. So, we used two trees for nuclear genes (Fig. 1 B and C). Molecular studies unambiguously support Chiroptera being monophyletic and thus flight most likely arose only once in the common ancestor of Chiroptera (20).
Fig. 1.
Phylogenies used for detection of positive selection. (A) Phylogeny of 60 mammals used for evolutionary analysis of mitochondrial genomes. (B and C) Phylogeny of 10 mammals used for evolutionary analysis of whole nuclear genomes.
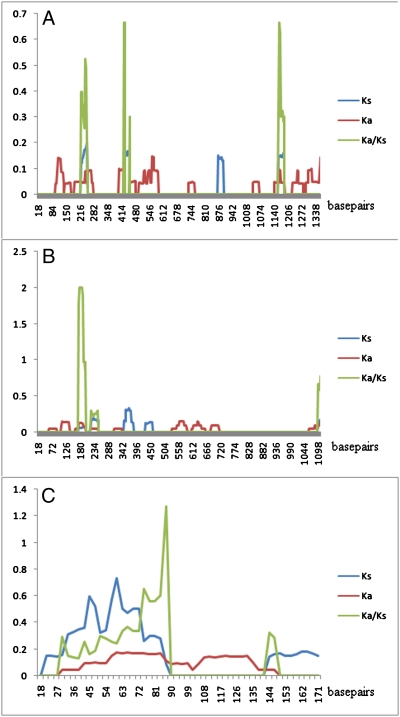
To determine whether there was evidence that adaptive evolution was occurring, we used the PAML package (21). This software package uses a maximum-likelihood approach to calculate nonsynonymous to synonymous rate ratios (ω = Ka/Ks or dN/dS). Ka/Ks > 1 indicates positive selection, Ka/Ks <1 negative selection, and Ka/Ks =1 neutrality. First, we used the one-ratio model (M0), a very strict model that allows only a single Ka/Ks ratio for all branches. The Ka/Ks ratios that we obtained for all 13 individual mitochondrial genes are significantly less than 1, providing good support for the expected presence of negative selection acting on all mitochondrial genes, showing that strong purifying selection plays a central role in the evolution of mtDNA to keep its important functions in energy metabolism (22–26). Purifying selection, by itself, cannot generate “good” genes as it only maintains a gene's function. Thus this phenomenon raises an interesting question: where do the present-day “good” (adapted) mitochondrial genes come from? Positive selection and gene duplication are two major mechanisms of adaptive evolution. Because mitochondrial genomes lack recom-bination and thus cannot generate duplicate genes, we deduced that positive selection must have occurred on some mtDNA genes during some lineages in the evolution of animals. Here, we focus on bats, the only mammalian group that have developed the ability to fly. As flight consumes much more energy than running or walking, and as mitochondria provide most of the energy needed for locomotion, we assumed that the mtDNA of flying mammals must be much adapted to this large energy demand. The common ancestral lineage leading to bats was the branch (marked b in Fig. 1) where flight originated and thus is the lineage of interest. The two-ratio model showed that ω for the ND2, ND3, ND4L, ND4, ND5, ND6, and Cox III genes on the lineage leading to bats (ω1) are significantly greater than ratio on all other mammalian lineages (ω0) (SI Appendix, Table S2). However, the two-ratio model was not significantly different from a two-ratio model where ω1 = 1. Consequently, although it is clear that the branch leading to common ancestor of bats has undergone selective pressure that is significantly different from that of other branches, and although these mitochondrial genes have accumulated more nonsynonymous than synonymous mutations, the test did not unambiguously support positive selection on this branch, as we cannot exclude the possibility of relaxation of selection. Typically, positive selection will act on only a few sites and for a short period of evolutionary time; thus the signal for positive selection usually is swamped by the continuous negative selection that occurs on most sites in a gene sequence (27). Even after a short period of positive selection, this is commonly followed by a long period of purifying selection, which would obscure the selective processes. These processes explain why it has been so difficult to detect positive selection in mtDNA, despite extensive studies (28–30). Branch-site models, in contrast to branch models in which ω varies only among branches, allows variation in the selective pressure to occur at both amino acid sites and on lineages; thus, these models are considered to be powerful in distinguishing positive selection from the relaxation of purifying selection (27). We therefore used branch-site models to further examine for possible positive selective pressures on the branch leading to bats (marked b in Fig. 1). ND4, CytB, and ATP8 show evidence for significant positive selection according to the LRTs. Eight, two, and one amino acid sites, respectively, were identified as candidate sites that had undergone positive selection (posterior probability ≥90%; SI Appendix, Table S2). These results were supported by a sliding window analyses that can intuitively show where rates of nonsynonymous substitution (Ka) exceed rates of synonymous substitution (Ks) (Fig. 2).
