Abstract
p53 maintains genome integrity either by regulating the transcription of genes involved in cell cycle, apoptosis, and DNA repair or by interacting with partner proteins. Here we provide evidence for a direct physical interaction between the tumor suppressors p53 and BRCA2. We found that the transactivation domain of p53 made specific interactions with the C-terminal oligonucleotide/oligosaccharide-binding-fold domains of BRCA2 (BRCA2CTD). A second distinct site situated on the p53 DNA-binding domain, bound to a region containing BRC repeats of BRCA2 (BRCA2[BRC1-8]) and may contribute synergistically for high affinity association of intact full-length proteins. Overexpression of BRCA2 and BRCA2CTD suppressed the transcriptional activity of p53 with a concomitant reduction in the expression of p53-target genes such as Bax and p21. Consequently, p53-mediated apoptosis was significantly attenuated by BRCA2. The observed physical association of p53 and BRCA2 may have important functional implications in the p53 transactivation-independent suppression of homologous recombination and suggests a possible interregulatory role for both proteins in apoptosis and DNA repair.
Tumor suppressor p53 is the most commonly mutated gene in human cancers. It is regarded as the guardian of the genome because of its multifunctional role in deciding the fate of the cells upon DNA damage (1). p53 maintains genome integrity by regulating the transcription of various target genes that are involved in cell-cycle arrest, apoptosis, or the DNA-repair pathway (1, 2). In response to DNA damage, p53 becomes stabilized, signaling the cells to undergo either cell-cycle arrest or apoptosis and thus preventing the perpetuation of potentially oncogenic mutations. How p53 decides between cell-cycle arrest, apoptosis, or DNA-repair pathway is not fully understood, although p53-mediated G1 arrest and apoptosis have been studied in detail (3–5).
In addition to the classical tumor-suppressor functions in cell-cycle control and apoptosis, p53 exhibits direct regulatory activities in DNA-repair: Chromosomal abnormalities such as gene amplification, translocations, inversions, and deletions are observed in tumor cells lacking wild-type p53 (6, 7). There is compelling evidence for direct involvement of p53 in DNA repair that is independent of its transcriptional activity and checkpoint control (8, 9). In particular, numerous studies have demonstrated the direct role of p53 in the fidelity control of homologous recombination (HR), which in turn may be one mechanism by which p53 maintains genomic integrity and stability (9–11). p53 can modulate HR through direct physical interactions with DNA or DNA-repair proteins. p53 binds to nascent HR intermediates carrying mismatches, three-stranded hetero-duplex joints, and four-stranded Holiday junctions (12). p53 suppresses inappropriate recombination between mispaired DNA sequences (13, 14). p53 physically interacts with and inhibits several key proteins implicated in HR such as RAD51, RecA, and replication protein A (RPA) (15, 16). Further, immunoprecipitation experiments have demonstrated that p53 exists in complexes in vivo containing BRCA2, an essential protein for the efficient repair of double-stranded DNA breaks (17).
BRCA2 regulates the function of the recombinase enzyme RAD51, which plays a crucial role in the DNA-repair pathway (18, 19). Germ-line mutations in BRCA2 are associated with increased susceptibility to breast and ovarian cancers and are responsible for one-third of hereditary breast cancer (20, 21). There is a high incidence of p53 somatic mutations in BRCA-related cancers, and the spectrum of these mutations is different from those found in sporadic cases (22–25). Moreover, recent studies have demonstrated that combined inactivation of BRCA2 and p53 mediates mammary tumorigenesis and that disruption of the p53 pathway is pivotal in BRCA2-associated breast cancer (26). Thus BRCA2 and p53 act synergistically as tumor suppressors, and simultaneous loss of BRCA2 and p53 results in genomic instability and an impaired ability to respond to radiation-induced DNA damage (8, 27).
p53 has a complex domain organization (Fig. 1A) (28). The N-terminal region of the protein, the so-called transactivation domain (TAD) consists of two subdomains (TAD1 and TAD2), both of which exhibit independent transactivation functions and associate with a variety of proteins regulating transcription (29). p53 TAD2 interacts with several DNA-binding domains such as RPA, positive cofactor 4 (PC4), mitochondrial single-stranded DNA-binding protein (mtSSB), and the TfB1 subunit of transcription factor II H (TFIIH), and we and others have demonstrated that it acts as a ssDNA mimetic (30, 31). Two of the aforementioned domains (RPA and mtSSB) adopt oligonucleotide/oligosaccharide-binding (OB) folds and are structurally very similar to the C-terminal DNA-binding domains of BRCA2.
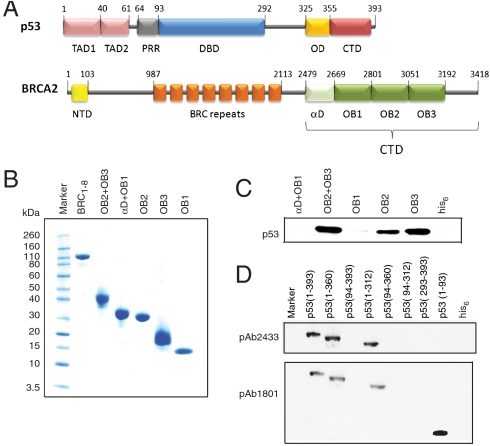
Fig. 1.
