Abstract
Crohn's disease (CD), a major form of human inflammatory bowel disease, is characterized by primary immunodeficiencies. The nuclear receptor peroxisome proliferator-activated receptor gamma (PPARγ) is essential for intestinal homeostasis in response to both dietary- and microbiota-derived signals. Its role in host defense remains unknown, however. We show that PPARγ functions as an antimicrobial factor by maintaining constitutive epithelial expression of a subset of β-defensin in the colon, which includes mDefB10 in mice and DEFB1 in humans. Colonic mucosa of Pparγ mutant animals shows defective killing of several major components of the intestinal microbiota, including Candida albicans, Bacteroides fragilis, Enterococcus faecalis, and Escherichia coli. Neutralization of the colicidal activity using an anti-mDefB10 blocking antibody was effective in a PPARγ-dependent manner. A functional promoter variant that is required for DEFB1 expression confers strong protection against Crohn's colitis and ileocolitis (odds ratio, 0.559; P = 0.018). Consistently, colonic involvement in CD is specifically linked to reduced expression of DEFB1 independent of inflammation. These findings support the development of PPARγ-targeting therapeutic and/or nutritional approaches to prevent colonic inflammation by restoring antimicrobial immunity in CD.
Keywords: β-defensin 1, Crohn's disease, microbiota, nutrition, PPAR-γ
Crohn's disease (CD) and ulcerative colitis (UC) are chronic inflammatory disorders of the gastrointestinal tract that are influenced by both genetic and environmental factors. As many as 1.4 million persons in the United States may suffer from these forms of inflammatory bowel disease (IBD) (1). No curative treatment is available for these lifelong and disabling disorders. Although UC lesions are limited to the colon and the rectum, CD lesions can affect any portion of the gastrointestinal tract. Antimicrobial peptides, including α- and β-defensins, are key effectors of the gastrointestinal innate immune response. Ileal CD is specifically characterized by reduced expression of Paneth cell–derived α-defensins, which is independently linked to the CD-associated NOD2 and TCF7L2 mutations (2, 3). Reduced antimicrobial activity against certain bacterial groups of the intestinal microbiota has been reported in the colonic mucosa of patients with CD compared with patients with UC and controls (4). The mechanisms underlying this phenomenon of reduced colonic antimicrobial immunity in CD remain poorly understood, however.
The nuclear receptor peroxisome proliferator-activated receptor-gamma (PPARγ) is expressed primarily in colonocytes. On recognition of either natural or synthetic ligands, a heterodimer of retinoid X receptor alpha (RXRα) and PPARγ is formed that allows the regulation of a specific set of genes involved in intestinal homeostasis through its binding to PPARγ-response elements (PPREs) (5). Genetic ablation of PPARγ was found to result in increased susceptibility to experimental colitis in rodents (5). Conversely, engagement of PPARγ-mediated signaling by its cognate agonists, such as rosiglitazone, attenuated the severity of inflammatory lesions in both experimental and spontaneous models of colitis (5) and might be effective in UC (6). Consequently, we evaluated the role of PPARγ in host defense through regulation of antimicrobial peptides in the intestinal mucosa of patients with IBD.
Results
PPARγ Directly Regulates DEFB1 Expression in Human Colonocytes.