Fig. 2.
Sliding windows of Nei-Gojobori estimates of Ks (blue), Ka (red), and Ka/Ks (green) on the common ancestral branch of bats. Window size was set as 30 and step size as 3. (A) ND4. (B) CytB. (C) ATP8.
Positive selection in mtDNA genes strongly suggests that adaptive evolution of energy metabolism genes played a critical role during the attainment of flight by bats by allowing elevated energy metabolism. Considered the dual (mitochondrial and nu-clear) genetic foundation of the respiratory chain, we took advantage of the 10 available mammalian genomes (including two bats; Materials and Methods), to examine the evolution of all of the nuclear-encoded OXPHOS genes. The nuclear-encoded OXPHOS genes for human were compiled from KEGG (31). We then used this dataset to BLAST (32) the other nine mammalian genomes to identify all known nuclear-encoded OXPHOS genes (SI Appendix, Table S3). As the relationships of Carnivora, Perissodactyla, and CetArtidactyla were not completely resolved (19), we used two topologies (Fig. 1 B and C) as guide trees, both of which yielded the same result. A total of five (ATP4B, ATP5B, ATP5L, COX4I1, and COX6A2) of the 102 (4.90%) nuclear encoded OXPHOS genes that were examined showed significant evidence for positive selection. Three of these five nuclear genes (ATP4B, ATP5B, and ATP5L) belong to Complex V, a complex that also includes the mitochondrial ATP8 gene, which also showed evidence of positive selection. The clustering of these mitochondrial and nuclear genes suggests that balancing cytonuclear coevolutionary constraint occurred during evolution of flight by bats. The complex V translocates protons from the intermembrane space to the matrix and phosphorylates ADP into ATP. The cytonuclear coevolution of this complex may reflect the greater functional importance of complex V in ATP generation.
The 102 nuclear-encoded OXPHOS genes represent only a small portion of the more than 1,000 nuclear-encoded proteins that function in mitochondria. Although these genes encode essential components of OXPHOS, other proteins also play important roles such as transporters, anchoring proteins, and cell channels (33). Therefore, to gain a global view of adaptive evolution of mitochondrial genes during the attainment of flight, we analyzed the evolution of all known nuclear-encoded mitochondrial genes during this time period. We identified 20 of the 888 (2.25%) nuclear-encoded mitochondrial protein genes as showing evidence of positive selection on the branch that leads to bats.
Previous studies have concluded that no more than 2% of orthologous genes in genome-wide studies show evidence for positive selection (34, 35). Here, in this study, we identified that 23.08% of mitochondrial and 4.90% of nuclear-encoded OXPHOS genes show evidence of positive selection, proportions that are greater than 2%. This observation supports our hypothesis that energy metabolism played a crucial role in the attainment of flight by bats. A caveat of our genome-wide analyses is that both of the bat genomes (little brown bat and flying fox) used to identify nuclear-encoded OXPHOS and mitochondrial protein genes are only of draft quality (1.7× and 2.63×, respectively); thus, a smaller number of complete genes could be identified and analyzed, and required additional care to avoid false-positive predictions (36). To control for the quality of the gene data in the analyses of the nuclear-encoded OXPHOS genes and mitochondrial protein genes, we used two approaches (i): measurement of the background rate of positive selection in the draft bat genomes, and (ii) resequencing of OXPHOS genes from several bat species and reanalysis.