(A) Domain organization of p53 and BRCA2. The functional domains of p53 comprise an intrinsically unstructured transactivation domain TAD (1–93), specific DNA-binding core domain DBD (94–292), oligomerization domain (325–355), and C-terminal negative regulatory domain (362–293) (28). BRCA2 is a 3,418 amino acid protein consisting of an N-terminal transactivation domain (23–105), eight central BRC repeats of ∼30 amino acids each (987–2113), and a conserved C-terminal domain BRCA2CTD (2479–3152). The latter comprises an α-helical domain and three OB domains. (B) SDS-PAGE showing the purity of different BRCA2 domains used in pull-down experiments. (C) Interaction of p53 and BRCA2. Nickel pull-down of his-tag purified BRCA2CTD domains with flp53. Immunoblotting by using PAb1801 showed that OB2 + OB3, OB2, and OB3 interact with p53. *D) Nickel pull-down of truncated domains of p53 with his-tagged OB2 + OB3 construct of BRCA2. Immunoblots using PAb2433 (Abcam, specific for 277–296 aa of p53) and PAb1801 (Abcam, specific for 46–55 aa of p53) identified that OB2 + OB3 interacts with the transactivation domain of p53 (p53 TAD, 1–93).
By using biophysical methods, we demonstrated here that the C-terminal domain of BRCA2 (BRCA2CTD) interacted with p53 TAD. Furthermore, we identified a second distinct binding site situated on the p53 DNA-binding domain (DBD) that bound to a 1,127-residue fragment of BRCA2 containing the eight BRC repeats (BRCA2[BRC1-8]). Both the full-length BRCA2 and BRCA2CTD reduced the transcriptional activity of p53. We also observed down-regulation of p53 target genes in H1299 cells with a concomitant reduction in p53-mediated apoptosis when BRCA2 was overexpressed. On the basis of our findings, we discuss the possible interregulatory role of p53 and BRCA2 in DNA-repair pathways and apoptosis.
Results
p53 TAD Interacts with BRCA2CTD.
Because the p53 TAD binds to DNA-binding domains that are structurally similar to BRCA2 DNA-binding domains, we examined whether p53 TAD or other domains of p53 could potentially interact with BRCA2CTD. We expressed and purified several recombinant constructs comprising different regions of BRCACTD (Fig. 1A), referred to as αD + OB1, OB2 + OB3, OB1, OB2, and OB3, to high homogeneity (Fig. 1B). Pull-down experiments using these constructs with full-length p53 (flp53) indicated that p53 interacts with OB2 and OB3 but not with αD + OB1 nor OB1 alone (Fig. 1C). We next examined the domains of p53 that interact with OB2 + OB3. Pull-down experiments were carried out by using constructs comprising distinct domains of p53 having OB2 + OB3 as bait. The interactions were monitored by using two different p53 antibodies: PAb2433 (specific for the DNA-binding domain) and pAb1801 (specific for p53 TAD). The results confirmed that p53 TAD (1–93) strongly interacted with OB2 + OB3, whereas constructs lacking p53 TAD exhibited no binding (Fig. 1D).
p53 TAD Binds to Isolated OB2 and OB3 Domain.
Pull-down assays have high false positive rates, and hence we used biophysical experiments such as fluorescence anisotropy, isothermal titration calorimetry (ITC), and NMR to confirm the binding interactions. Having identified that p53 TAD associates with OB2 + OB3 by pull-down experiments, we carried out fluorescence titrations by using labeled p53 TAD with OB2 + OB3, OB2, and OB3 domains in individual experiments. Labeled p53 TAD was titrated with the OB domains, the change in polarization was measured, and the data were globally fitted to a one-state binding model. OB2 + OB3 bound p53 TAD with a Kd of ∼3 μM (Fig. 2A). Stoichiometric titrations indicated that each OB-fold domain in OB2 + OB3 bound one p53 TAD molecule (Fig. S1). Isolated OB2 and OB3 domains bound p53 TAD with a dissociation constant of ∼5.2 μM and ∼2.5 μM, respectively (Fig. 2A).
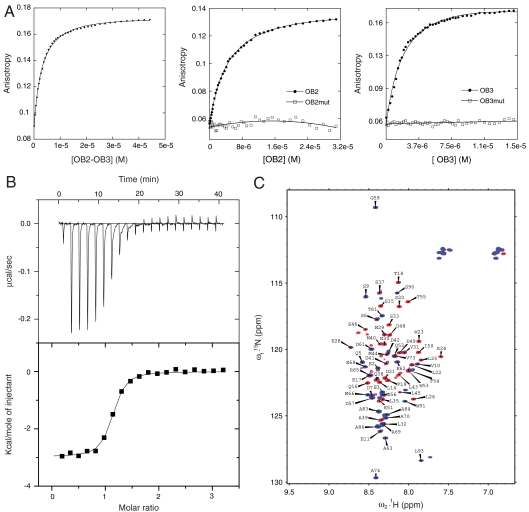
Fig. 2.
Binding of OB2, OB3, and OB2 + OB3 to p53 TAD. (A) Alexa fluor 546-labeled p53 TAD was titrated with OB2 + OB3, OB2, and OB3 in separate experiments. The data were fitted to a simple one-state binding model. (B) Binding isotherm of full-length p53 with OB3 as measured by using ITC in a buffer containing 25 mM Hepes, 150 mM NaCl, 3 mM DTT, pH 7.4 at 293 K. (C) Overlay of 1H15N HSQC of labeled p53 TAD (1–93) in the presence (Blue) and absence (Red) of OB3.