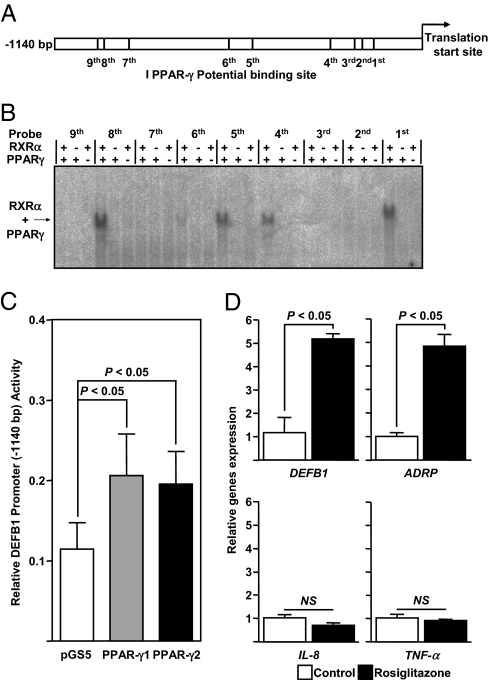
We first searched for potential binding sites of PPARγ within the promoter of human β-defensins expressed primarily in the colon, namely the β-defensin 1–4 (DEFB1-4) (3, 7). In silico approaches were performed within the 5′ vicinity to the starting codon of the DEFB1-4–encoding genes. We failed to detect putative PPRE within the 0.3-kb downstream untranslated region of DEFB2-4. In contrast, the promoter region of DEFB1 had four sites with a potential binding affinity to PPARγ (Fig. 1A). Next, by combining two in silico approaches, we identified five additional putative PPREs within the promoter region of DEFB1 (Fig. 1A). Among the total of nine potential PPREs, three were of DR1 motif and six were of DR2 motif (Table S1). We then systematically and formally assessed the binding activity of RXRα/PPARγ heterodimer to each of these potential PPREs through an electrophoretic mobility shift assay. Four potential sites bound avidly to recombinant RXRα and PPARγ protein (Fig. 1B). Up to a 2-fold increase in luciferase activity of the DEFB1 promoter was consistently observed in Caco-2 cells after cotransfection with a DEFB1 reporter plasmid vector and a pSG5-PPARγ1 or pSG5-PPARγ2 expression construct (Fig. 1C). Treatment of Caco-2 cells with the synthetic thiazolidinedione rosiglitazone caused a significant increase in mRNA levels in both DEFB1 and ADRP, a known PPARγ target gene, compared with mock-stimulated cells. In contrast, IL-8 and TNF-α expression was unaffected by PPARγ activation in vitro (Fig. 1D). Collectively, these in vitro results suggest that certain exogenous and endogenous signals might be involved in the regulation of epithelial expression of DEFB1 through PPARγ activation.
Fig. 1.
PPARγ binds to the human DEFB1 promoter and transactivates DEFB1 expression in human colonic epithelial cells. (A) Potential PPARγ-binding sites within the DEFB1 promoter. (B) Systematic EMSA analysis of the heterodimer RXRα/PPARγ within the DEFB1 gene promoter. Complexes of recombinant proteins with radiolabeled oligonucleotide are indicated by arrow. (C) Relative luciferase activity for the empty control vector (white bar), PPARγ-1 (gray bar), and PPARγ-2 (black bar) isoforms expressing vectors (23). P values were determined by the unpaired Student t test. (D) Relative expression of DEFB1, ADRP, IL-8, and TNF-α in rosiglitazone-treated Caco-2 cells (100 nM) compared with mock-treated cells. Values represent the mean of normalized data ± SEM, as measured by real-time qPCR. P values were determined by the Mann-Whitney test. NS, not significant. All experiments were performed in triplicate and repeated independently at least three times.
PPARγ Is Essential for Colonic Expression of a Subset of β-Defensins in Mice.