Firstly, we analyzed 7,164 nuclear-encoded orthologous (nonmitochondrial) genes to identify the general background pattern of positive selection on the bat lineage (data available upon request). If the low-coverage genome data were responsible for the evidence for increased levels of positive selection in the nuclear-encoded OXPHOS genes, then we should also expect an increase in a random subset of genes that have no function in the mitochondria. Of the 7,164 genes examined, only 72 (1.005%) showed evidence for positive selection; this percentage is similar to that found in a recent studies of better assembled genomes (34, 35), suggesting that the relative low coverage assemblies are not responsible for the increased evidence for positive selection. In addition to the fact that no significant difference in the level of positive selection in background genes was observed between genomes, the higher positive selection ratio in nuclear-encoded OXPHOS genes strengthen our conclusion that the higher levels of adaptive evolution detected for energy metabolism genes is not simply due to a general phylogenetic pattern or sequencing errors in the draft bat genomes.
Second, we used a direct method to confirm our result: we resequenced 77 OXPHOS genes from four species of bats (SI Appendix, Table S4) and redid our analyses. Clearly, our approach would detect some false-positive results; in fact, we found that the ATP5B gene, which had been suggested to have undergone positive selection with the draft genome sequence data, did not show evidence of positive selection when analyzed using the resequencing data. However, sequences from 21 of the 77 genes (27.27%) examined showed evidence of accumulating greater numbers of nonsynonsymous mutations on the branch leading to the common ancestor of bats (SI Appendix, Table S5). These conclusions were also supported by the sliding window analyses. Our previous analyses showed that seven of 13 (53.85%) mitochondrial genes (ND2, ND3, ND4L, ND4, ND5, ND6, and Cox III) accumulate a greater proportion of nonsynonsymous mutations (SI Appendix, Table S2); and, considering that mitochondrial genes have much higher mutation rate than nuclear genes, the larger number of changes in mitochondrial genes may have driven the changes in the nuclear-encoding OXPHOS genes, suggesting a mito-nuclear interaction during the attainment of flight by bats.
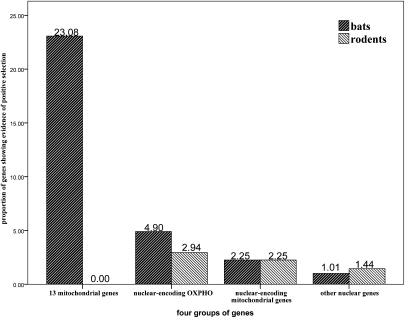
Our multiple approaches strengthen our conclusion that energy metabolism genes have undergone adaptive evolution during the attainment of flight by bats to adapt to the much greater energy demand of flight. Although an explanation that the adaptation to the greater energy demand imposed by flight would appear to be the best explanation, another possibility is that this phenomenon is simply a general pattern observed in mammals. To address this question, we also computed the level of positive selection on the common ancestral lineage leading to rodents (marked as r in Fig. 1) using the same datasets and with same analysis as described above for bats. On the common ancestral rodent lineage, we failed to detect any positive selection in all of the 13 mitochondrial genes. The proportion of nuclear-encoded OXPHOS genes showing evidence for positive selection along the common ancestor branch of rodents (1.96%) is lower than that observed on the bat lineage (4.90%). For the nuclear-encoded mitochondrial genes (excluding OXPHOS genes) and background genes, the levels of positive selection on the rodent lineage were similar to those observed on the bat lineage (Fig. 3). In addition, because Insectivora have a closer relationship with bats and have elevated metabolic rates, we made a similar com-parison with the two available Insectivora genomes (Erinaceus europaeus and Sorex araneus). However, they are of only of draft quality (1.86× and 1.9×, respectively), which limited our ability to identify their gene sequences. We were able to obtain sequences of only 48 of the 102 nuclear-encoding OXPHOS genes for all insectivores and bats. Of these 48 genes, positive selection was detected on the common ancestor of bats for two genes. However, for insectivores, none of the genes showed evidence of positive selection on their common ancestral lineage. The conclusions from the insectivore data, although limited, are in agree-ment with those derived from the rodent data. We also examined the two distinct lineages of bats. Here, we found that only three of the 102 OXPHOS genes (ATP6V0A1, NDUFB10 and SDHD) were under positive selection on the little brown bat lineage. However, our