We next carried out ITC experiments to investigate the binding interactions of OB2 + OB3, OB2, and OB3 with flp53. A Kd of 3 μM for OB3 binding to flp53 was obtained by using ITC (Fig. 2B). The binding affinities derived for OB2 and OB2 + OB3 to flp53 were 5.5 and 4.2 μM, respectively (data not shown). The Kd values deduced from ITC were slightly higher than those obtained by using fluorescence titrations on isolated p53 TAD. The higher Kd values obtained for OB2 + OB3 compared with isolated OB3 probably resulted from steric hindrance and reduced accessibility of the binding sites when both the OB domains are fused together.
15N,1H heteronuclear single quantum coherence (HSQC) NMR spectra of labeled p53 TAD (1–93) in the absence and presence of the OB3 domain revealed significant chemical shift perturbations and bleaching of resonances in the TAD2 (35–57) region of p53 (Fig. 2C). In particular, resonances from residues such as E51, W53, and F54 showed chemical shift changes, whereas those for the residues S46, D48, and T55 completely disappeared (Fig. S2). Line broadening and disappearance of resonances might occur because of the exchange between free and complexed p53 TAD-OB3 with low micromolar Kd. HSQC peaks from a few residues in the TAD1 domain such as L22, W23, and K24 also disappeared in the bound complex. NMR experiments confirmed that the TAD2 region of p53 TAD is the primary interaction site of OB3. Experiments carried out by using OB2 gave similar results (data not shown).
Cooperative Binding of OB2 + OB3 to ssDNA.
The structural and DNA-binding properties of the mouse homologue of BRCA2 have been characterized in complex with DNA and DSS1 (Deleted in Split-hand/Split-foot Syndrome) (32). However, no ssDNA-binding affinity data on the isolated OB domains of human BRCA2 are known. We probed, therefore, the ssDNA (dT)15 binding activities of isolated OB domains in BRCA2CTD by using fluorescence titrations. We observed cooperative DNA binding of OB2 + OB3 having a dissociation constant of 40 nM and a Hill constant of ∼2.2 (Fig. S3). Furthermore, OB2 + OB3 bound DNA in a 1∶1 ratio as observed by stoichiometric titration, in contrast to that observed for the binding of p53 TAD to OB2 + OB3 (Fig. S4). Isolated OB2 and OB3 domains bound ssDNA in a 1∶1 ratio with lower affinities of 150 and 110 nM, respectively, and showed no cooperativity (Figs. S5 and S6). OB1 bound ssDNA with a much weaker affinity of ∼10 μM (Fig. S7). Table 1 lists the dissociation constants obtained for the binding of different BRCA2CTD constructs to p53 TAD and ssDNA.
Table 1.
Dissociation constants of BRCA2 OB domains and ssDNA or p53 TAD
| BRCA2 domains | ssDNA (nM) | p53N (μM) | p53 TAD2WT (μM) | p53 TAD2Mut1 (μM) | p53 TAD2Mut2 (μM) |
| OB1 | 10,000 ± 1,000 | n.q.* | n.q.* | n.q.* | n.q.* |
| OB2 | 150 ± 7 | 5.2 ± 0.3 | 6.0 ± 0.5 | 50 ± 4 | n.q.* |
| OB3 | 110 ± 6 | 2.5 ± 0.2 | 3.2 ± 0.4 | 43 ± 3 | n.q.* |
| OB2 + OB3 | 40 ± 5 | 3.0 ± 0.4 | 4.0 ± 0.3 | 39 ± 5 | 120 ± 10 |
ssDNA (dT)15, p53 TAD (1–93), p53TAD2 WT (35–57), p53TAD2 Mut1 (35–57): L43A/L45A/I50A/W53A/F54A and p53TAD2 Mut2 (35–57): D41K/D42K/D48K/D49K/E51K were used. Experiments were carried out in a buffer containing 25 mM Hepes, 150 mM NaCl, 3 mM DTT, pH 7.4 at 20 °C. The dissociation constant was obtained by fitting to a cooperative binding model and gave a Hill constant of ∼2.2.
*n.q.: not quantified (> 250 μM).
p53 TAD and ssDNA Share Similar Binding Site on the BRCA2 OB Domains.
p53 TAD2 interacts with RPA, PC4, Tfb1, and mtSSB (31, 33, 34) and competes with ssDNA for binding to these domains. We hypothesized that p53 TAD2 could similarly bind to the DNA-binding sites of BRCA2 OB2 and OB3 domains. We carried out fluorescence competition titrations to test whether ssDNA competes with p53 TAD2 for binding to OB domains. Coumarin-labeled p53-TAD2 peptide (35–57) was mixed with OB3 and was titrated with ssDNA. The decay in polarization with increasing ssDNA concentration was followed until complete displacement of p53 TAD by ssDNA was reached. A Kd value of 125 nM for ssDNA binding to OB3 was obtained from competition experiments, which is similar to that found above by direct titrations. The results confirmed that binding of DNA and p53 TAD2 to OB3 was mutually exclusive and that p53 TAD2 competes with DNA for binding to the same or a similar site (Fig. 3A). Similar competition between ssDNA and p53 TAD2 was observed in the binding to OB2 (data not shown). Taken together, these findings reinforce the notion that p53 TAD2 acts as a ssDNA mimetic and that this property serves as a general function of this region of p53.