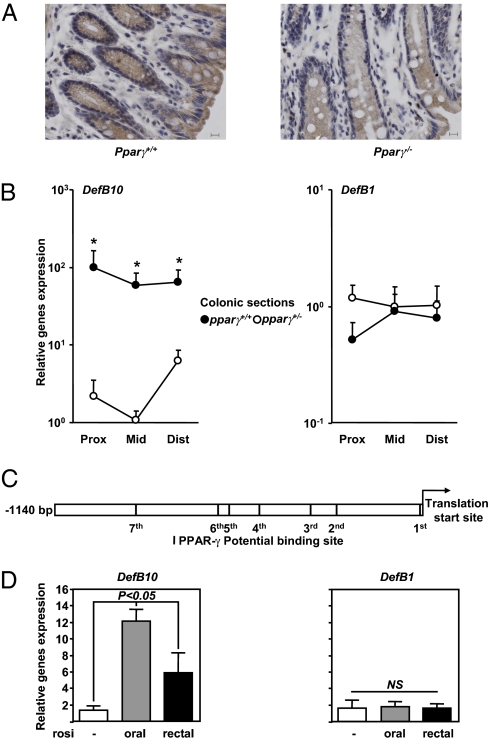
To further assess the regulatory role of PPARγ in β-defensin expression in vivo, we used two mice models of PPARγ deficiency. Among the 21 mouse β-defensins encoded in the Mus musculus C57BL6/J genome reference assembly (build 37.1), the colonic mucosa of both Pparγ+/− and Pparγ−/− mice showed significantly reduced expression of certain β-defensins, including mDefB10 (Fig. 2 A and B), compared with WT littermates. The in silico analyses consistently revealed seven putative PPREs within the promoter region of mDefB10 (Fig. 2C and Table S2). The mDefB10 gene expression pattern remained broadly unchanged along the colon (Fig. 2B). In contrast, we found that the colonic expression of other β-defensins, including mDefB1, was regulated independently of PPARγ (Fig. 2B). Rosiglitazone given orally or rectally for 14 days induced a 6- to 12-fold induction of mDefB10 mRNA expression throughout the colon compared with untreated mice, whereas mDefB1 mRNA expression was unaffected by rosiglitazone treatment (Fig. 2D). Taken together, these findings indicate that PPARγ activation by both exogenous and endogenous signals may be required to maintain the constitutive physiological expression of certain β-defensins in the colon.
Fig. 2.
PPARγ activation is required for expression of mDefB10 in the colon. (A) Immunohistochemical localization of mDefB10 in Pparγ+/+ (Left) and Pparγ−/− (Right) mice. (Scale bar: 10 μm.) (B) To avoid estrus variation, the proximal colon of 8-week-old Pparγ mutant males and their WT littermates (n = 4) were dissected out, flushed with cold PBS, and processed for expression analysis. (C) Potential PPARγ-binding sites within the mDefB10 promoter. (D) Relative gene expression in 8-week-old WT C57BL6/J female mice treated orally or rectally with rosiglitazone at a single or double dose of 10 mg/kg for 14 days compared with untreated mice (n = 4). Values represent the mean of normalized data ± SEM, as measured by real-time qPCR. P values were determined by the Mann-Whitney test. NS, not significant.
PPARγ Deficiency Impairs Innate Antimicrobial Immunity in the Mouse Colon.
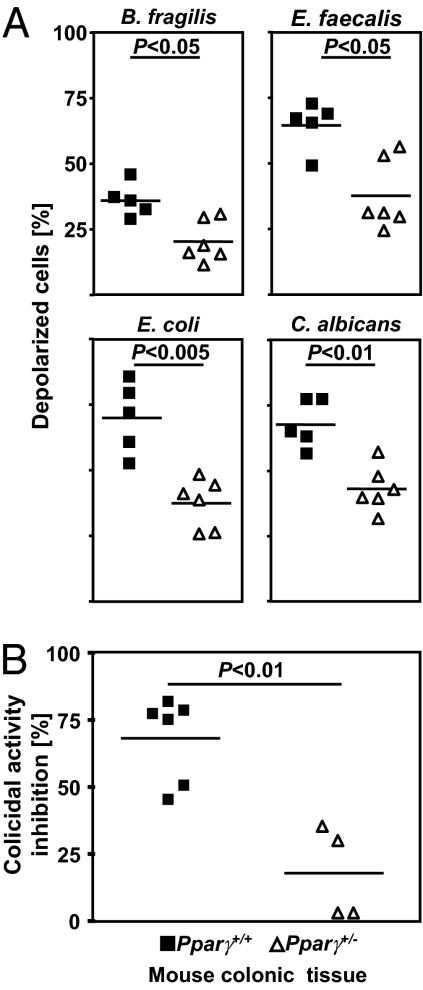
Human colon coexists in intimate contact with up to 100 trillion microorganisms (8). Imbalance in the gut microbiome has been implicated in the pathogenesis of CD. PPARγ is thought to be involved in the innate immune response to microbial infection, but the mechanisms remain poorly understood (9, 10). Thus, we investigated the antimicrobial activity of PPARγ against microbes linked to IBD pathogenesis (8). Interestingly, cationic peptides extracted from colonic mucosa of Pparγ+/− mice exhibited defective killing of cultured Bacteroides fragilis, Enterococcus faecalis, and Candida albicans compared with WT animals (Fig. 3A). The viability of a clinical isolate of Escherichia coli also was significantly decreased after a 90-min exposure to colonic biopsy extracts of controls compared with Pparγ mutant animals (Fig. 3A). We then evaluated the regulatory role of PPARγ on the mDefB10-mediated mucosal antibacterial activity. Most of the colicidal activity of colonic cationic extracts isolated from WT mice was blocked using an anti-mDefB10 antibody (Fig. 3B). Conversely, the blocking activity of anti-mDefb10 on colonic biopsy extracts isolated from Pparγ+/− mice was decreased significantly (Fig. 3B), providing a link between the mDefB10 expression deficiency and impaired antimicrobial immunity in the colon of Pparγ mutant mice.