resequencing data demonstrated that the NDUFB10 result was due to a sequence error. Two genes (NDUFB7 and SDHB) showed evidence of positive selection on the flying fox lineage. The proportions of nuclear-encoded OXPHOS genes showing evidence for positive selection on the little brown bat (1.96%) and flying fox (1.96%) lineages are the same, and lower than that observed on the common ancestral lineage for all bats (4.90%). Previous studies suggested that bats have “flying DNA,” DNA that is composed of elevated levels of adenine and thymine (37); therefore, we tested the bias of base frequencies and codon usage for the genes that we studies. Consistent with a previous study (38), we failed to find the common trend of elevated levels of adenine and thymine or codon bias for bats (data available upon request). Thus, our observation of elevated levels of positive selection for OXPHOS related genes is not simply due to changes in base frequency or codon bias in bats. These results further strengthen our conclusion that energy metabolism genes played an important role in the attainment of flight by bats by adapting to greater energy demands.
Fig. 3.
Comparison of proportions of genes showing evidence of positive selection on the common ancestral lineages for bats and rodents. Group 1, 13 mitochondria-encoded genes. Group 2, nuclear-encoded OXPHOS genes. Group 3, nuclear-encoded mitochondrial proteins. Group 4, other nuclear genes (background genes).
Primates have a larger brain than most mammals, and the brain consumes more energy than other tissues, which has resulted in genes involved in aerobic energy metabolism undergoing adaptive evolution (29, 39–41). Similarly, the elephant also has larger brain, and a convergent pattern of adaptive evolution (higher proportions of adaptive evolution in aerobic energy metabolism genes) is supported (42). Here we also show adaptive evolution of genes involved in aerobic energy metabolism; however, in this case, it is due to the energetic demands of flight. Adaptation of energy demands is clearly one of the important factors that influence animals’ evolution.
A recent study has questioned the reliability of identifying positive selection by the branch-site method compared with parsimon- based methods, as it often generates false-positive results when the number of nucleotide substitutions is very small (43). However, the false-positive rate of branch-site method is much lower than the nominal significance level (<5%) (44), and parsimony-based methods have little power (45); thus, the branch-site method is valuable and has been widely used for detecting evidence for positive selection in comparative analyses of genomic data (44). Although branch site models do yield some false-positive predictions, a similar level should be observed in analyses of bat, insectivore, and rodent data; thus, the observation that bats have a much higher proportion of OXPHOS genes that show evidence of positive selection suggests that they have undergone a greater amount of positive selection.
Bats are unique in being the only mammals capable of powered flapping flight. As in birds, bat flight is a highly energetically expensive form of locomotion (2, 6). However, it is also a very efficient mode of transport and assists flyers in feeding and breeding as well as avoidance of predators. The evolution of flight in bats was a major factor leading to the success of this amazing group of mammals, although the evolution of this ability has required complex changes in the anatomy of these animals. In addition to other important factors, such as changes in bone density and development of the wings, bat flight also requires a significantly higher metabolic rate, a rate well above the maximum capable by other similar-sized terrestrial mammals during exercise (2, 6, 46). Aerobic metabolism by mitochondria plays a vital role as the energy production centers of cells The OXPHOS pathway of mitochondria has adaptively evolved to meet the demands of changing environmental and physiological conditions. Because the mitochondrial respiratory chain has a dual genetic foundation (mitochondria and nuclear genomes), here we examined both genomes to obtain insights into the evolution of flight by mammals. Both mitochondrial genes and nuclear-encoded OXPHOS genes showed greater evidence for adaptive evolution; this result supports our hypothesis that energy metabolism genes were targets of natural selection that included a balancing cytonuclear coevolutionary constraint, which allowed adaptive changes in energy demands and thus played a crucial role in attainment of flight by bats.