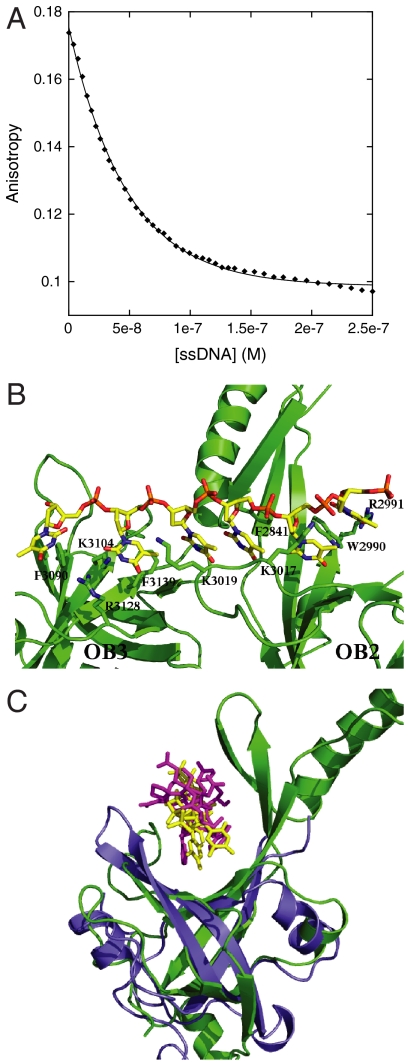
Fig. 3.
Binding of p53 TAD2 to BRCA2CTD. (A) Competition titration of a complex of coumarin-labeled p53 TAD2 (35–57)-OB3 with unlabeled ssDNA. (B) Homology model of human BRCA2 OB2 and OB3 domains with DNA placed in the same position as in the structure of the mouse BRCA2. Residues that can make a contact with DNA and have been mutated in this study are shown as sticks. (C) Superimposition of modeled human BRCA2-OB2 bound to ssDNA with RPA bound to p53 TAD. The color scheme is as follows: BRCA2-OB2 shown in green, RPA in blue, DNA in yellow, and p53 TAD in pink. DNA is placed in the same position as in the structure of the mouse BRCA2.
Sequence Determinants of p53 Involved in TAD2-BRCA2 Recognition.
Several attempts to cocrystallize OB domains with p53 TAD2 did not yield crystals. Because human and mouse BRCA2CTD share 68% sequence identity, we were able to build a homology model by using the crystal structure of the mouse BRCA2CTD, bound to ssDNA as a template [Protein Data Bank (PDB) ID code 1MJE] (32). This model was used to identify residues in human BRCA2 that might be implicated in the binding of DNA and p53 TAD2 (Fig. 3B and Fig. S8). The mode of interaction of BRCA2 with the latter was also deduced from the superimposition of BRCA2 OB domains bound to DNA and RPA bound to p53 TAD2 (Fig. 3C). On the basis of our analysis, we designed mutant OB domains, referred to as OB2mut and OB3mut, containing the following substitutions: F2814A, W2990A, R2991A, K3017A, and K3019A in OB2 and F3090A, K3104A, R3128A, and F3139A in OB3. Circular dichroism measurements showed no significant perturbation in the secondary structures of OB2mut and OB3mut (data not shown). DNA-binding experiments revealed that both OB2mut and OB3mut bound DNA significantly more weakly than did the wild-type domains (Figs. S5 and S6). p53 TAD binding to OB2mut and OB3mut was completely abolished, confirming that its interaction site overlaps with that of DNA (Fig. 1A).
p53 TAD is intrinsically unstructured in its free state. Upon binding to different target proteins, TAD undergoes disorder-to-order conformational change (33). This phenomenon is well documented not only for p53 but also for other acidic activation domains (33). Two major forces have been attributed to this process: electrostatic and hydrophobic interactions. Whereas electrostatics is essential for the mutual attraction of the partner domains during the initial phase of binding, the hydrophobic interactions are probably associated with the folding of the participating activation domain (33). Accordingly, we designed mutations that targeted both hydrophobic and acidic residues of p53 TAD2, some of which underwent significant chemical shift perturbation during the NMR titration experiment described above. We tested the effect of two mutant peptides: p53TAD2 mut1 (35–57) in which hydrophobic residues were replaced by small hydrophobic residues (L43A, L45A, I50A, W53A, and F54A) and p53TAD2 mut2 (35-57) in which the acidic residues were replaced by positively charged residues (D41K, D42K, D48K, D49K, and E51K). The introduction of positive charge in TAD2 is expected to result in repulsion between p53 TAD and BRCA2 OB domains and subsequently to affect the initial phase of binding. The aforementioned mutations do not affect the structure of unbound TAD2 (disordered region) as shown in our previous analysis (33). Table 1 lists the results from the binding experiments. Whereas binding of p53 TAD mut1 to OB2 and OB3 had a > 10-fold decrease, the binding of p53 TAD mut2 to OB2 and OB3 was completely abolished, suggesting that the contribution of both electrostatic and hydrophobic interactions is essential for p53-BRCA2 OB complex formation.
Secondary Binding Site of BRCA2 in the DNA-Binding Domain of p53.