Fig. 3.
PPARγ activation is required for microbicidal activity in the colon. (A) Cationic proteins were extracted from colonic mucosal biopsy specimens from Pparγ+/− mice (n = 6) and controls (n = 5). Values represent the mean and normalized antimicrobial activity of each sample against B. fragilis (ATCC 25285), E. faecalis (clinical isolate 404), E. coli (clinical isolate 304446), and C. albicans (clinical isolate 526). (B) Anti-mDefB10 antibody was used to inhibit the antimicrobial activity of the colon of Pparγ+/− mice (n = 4) and their WT littermates (n = 6) against E. coli (clinical isolate 304446). All experiments were performed in duplicate, and P values were determined by the Mann-Whitney test. NS, not significant.
PPARγ Is Dispensable for Innate Antimicrobial Immunity in the Mouse Ileum.
Given our previous results and the fact that PPARβ regulates Paneth cell differentiation (10), we next explored the hypothesis that reduced expression of PPARγ might be linked to Crohn's ileitis by failing to regulate antimicrobial immunity in the ileum. In contrast to the expression of DefB10 in the colon, the expression of Paneth cell–derived antimicrobial peptides remained unaffected in Pparγ+/− mice (Fig. S1). Moreover, no significant difference in small intestine antimicrobial activity was seen between the WT and Pparγ mutant mice (Fig. S2). Similarly, the diversity of the fecal-associated microbiota was similar in Pparγ+/− and control littermates (Fig. S3), as determined by real-time quantitative PCR (qPCR) on bacterial 16S rDNA of the major bacterial phyla of the fecal flora (9). Collectively, these results demonstrate that PPARγ is essential for maintaining optimal expression of a subset of β-defensins in the mouse colon, providing a possible mechanism for the impaired microbial killing of the colon in Pparγ-deficient mice and mucosal adherence of certain microorganisms in CD (11).
DEFB1 Expression Is Reduced in CD with Colonic Involvement.
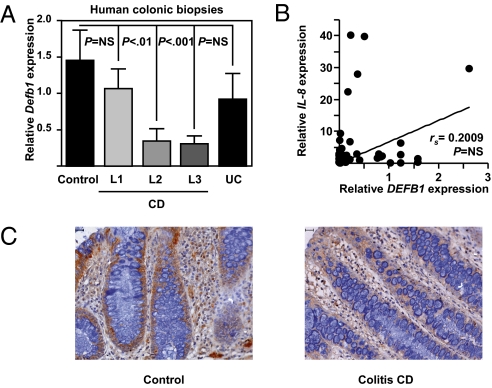
We next measured the mucosal level of DEFB1 in patients with CD, patients with UC, and controls. DEFB1 expression was specifically reduced at the mRNA level in the macroscopically and histologically noninflamed colonic mucosa of patients with colonic involvement (L2 and L3) (P < 0.001 by one-way ANOVA) (Fig. 4A), resulting in decreased protein expression of DEFB1 (Fig. 4C). The colonic expression of DEFB1 in colonic biopsy specimens from patients with pure ileal CD (L1) or UC did not differ significantly from that in controls (Fig. 4A). No correlation between DEFB1 and IL-8 transcript levels was observed in colonic biopsy specimens from control and CD subjects (Fig. 4B), suggesting that impaired DEFB1 expression in colonic CD is not linked to inflammation-associated tissue damage. In contrast, and as reported previously (3), constitutive DEFB1 expression was unchanged in noninflamed colonic mucosa of UC patients expressing low transcript levels of PPARγ (5). These results indicate PPARγ-independent regulatory mechanisms of colonic DEFB1 expression in UC that remain to be identified.