Materials and Methods
Source of Data and Primary Treatments.
Complete mitochondrial genomes were collected form GenBank for 60 species representing the major groups of mammals (SI Appendix, Table S1).
Human nuclear-encoded OXPHOS proteins were identified from the KEGG database (31). Genome sequences from 10 mammalian species (cow, dog, horse, macaque, mouse, little brown bat, chimpanzee, rat, flying fox, and human) in the ENSEMBL public database (version 52, December 2008) were searched for all of the known human nuclear-encoded OXPHOS genes (SI Appendix, Table S3).
Nuclear-encoded mitochondrial protein genes (excluding OXPHOS genes) were identified from the Human Mitochondrial Protein Database (http://bioinfo.nist.gov/), MitoProteome (47), MITOMAP (48), and MitoCarta (33). Orthologs were identified from the 10 mammalian genomes in the ENSEMBL public database (data available upon request).
To confirm the orthology of background genes, we used the Ensembl ortholog_one2one genes database (version 52, December 2008) (49) for each pair of the 10 genomes that we used. Only those genes that were in a one-to-one orthologs for every pair of genomes of the 10 species were used in our analyses. A total of 7,644 orthologus genes that had no identified mitochondrial function were used as “background genes.” For genes that have more than one transcript, we aligned all of the possible transcript pairs for all 10 species and retained those that provided the highest alignment scores. The two bat genomes (little brown bat and flying fox) are only of draft quality (1.7× and 2.63×, respectively), suggesting that sequencing or assembly errors may interfere with the ability to detect genes that have experienced positive selection. To reduce the rate of false-positive prediction, we deleted all gaps and “N” in the alignments. To exclude sequencing errors, incorrect alignments and nonorthologus regions in the alignments, we used a 15-bp sliding window on each alignment, and moved the sliding window by one codon for each step to the end of the alignment. For each window, we calculated two types of similarity; one is the average similarity of the 10 species within a sliding window, whereas the second is the lowest similarity of an alignment pair of the 10 species within the sliding window. Alignment regions with average similarity <80% or lowest similarity <60% were discarded, as these may included errors in sequence or assembly. After the deletion step, if the remaining alignment was shorter than 100 bp, then the entire alignment was discarded. Our final data set contained 7,164 genes.
RNA Isolation and Sequencing.
To confirm the analysis of the OXPHOS genes in bats we amplified and sequenced 77 OXPHOS genes from four species of bats, including Yinpterochiroptera (Old-World fruit bats, Rousettus leschenaultia and Cynopterus sphinx) and Yangochiroptera (Miniopterus fuliginosus and Scotophilus kuhlii) (SI Appendix, Table S4) (accession nos. GQ427677-GQ427913, and GU292797-GU292809). RNA was isolated with the RNAiso Plus Kit (Takara). RT-PCR was performed on these RNAs using the PrimeScript RT-PCR Kit (Takara).
Phylogenetic Analysis.
A phylogenetic tree of 60 mammalian species was reconstructed from concatenated 13 protein-coding genes using two algorithms (i): maximum likelihood in PAUP 4.0b10 (50), with the best tree being identified using a heuristic method with nearest-neighbor interchange and node support was evaluated by bootstrapping with 1,000 replicates; and (ii) Bayesian analysis in MrBayes 3.12 (51).
Phylogenetic trees for each of the genes from the 10 mammalian genomes were based on previous phylogenetic studies (19, 52) (Fig. 1 B and C).
Selection Analyses.