BRCA2 contains eight BRC repeats (BRC1-8) that mediate the interaction of BRCA2 with RAD51. BRC1-8 facilitates the complex formation of RAD51-ssDNA while it slows down RAD51 assembly on dsDNA (35). Point mutations within the region of BRCA2 encoding BRC1-8 that disrupt RAD51 interactions are associated with a predisposition to cancer. More importantly, the BRC repeats are highly conserved, indicating their importance to biological activity. We expressed and purified BRCA2[BRC1-8] (spanning residues 987–2113) as described by Shivji et al. (36) to test whether this region of BRCA2 interacts with p53. Nickel pull-down experiments, as described above, showed that the DNA-binding domain of p53 (p53 DBD) interacts with BRCA2[BRC1-8] (Fig. 4A). 15N,1H HSQC analysis of labeled p53 DBD (94–312) in the absence and presence of BRCA2[BRC1-8] was used to identify the interaction sites on p53 DBD. A clear indication of specific binding was observed in NMR with severe line broadening and bleaching effects, probably because of the large size of the complex. Several residues such as G117, R174, R181, R267, and L265 showed large chemical shift perturbations (Fig. 4B). Importantly, residues such as R248, S241, and R280 that make direct contact with DNA disappeared in the complex. Resonances for the residues G245, R273, and K120 had significant chemical shift perturbations, and these residues are located in the DNA-binding loop of p53 DBD (Fig. S2). Mapping the perturbed residues onto the crystal structure of p53 DBD revealed that the interaction site of BRCA2[BRC1-8] overlaps with the DNA-binding surface of p53 (Fig. 4C). However, this interaction is presumably weak because complete binding saturation could not be obtained because of solubility problems at higher concentrations. However, this interaction is specific because no binding of p53 DBD to the OB domains of BRCA2 was observed. In the context of the full-length BRCA2 and p53, this second binding site may act synergistically, resulting in the high affinity association of the proteins.
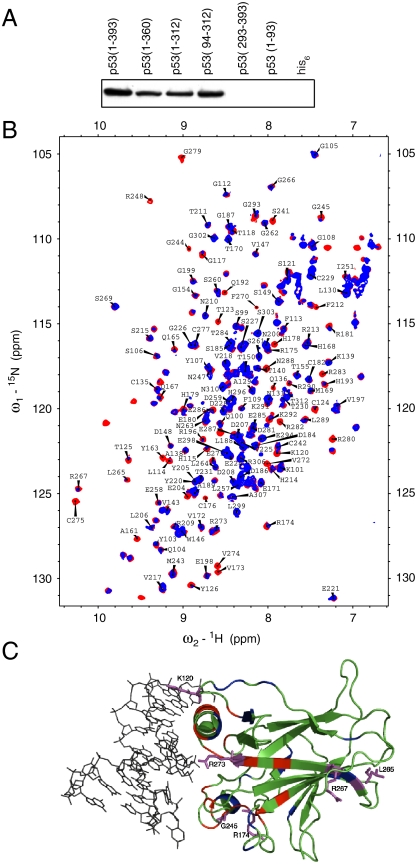
Fig. 4.
Interaction of BRCA2[BRC1-8] with the DNA-binding domain of p53. (A) Nickel pull-down experiment of his6 BRCA2[BRC1-8] with different domains of p53. Immunoblots were developed by using ab2957 (Abcam, specific for 1324–1347 of human BRCA2). (B) Overlay of 1H15N HSQC of labeled p53 DNA-binding domain (94–312) in the presence (Blue) and absence (Red) of BRCA2[BRC1-8]. (C) Chemical shift mapping of the residues on the p53 DNA-binding domain (PDB ID code 2AC0) that are perturbed upon BRCA2[BRC1-8] binding. p53 DBD is shown in green and DNA in black. Residues that showed chemical shift changes and those that disappeared are shown in red and blue, respectively. Selected residues that showed significant chemical shift perturbations are highlighted in magenta.
BRCA2 Down-Regulates the Expression of p53 Target Genes.
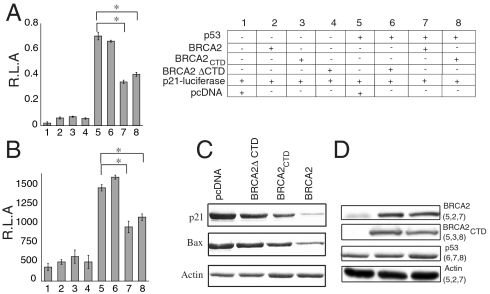
BRCA1, BRCA2, and RAD51 show similar expression profiles, and mouse mutants of each of these genes exhibit embryonic lethality, which is partially rescued in a p53-null background (37, 38). Furthermore, when both BRCA2 and p53 are overexpressed by transient transfection, inhibition of the transcriptional activity of p53 is observed, and coexpression with RAD51 enhances this inhibitory effect (17). In this study, we investigated the effect of BRCA2 on the transactivation function of p53 by limiting the expression levels of p53. In contrast to overexpression, a constant level of p53 was achieved in H1299 cell lines in which p53 can be induced upon removal of tetracycline from the growth medium. We tested the effect of BRCA2 and BRCA2CTD (2281–3418) on the transcriptional activity of p53 by using a luciferase reporter assay. BRCA2CTD contains a nuclear localization signal, which localizes it to the nucleus (39). Exogenous expression of BRCA2 reduced the transcriptional activity of p53 (Fig. 5A). A significant level of inhibition was also observed when BRCA2CTD was transfected (Fig. 5A). No reduction in the transcriptional activity of p53 was observed when transfected with empty vector or BRCA2 ΔCTD (1–2151). This experiment confirmed that the inhibition of p53 transcriptional activity was BRCA2-mediated and furthermore that the CTD region is responsible for this inhibition (Fig. 5A). Expression levels of p53 were unaffected upon BRCA2 transfection. H1299 cells that were transfected with BRCA2, BRCA2CTD, BRCA2 ΔCTD, or empty vectors were assayed for apoptosis after 24 and 48 h posttransfection. A significant percentage of cells underwent apoptosis upon p53 induction by 24 h. After 48 h, a majority of the cells had undergone apoptosis. Attenuation in the apoptotic induction by p53 was observed in the presence of BRCA2 or BRCA2CTD, whereas no effect was observed with BRCA2 ΔCTD or empty vector (Fig. 5B). We also observed a notable percentage of cells restored from p53-mediated G1/S arrest when transfected with BRCA2 (Fig. S9).