Fig. 4.
Reduced expression of DEFB1 is specifically linked to colonic involvement in CD. (A) Relative gene expression in macroscopically noninflamed colonic biopsy specimens from ileal (n = 21), colonic (n = 16), and ileocolonic (n = 21) CD patients compared with UC patients (n = 8) and controls (n = 17) who underwent colonoscopy. Values represent the mean of normalized data ± SEM, as measured by real-time qPCR. NS, not significant. P values were determined by the nonparametric Kruskall-Wallis and Mann-Whitney tests. (B) Correlation between DEFB1 and IL-8 transcript levels in CD. P values were determined by the nonparametric Spearman test. (C) Immunohistochemical localization of DEFB1 in control (Left) and Crohn's colitis patients (Right). (Scale bar: 10 μm.)
Colonic Involvement in CD Is Associated with a Functional Variant of DEFB1 Promoter.
To test the hypothesis that the reduced DEFB1 expression in L2 and L3 patients may be related to the CD-associated variants within the promoter of DEFB1 (12), we next performed a genotype-phenotype analysis in CD. No significant differences were observed between the allele frequencies in the Hungarian cohort (13) and our French cohort of CD patients (Table 1). Consistently, we confirmed that the genetic promoter variation in DEFB1, namely rs1800972, was solely associated with colonic involvement in CD (13) (Table 1). Notably, the rs1800972 G allele had a significantly lower frequency in patients with pure colonic disease (L2) compared with L1 patients [odds ratio (OR), 0.524; 95% confidence interval (CI), 0.286–0.961; P = 0.035], as well as in patients with colonic involvement (L2 + L3) (OR, 0.559; 95% CI, 0.344–0.909; P = 0.018). Unlike rs1800972, the SNP rs11362 G allele was found with a lower frequency in patients with colonic involvement, but this effect was not statistically significant (OR, 0.710; 95% CI, 0.473–1.064; P = 0.096).
Table 1.
Genotype and allele frequencies of DEFB1 rs1800972 and rs11362 in CD
| SNP | Crohn´s disease, n (%) | Ileal (L1), n (%) | Colonic (L2), n (%) | Ileocolonic (L3), n (%) | ||
| rs1800972 | L1 vs. L2 | L1 vs. L2 + L3 | ||||
| CC | 170 (68.27) | 40 (58.82) | 53 (72.60) | 77 (71.30) | ||
| CG | 70 (28.11) | 23 (33.82) | 19 (26.03) | 28 (25.93) | ||
| GG | 9 (3.61) | 5 (7.35) | 1 (1.37) | 3 (2.78) | ||
| C | 410 (82.33) | 103 (75.74) | 125 (85.62) | 182 (84.26) | C vs. G | C vs. G |
| G | 88 (17.67) | 33 (24.26) | 21 (14.38) | 34 (15.74) | P = 0.035; OR = 0.524 | P = 0.018; OR = 0.559 |
| rs11362 | ||||||
| GG | 82 (33.06) | 27 (40.30) | 24 (32.88) | 31 (28.70) | ||
| GA | 113 (45.56) | 29 (43.28) | 34 (46.58) | 50 (46.30) | ||
| AA | 53 (21.37) | 11 (16.42) | 15 (20.55) | 27 (25.00) | ||
| G | 277 (55.85) | 83 (61.94) | 82 (56.16) | 112 (51.85) | G vs. A | G vs. A |
| A | 219 (44.15) | 51 (38.06) | 64 (43.84) | 104 (48.15) | P = 0.326; OR = 1.270 | P = 0.096; OR = 1.409 |
Discussion
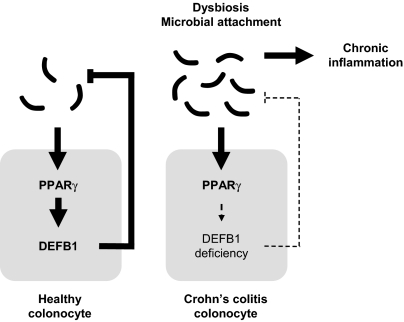