Alignments and consensus trees were used for posterior molecular evolutionary analysis. Positive selection analysis was restricted to the branch of interest (the branch leading to the most recent reconstructed ancestor of bats, marked b, or the common ancestor or rodents, marked r in Fig. 1). We used a gene-level approach based on the ratio (ω) of nonsynonymous (Ka) to synonymous (Ks) substitutions rate (ω = Ka/Ks) to identify potential positive selection, using likelihood ratio tests using the CODEML algorithm from the PAML package (21). First, we tested branch models M0, the most simple model, which has a single ω ratio for the entire tree. Subsequently, we used two-ratio models that allow a background ω ratio and a different ω on the branch of interest. For null hypotheses, we used the one-ratio model and a two-ratio model with a fixed ω = 1 on the branch under analysis. The level of significance for these LRTs was calculated using a χ2 approximation, where twice the difference of log likelihood between the models (2ΔlnL) would be asymptotic to a χ2 distribution, with the number of degree of freedom corresponding to the difference in number of parameters between the nested models. We then used the branch site model to detect positively selected sites on the branch leading to bats. Test 1 and test 2, developed by Zhang et al., were used as controls, as these two tests have been shown to have power to differentiate positive selection from the relaxation of selective constraints (27).
To intuitively show where rates of nonsynonymous substitution (Ka) exceeded rates of synonymous substitution (Ks) along the branch leading to the common ancestor of bats, we further performed sliding-window analyses (window size, 30 codons; step size, 3 codons) of Nei-Gojobori estimates of synonymous (Ks) and nonsynonymous substitutions per site (Ka) and the Ka/Ks in SWAAP 102 (53).
Supplementary Material
Acknowledgments
The authors thank Peng Shi, Scott Groom, and two anonymous reviewers for helpful comments. This work was supported by grants from the National Basic Research Program of China (973 Program, 2007CB411600), National Natural Science Foundation of China, and Bureau of Science and Technology of Yunnan Province (to Y.-P.Z.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. GQ427677–GQ427913 and GU292797-GU292809).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912613107/DCSupplemental.
References
- 1.Nowak RM. Walker's Mammals of the World. 6th Ed. Baltimore: Johns Hopkins University Press; 1999. [Google Scholar]
- 2.Thomas SP, Suthers RA. The physiology and energetics of bat flight. J Exp Biol. 1972;57:317–335. [Google Scholar]
- 3.Norberg UM. Wing design, flight performance, and habitat use in bats. In: Wainwright PC, Reilly SM, editors. Ecological Morphology: Integrative Organismal Biology. Chicago: University of Chicago Press; 1994. pp. 205–239. [Google Scholar]
- 4.Darwin C. On the Origin of Species by Means of Natural Selection. London: John Murray; 1859. [Google Scholar]
- 5.Bishop KL. The evolution of flight in bats: Narrowing the field of plausible hypotheses. Q Rev Biol. 2008;83:153–169. doi: 10.1086/587825. [DOI] [PubMed] [Google Scholar]
- 6.Maina JN. What it takes to fly: The structural and functional respiratory refinements in birds and bats. J Exp Biol. 2000;203:3045–3064. doi: 10.1242/jeb.203.20.3045. [DOI] [PubMed] [Google Scholar]
- 7.Das J. The role of mitochondrial respiration in physiological and evolutionary adaptation. Bioessays. 2006;28:890–901. doi: 10.1002/bies.20463. [DOI] [PubMed] [Google Scholar]
- 8.Shen YY, Shi P, Sun YB, Zhang YP. Relaxation of selective constraints on avian mi-tochondrial DNA following the degeneration of flight ability. Genome Res. 2009;19:1760–1765. doi: 10.1101/gr.093138.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bayona-Bafaluy MP, Müller S, Moraes CT. Fast adaptive coevolution of nuclear and mitochondrial subunits of ATP synthetase in orangutan. Mol Biol Evol. 2005;22:716–724. doi: 10.1093/molbev/msi059. [DOI] [PubMed] [Google Scholar]