Fig. 5.
Transcriptional and apoptosis assays. (A) H1299 cells with inducible p53 expression were transfected with BRCA2, BRCA2CTD, BRCA2 ΔCTD, or pcDNA (+)(empty) vectors as indicated. Expression of p53 was induced by placing the cells in medium containing no tetracycline. Bars 1–4 represent experiments where p53 was not induced (∗P < 0.05). (B) Caspase-3 activity assay monitoring the apoptosis in H1299 cells transfected with BRCA2, BRCA2CTD, BRCA2 ΔCTD, and pcDNA (+)vectors as indicated in A (∗P < 0.05). (C) Expression levels of p53 target genes, p21 and Bax, when transfected with empty vector, BRCA2CTD, BRCA2, and BRCA2 ΔCTD. (D) Western blot showing the overexpression of BRCA2 when tranfected in H1299 cells. p53 levels remained unaffected by BRCA2 overexpression as indicated by Western blot. The numbers represent the cell extracts from the table in A.
We next looked at the levels of the p53 target genes, p21 and Bax, upon transfection of BRCA2 in H1299 cell lines. Reductions in p21 and Bax levels were observed, but there was no significant change upon transfecting with BRCA2 ΔCTD or empty vector (Fig. 5C). These findings are in agreement with previous observations that BRCA2 mutant mouse embryos exhibit growth arrest phenotype that is less severe in a p53-null background and that BRCA2 mutant embryo cells show elevated p21 levels (19, 27, 40).
Discussion
In the present study, we mapped a moderate affinity interaction between the transactivation domain (35–57) of p53 and C-terminal OB domains (OB2 and OB3) of BRCA2. A secondary weak interaction was identified between the p53 DNA-binding domain and BRCA2[BRC1-8], which may act synergistically to form a head-to-tail complex between p53 and BRCA2.
Simultaneous loss of BRCA2 and p53 synergistically drives genomic instability and down-regulation of apoptosis. Strikingly, loss of BRCA2 in T cells leads to accumulation of chromosomal aberrations resulting in increased p53-mediated apoptosis (41). We found that overexpression of BRCA2 and BRCA2CTD suppresses the transcriptional activity of p53 with a concomitant reduction in the expression levels of p53-target genes. Furthermore, a significant inhibition in p53-mediated apoptosis was observed when BRCA2 and BRCA2CTD were overexpressed. The inhibition of p53 transactivation by BRCA2 is probably mediated by the direct physical association between the p53 TAD2 and OB domains of BRCA2. The fact that TAD2 is important for transactivation function of p53 is evident from the reports that just TAD2 alone in flp53(Δ1-39) is found to be sufficient for transcriptional activation (29). Furthermore, several transcriptional coactivators such as PC4 and the p62/Tfb1 subunit of TFIIH interact with the p53 TAD2 domain and regulate its function (33, 34). For example, interaction between p53 TAD2 and the p62/Tfb1 subunit of TFIIH is important for the recruitment of p53 to the TFIIH complex and for stimulation of transcriptional elongation (34). Hence, BRCA2 OB domains probably compete for binding with TFIIH (or other coactivators of basal transcription machinery) to p53 TAD2, thereby suppressing the transcriptional activity of p53.