Taken together, our findings suggest a key role of PPARγ in the maintenance of DEFB1 expression, thus contributing to the bactericidal and candidacidal activity of the colonic mucosa (4). In CD with colonic involvement, constitutive deficiency of DEFB1 expression might contribute to diminished microbial killing by the colonic mucosa that subsequently results in increased mucosal adherence of certain microorganisms (11), excessive inflammation, and enhanced antibody response to microbial antigens in CD (14) (Fig. 5). Finally, in line with previous in vitro findings (15–17), DEFB1 expression was inversely correlated with the carriage of C. albicans (18) and the humoral response to mannan, a major epitope for anti-Saccharomyces cerevisiae antibody (ASCA) production (17). C. albicans colonization was significantly increased in CD patients and was identified as an immunogen for ASCA (19), a serologic marker associated mainly with colonic involvement in CD (23). In line with the findings of a recent study (20), we provide a mechanism whereby the rs1800972 G allele might be linked to transactivation of DEFB1 expression through PPARγ. It also might account for the inefficacity of PPARγ-based therapy, such as 5-aminosalicylates, in the colon of CD patients with colonic involvement compared with UC patients. Whether a maintained DEFB1 expression level might be necessary to account for the protective effect of PPARγ on the development of colorectal cancer will require additional investigation (21, 22). In summary, we believe that restoring PPARγ-dependent antimicrobial barrier function might prevent and/or cure inflammatory lesions in the colon of patients with CD.
Fig. 5.
Model of PPARγ-mediated antimicrobial immunity in the colon.
Materials and Methods
Patients.
Through colonoscopy, human colonic biopsy specimens were obtained from macroscopically noninflamed colonic mucosa of healthy individuals (controls, n = 17), CD patients with pure ileal disease (L1 according to the Montreal classification; n = 21) (Table S4), CD patients with solely colonic disease (L2; n = 16) (Table S5), CD patients with ileocolitis (L3; n = 21) (Table S6), and UC patients (n = 8). The diagnoses of CD and UC were based on standard criteria using clinical, radiologic, endoscopic, and histopathologic findings. All clinical investigations were conducted according to the principles expressed in the Declaration of Helsinki.
Animals.
Animal experiments were performed in accredited establishments (B59-108 and B67-218-5) according to European guidelines 86/609/CEE. Age- and sex-matched animals were housed five per cages and had free access to a standard laboratory chow diet in a temperature-controlled specific pathogen-free environment and a half-day light cycle exposure. To avoid estrus variation, 8-week-old PPARγ mutant males and their WT littermates were bred in a specific pathogen-free environment as reported previously (23). The Pparγ-null mouse strain was provided by Prof. Béatrice Desvergne. The proximal colon and terminal ileum were dissected out, flushed with cold PBS, and kept frozen in liquid nitrogen until further gene expression analysis. Rosiglitazone was administered orally or rectally to 8-week-old C57BL/6J mice at a dose of 10 mg/kg for 14 days once or twice daily. All animal studies were approved by the local institutional review board.
Plasmids.
The DEFB1 promoter–containing luciferase reporter constructs DEFB1-1140 (started from translation codon ATG) were kindly provided by Dr. John A. Petros (Emory University). The human PPARγ-expressing plasmids pSG5-h-PPARγ-1 and pSG5-h-PPARγ-2 contain the cDNA of the human PPARγ-1 and PPARγ-2 genes, respectively (23). The pCDNA3.1-RXRα was kindly provided by Dr. Oliver Burk (Dr. Margarete Fischer-Bosch-Institute of Clinical Pharmacology and University of Tübingen).
Promoter Analysis.