- 10.Rand DM, Haney RA, Fry AJ. Cytonuclear coevolution: The genomics of cooperation. Trends Ecol Evol. 2004;19:645–653. doi: 10.1016/j.tree.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 11.van den Heuvel L, Smeitink J. The oxidative phosphorylation (OXPHOS) system: Nuclear genes and human genetic diseases. Bioessays. 2001;23:518–525. doi: 10.1002/bies.1071. [DOI] [PubMed] [Google Scholar]
- 12.Edmands S, Burton RS. Cytochrome c oxidase activity in interpopulation hybrids of a marine copepod: A test for nuclear-nuclear or nuclear-cytoplasmic coadaptation. Evolution. 1999;53:1972–1978. doi: 10.1111/j.1558-5646.1999.tb04578.x. [DOI] [PubMed] [Google Scholar]
- 13.Ellison CK, Burton RS. Disruption of mitochondrial function in interpopulation hybrids of Tigriopus californicus. Evolution. 2006;60:1382–1391. [PubMed] [Google Scholar]
- 14.Murphy WJ, et al. Molecular phylogenetics and the origins of placental mammals. Nature. 2001;409:614–618. doi: 10.1038/35054550. [DOI] [PubMed] [Google Scholar]
- 15.Waddell PJ, Kishino H, Ota R. A phylogenetic foundation for comparative mammalian genomics. Genome Inform. 2001;12:141–154. [PubMed] [Google Scholar]
- 16.Corneli PS. Complete mitochondrial genomes and eutherian evolution. J Mammal Evol. 2002;9:281–305. [Google Scholar]
- 17.Nikaido M, Cao Y, Harada M, Okada N, Hasegawa M. Mitochondrial phylogeny of hedgehogs and monophyly of Eulipotyphla. Mol Phylogenet Evol. 2003;28:276–284. doi: 10.1016/s1055-7903(03)00120-9. [DOI] [PubMed] [Google Scholar]
- 18.Springer MS, et al. The adequacy of morphology for reconstructing the early history of placental mammals. Syst Biol. 2007;56:673–684. doi: 10.1080/10635150701491149. [DOI] [PubMed] [Google Scholar]
- 19.Prasad AB, Allard MW, NISC Comparative Sequencing Program, Green ED Confirming the phylogeny of mammals by use of large comparative sequence data sets. Mol Biol Evol. 2008;25:1795–1808. doi: 10.1093/molbev/msn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teeling EC, et al. Molecular evidence regarding the origin of echolocation and flight in bats. Nature. 2000;403:188–192. doi: 10.1038/35003188. [DOI] [PubMed] [Google Scholar]
- 21.Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 22.Vermulst M, et al. DNA deletions and clonal mutations drive premature aging in mitochondrial mutator mice. Nat Genet. 2008;40:392–394. doi: 10.1038/ng.95. [DOI] [PubMed] [Google Scholar]
- 23.Stewart JB, et al. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 2008;6:e10. doi: 10.1371/journal.pbio.0060010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart JB, Freyer C, Elson JL, Larsson NG. Purifying selection of mtDNA and its implications for understanding evolution and mitochondrial disease. Nat Rev Genet. 2008;9:657–662. doi: 10.1038/nrg2396. [DOI] [PubMed] [Google Scholar]
- 25.Popadin K, Polishchuk LV, Mamirova L, Knorre D, Gunbin K. Accumulation of slightly deleterious mutations in mitochondrial protein-coding genes of large versus small mammals. Proc Natl Acad Sci USA. 2007;104:13390–13395. doi: 10.1073/pnas.0701256104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mamirova L, Popadin K, Gelfand MS. Purifying selection in mitochondria, free-living and obligate intracellular proteobacteria. BMC Evol Biol. 2007;7:17–28. doi: 10.1186/1471-2148-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 28.Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303:223–226. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- 29.Grossman LI, Wildman DE, Schmidt TR, Goodman M. Accelerated evolution of the electron transport chain in anthropoid primates. Trends Genet. 2004;20:578–585. doi: 10.1016/j.tig.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Mishmar D, et al. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 33.Pagliarini DJ, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bakewell MA, Shi P, Zhang J. More genes underwent positive selection in chimpanzee evolution than in human evolution. Proc Natl Acad Sci USA. 2007;104:7489–7494. doi: 10.1073/pnas.0701705104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kosiol C, et al. Patterns of positive selection in six Mammalian genomes. PLoS Genet. 2008;4:e1000144. doi: 10.1371/journal.pgen.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider A, et al. Estimates of positive Darwinian selection are Inflated by errors in sequencing, annotation, and alignment. Genome Biol Evol. 2009;2009:114–118. doi: 10.1093/gbe/evp012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettigrew JD. Genomic evolution. Flying DNA. Curr Biol. 1994;4:277–280. doi: 10.1016/s0960-9822(00)00065-8. [DOI] [PubMed] [Google Scholar]
- 38.Van Den Bussche RA, Baker RJ, Huelsenbeck JP, Hillis DM. Base compositional bias and phylogenetic analyses: A test of the “flying DNA” hypothesis. Mol Phylogenet Evol. 1998;10:408–416. doi: 10.1006/mpev.1998.0531. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt TR, et al. Rapid electrostatic evolution at the binding site for cytochrome c on cytochrome c oxidase in anthropoid primates. Proc Natl Acad Sci USA. 2005;102:6379–6384. doi: 10.1073/pnas.0409714102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doan JW, et al. Coadaptive evolution in cytochrome c oxidase: 9 of 13 subunits show accelerated rates of nonsynonymous substitution in anthropoid primates. Mol Phylogenet Evol. 2004;33:944–950. doi: 10.1016/j.ympev.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 41.Goldberg A, et al. Adaptive evolution of cytochrome c oxidase subunit VIII in anthropoid primates. Proc Natl Acad Sci USA. 2003;100:5873–5878. doi: 10.1073/pnas.0931463100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodman M, et al. Phylogenomic analyses reveal convergent patterns of adaptive evolution in elephant and human ancestries. Proc Natl Acad Sci USA. 2009;106:20824–20829. doi: 10.1073/pnas.0911239106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nozawa M, Suzuki Y, Nei M. Reliabilities of identifying positive selection by the branch-site and the site-prediction methods. Proc Natl Acad Sci USA. 2009;106:6700–6705. doi: 10.1073/pnas.0901855106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Z, Nielsen R, Goldman N. In defense of statistical methods for detecting positive selection. Proc Natl Acad Sci USA. 2009;106:E95. doi: 10.1073/pnas.0904550106. author reply E96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong WSW, Yang Z, Goldman N, Nielsen R. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics. 2004;168:1041–1051. doi: 10.1534/genetics.104.031153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pasquis P, Lacaisse A, Dejours P. Maximal oxygen uptake in four species of small mammals. Respir Physiol. 1970;9:298–309. doi: 10.1016/0034-5687(70)90078-2. [DOI] [PubMed] [Google Scholar]
- 47.Cotter D, Guda P, Fahy E, Subramaniam S. MitoProteome: Mitochondrial protein sequence database and annotation system. Nucleic Acids Res. 2004;32(Database issue):D463–D467. doi: 10.1093/nar/gkh048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brandon MC, et al. MITOMAP: A human mitochondrial genome database—2004 update. Nucleic Acids Res. 2005;33(Database issue):D611–D613. doi: 10.1093/nar/gki079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vilella AJ, et al. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swofford DL. PAUP* 4.06. 10: Phylogenetic Analysis Using Parsimony. Sunderland, MA: Sinauer Associates; 2003. [Google Scholar]
- 51.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 52.Murphy WJ, et al. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- 53.Pride DT. SWAAP Version 1.0. 0—sliding windows alignment analysis program: A tool for analyzing patterns of substitutions and similarity in multiple alignments. 2000.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.