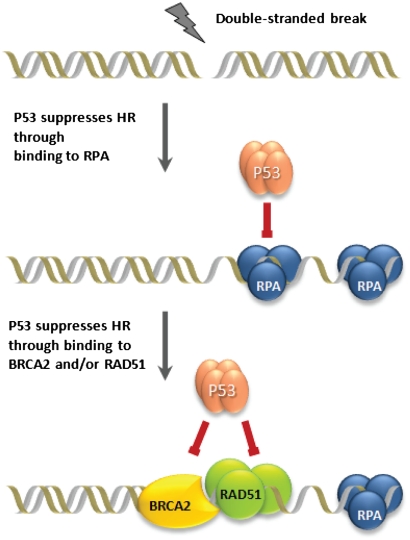
The physical association between p53 and BRCA2 may also have important implications in the control of HR. Precise control of HR is essential to ensure genetic stability because excess HR can be deleterious and induces chromosomal aberrations. The initial steps in HR involve recognition and preprocessing of the DNA lesions to form ssDNA tails. The resulting 3′- ssDNA is then coated with RPA followed by assembly of RAD51 that forms polymers along ssDNA–nucleoprotein filaments. This process is assisted by BRCA2, which binds to DNA and RAD51 and delivers RAD51 to the sites with DNA breaks (42). p53 is known to suppress HR as evidenced by several analyses demonstrating that p53-/- mouse embryonic fibroblasts (MEFs) have higher levels of HR than p53+/+ MEFs and that p53+/- MEFs show an intermediate response (12). p53 binds to each of the factors involved at the early steps of HR such as RAD51, RPA, and BRCA2 (15–17). Furthermore, whereas cells expressing both wild-type and transactivation deficient (L22Q/W23S) p53 show HR suppression, mutations affecting p53 TAD2 (D48H/D49H and W53S/F54S) have no effect on HR, suggesting that this region of p53 plays an important role in HR regulation (16). Most importantly, as we demonstrate here, p53 TAD2 is not only involved in interaction with RPA but also with BRCA2CTD. Because p53 interacts with both BRCA2CTD and BRCA2[BRC1-8], each of which is essential for HR, it is tempting to speculate that p53 might directly interfere in HR through its association with BRCA2. Moreover, BRCA2 exists in complex with RAD51, which is yet another target for p53 binding (15). p53 might associate with BRCA2-RAD51 complex and regulate HR by either disrupting this complex or preventing its binding to DNA. Thus HR modulation by p53 may occur at different stages through its binding to RPA, RAD51, and BRCA2, thereby ensuring the fidelity of DNA repair and maintenance of genome integrity (Fig. 6). While this is speculative at the moment, further investigations would shed more light on the mechanism by which p53 interferes with BRCA2-mediated HR.
Fig. 6.
Role of p53 in HR repression. p53 can suppress the early stage of HR by physical association with HR proteins such as RPA, BRCA2, and RAD51. Both p53-mediated cell-cycle arrest (p53-transactivation-dependent) and repression of HR (p53-transactivation-independent) may prevent cell proliferation under circumstances where DNA damage is left unrepaired.
Methods
Details of protein purification, binding experiments, and cell biology assays are provided in SI Text.
Supplementary Material
Acknowledgments.
We thank Dr. A.R. Venkitaraman for the kind gift of BRCA2 gene. We acknowledge the kind help of C.M. Blair in molecular biology. S.R is supported by a fellowship from Medical Research Council.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003689107/-/DCSupplemental.
References
- 1.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 2.Vousden KH. p53: Death star. Cell. 2000;103:691–694. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 4.Vousden KH, Lu X. Live or let die: The cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 5.Das S, Boswell SA, Aaronson SA, Lee SW. P53 promoter selection: Choosing between life and death. Cell Cycle. 2008;7:154–157. doi: 10.4161/cc.7.2.5236. [DOI] [PubMed] [Google Scholar]
- 6.Yin Y, Tainsky MA, Bischoff FZ, Strong LC, Wahl GM. Wild-type p53 restores cell cycle control and inhibits gene amplification in cells with mutant p53 alleles. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]
- 7.Livingstone LR, et al. Altered cell cycle arrest and gene amplification potential accompany loss of wild-type p53. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 8.Bertrand P, et al. Increase of spontaneous intrachromosomal homologous recombination in mammalian cells expressing a mutant p53 protein. Oncogene. 1997;14:1117–1122. doi: 10.1038/sj.onc.1200931. [DOI] [PubMed] [Google Scholar]
- 9.Mekeel KL, et al. Inactivation of p53 results in high rates of homologous recombination. Oncogene. 1997;14:1847–1857. doi: 10.1038/sj.onc.1201143. [DOI] [PubMed] [Google Scholar]
- 10.Sengupta S, Harris CC. p53: Traffic cop at the crossroads of DNA repair and recombination. Nat Rev Mol Cell Biol. 2005;6:44–55. doi: 10.1038/nrm1546. [DOI] [PubMed] [Google Scholar]
- 11.Gatz SA, Wiesmuller L. p53 in recombination and repair. Cell Death Differ. 2006;13:1003–1016. doi: 10.1038/sj.cdd.4401903. [DOI] [PubMed] [Google Scholar]
- 12.Bertrand P, Saintigny Y, Lopez BS. p53’s double life: Transactivation-independent repression of homologous recombination. Trends Genet. 2004;20:235–243. doi: 10.1016/j.tig.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 13.Gebow D, Miselis N, Liber HL. Homologous and nonhomologous recombination resulting in deletion: Effects of p53 status, microhomology, and repetitive DNA length and orientation. Mol Cell Biol. 2000;20:4028–4035. doi: 10.1128/mcb.20.11.4028-4035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akyuz N, et al. DNA substrate dependence of p53-mediated regulation of double-strand break repair. Mol Cell Biol. 2002;22:6306–6317. doi: 10.1128/MCB.22.17.6306-6317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchhop S, et al. Interaction of p53 with the human Rad51 protein. Nucleic Acids Res. 1997;25:3868–3874. doi: 10.1093/nar/25.19.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romanova LY, Willers H, Blagosklonny MV, Powell SN. The interaction of p53 with replication protein A mediates suppression of homologous recombination. Oncogene. 2004;23:9025–9033. doi: 10.1038/sj.onc.1207982. [DOI] [PubMed] [Google Scholar]
- 17.Marmorstein LY, Ouchi T, Aaronson SA. The BRCA2 gene product functionally interacts with p53 and RAD51. Proc Natl Acad Sci USA. 1998;95:13869–13874. doi: 10.1073/pnas.95.23.13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorslund T, West SC. BRCA2: A universal recombinase regulator. Oncogene. 2007;26:7720–7730. doi: 10.1038/sj.onc.1210870. [DOI] [PubMed] [Google Scholar]
- 19.Patel KJ, et al. Involvement of Brca2 in DNA repair. Mol Cell. 1998;1:347–357. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 20.Martin AM, et al. Germline mutations in BRCA1 and BRCA2 in breast-ovarian families from a breast cancer risk evaluation clinic. J Clin Oncol. 2001;19:2247–2253. doi: 10.1200/JCO.2001.19.8.2247. [DOI] [PubMed] [Google Scholar]
- 21.The Breast Cancer Linkage Consortium. Cancer risks in BRCA2 mutation carriers. J Natl Cancer Inst. 1999;91:1310–1316. doi: 10.1093/jnci/91.15.1310. [DOI] [PubMed] [Google Scholar]
- 22.Smith PD, et al. Novel p53 mutants selected in BRCA-associated tumours which dissociate transformation suppression from other wild-type p53 functions. Oncogene. 1999;18:2451–2459. doi: 10.1038/sj.onc.1202565. [DOI] [PubMed] [Google Scholar]
- 23.Greenblatt MS, Chappuis PO, Bond JP, Hamel N, Foulkes WD. TP53 mutations in breast cancer associated with BRCA1 or BRCA2 germ-line mutations: Distinctive spectrum and structural distribution. Cancer Res. 2001;61:4092–4097. [PubMed] [Google Scholar]
- 24.Gretarsdottir S, et al. BRCA2 and p53 mutations in primary breast cancer in relation to genetic instability. Cancer Res. 1998;58:859–862. [PubMed] [Google Scholar]
- 25.Ramus SJ, et al. Increased frequency of TP53 mutations in BRCA1 and BRCA2 ovarian tumours. Genes Chromosomes Cancer. 1999;25:91–96. doi: 10.1002/(sici)1098-2264(199906)25:2<91::aid-gcc3>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 26.Jonkers J, et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 27.Connor F, et al. Tumorigenesis and a DNA repair defect in mice with a truncating Brca2 mutation. Nat Genet. 1997;17:423–430. doi: 10.1038/ng1297-423. [DOI] [PubMed] [Google Scholar]
- 28.Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53. Annu Rev Biochem. 2008;77:557–582. doi: 10.1146/annurev.biochem.77.060806.091238. [DOI] [PubMed] [Google Scholar]
- 29.Candau R, et al. Two tandem and independent sub-activation domains in the amino terminus of p53 require the adaptor complex for activity. Oncogene. 1997;15:807–816. doi: 10.1038/sj.onc.1201244. [DOI] [PubMed] [Google Scholar]
- 30.Rajagopalan S, Jaulent AM, Wells M, Veprintsev DB, Fersht AR. 14-3-3 activation of DNA binding of p53 by enhancing its association into tetramers. Nucleic Acids Res. 2008;36:5983–5991. doi: 10.1093/nar/gkn598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bochkareva E, et al. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc Natl Acad Sci USA. 2005;102:15412–15417. doi: 10.1073/pnas.0504614102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang H, et al. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 33.Rajagopalan S, Andreeva A, Teufel DP, Freund SM, Fersht AR. Interaction between the transactivation domain of p53 and PC4 exemplifies acidic activation domains as single-stranded DNA mimics. J Biol Chem. 2009;284:21728–21737. doi: 10.1074/jbc.M109.006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Lello P, et al. Structure of the Tfb1/p53 complex: Insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol Cell. 2006;22:731–740. doi: 10.1016/j.molcel.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 35.Shivji MK, et al. The BRC repeats of human BRCA2 differentially regulate RAD51 binding on single- versus double-stranded DNA to stimulate strand exchange. Proc Natl Acad Sci USA. 2009;106:13254–13259. doi: 10.1073/pnas.0906208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shivji MK, et al. A region of human BRCA2 containing multiple BRC repeats promotes RAD51-mediated strand exchange. Nucleic Acids Res. 2006;34:4000–4011. doi: 10.1093/nar/gkl505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharan SK, et al. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 38.Ludwig T, Chapman DL, Papaioannou VE, Efstratiadis A. Targeted mutations of breast cancer susceptibility gene homologs in mice: Lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev. 1997;11:1226–1241. doi: 10.1101/gad.11.10.1226. [DOI] [PubMed] [Google Scholar]
- 39.Schoenfeld AR, Apgar S, Dolios G, Wang R, Aaronson SA. BRCA2 is ubiquitinated in vivo and interacts with USP11, a deubiquitinating enzyme that exhibits prosurvival function in the cellular response to DNA damage. Mol Cell Biol. 2004;24:7444–7455. doi: 10.1128/MCB.24.17.7444-7455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki A, et al. Brca2 is required for embryonic cellular proliferation in the mouse. Genes Dev. 1997;11:1242–1252. doi: 10.1101/gad.11.10.1242. [DOI] [PubMed] [Google Scholar]
- 41.Cheung AM, et al. Loss of Brca2 and p53 synergistically promotes genomic instability and deregulation of T-cell apoptosis. Cancer Res. 2002;62:6194–6204. [PubMed] [Google Scholar]
- 42.Shivji MK, Venkitaraman AR. DNA recombination, chromosomal stability and carcinogenesis: Insights into the role of BRCA2. DNA Repair. 2004;3:835–843. doi: 10.1016/j.dnarep.2004.03.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.