To screen for potential PPRE within the human DEFB1-4 promoter region, we analyzed the nucleotide sequence using two different software types designed to predict transcription factor binding in silico. The 1,140 bp upstream of the DEFB1 promoter region was screened using MatInspector (http://www.genomatix.de/products/MatInspector/index.html) and NUBIScan (http://www.nubiscan.unibas.ch/).
Electrophoretic Mobility Shift Assay.
Gel mobility shift assays were performed as described previously (24). In brief, human PPARγ and RXRα protein were synthesized using pSG5-h-PPARγ-1, pSG5-h-PPARγ-2, and pCDNA3.1-RXRα according to the protocol of the TNT T7 Quick-Coupled Transcription/Translation System (Promega). Nuclear response elements were prepared by annealing 10 μL each of two complementary oligonucleotide stocks (100 μM) in 180 μL of 25 mM NaCl, 25 mM Tris-HCl (pH 7.5), and 5 mM MgCl2. For radioactive labeling, 2 μL of the annealed oligonucleotides; 5 μL of 10× buffer [500 mM NaCl, 500 mM Tris-Cl (pH 7.5), and 100 mM MgCl2]; 25 μCi of (α-32P)dCTP; 5 μL of 2 mM dATP, dGTP, and dTTP; 2 U of Klenow fragment; and water to a final volume of 50 μL were incubated at 37 °C for 1 h and purified through Sephadex columns (MicroSpin G-25; GE Healthcare). The binding reaction contained 10 mM Hepes (pH 7.8), 60 mM KCl, 0.2% Nonidet P-40, 6% glycerol, 2 mM DTT, 0.25 μg of poly(dI-dC), 2 μL of 10 μM nonspecific oligonucleotides (5′-AGC TTG CGA AAA TTG TCA CTT CCT GTG TAC ACC CA-3′), 50,000 cpm labeled probe, and 2 μL of full-length synthesized PPARγ and/or RXRα in a final volume of 20 μL. Samples were incubated on ice for 20 min after addition of the labeled probe. Protein–DNA complexes were resolved on a pre-electrophoresed 5% polyacrylamide gel in 44.5 mM boric acid and 1 mM EDTA (pH 8.3) at 200 V at 4 °C. Gels were dried and autoradiographed overnight at room temperature and analyzed with a Fuji BAS-1800 II phosphor-storage scanner and AIDA software (Raytest).
Cell Culture and Transient Transfection Assay.
Human intestinal epithelium cell line Caco-2 (German Collection of Microorganisms and Cell Cultures, ACC 169), were grown in Dulbecco's modified Eagle's medium containing 25 mM Hepes and 2 mM glutamine supplemented with 10% FCS, 50 μg of gentamicin/mL, and 5% of nonessential amino acids. Caco-2 cells were stimulated for 24 h with a synthetic activator of the RXRα/PPARγ heterodimer, rosiglitazone, at 100 nM.
Transient transfections were performed using FuGENE 6 (Roche Diagnostics) according to the manufacturer's protocol. In brief, 1 day before transfection, Caco-2 cells were seeded into the 24-well plates (1.0 × 105 cells/well). Twenty-four hous later, Caco-2 cells (80% confluence) cells were cotransfected with 0.2 μg of the indicated reporter plasmids plus 0.2 μg of pSG5-h-PPARγ-1 or pSG5-h-PPARγ-2 and 50 ng of Renilla luciferase expression plasmid as an internal control. Total amounts of plasmids were kept constant by adding the empty DNA vector when necessary. The cells were incubated for 48 h and then washed, lysed, and harvested using 100 μL of passive lysis buffer (Promega) per well. Firefly luciferase and Renilla luciferase activity were analyzed with the Promega Dual-Luciferase reporter assay system using a Berthold luminometer. All experiments were performed in triplicate and repeated independently at least three times by two independent investigators.
Gene Expression Analysis.
For gene expression analyses, colonic biopsy specimens were immediately frozen in liquid nitrogen and stored at −80 °C. Total RNA from cells and colonic specimens was extracted using the Nucleospin II tissue extraction kit (Macherey Nagel) and reverse-transcribed with the High-Capacity cDNA archive kit (Applied Biosystems), according to the manufacturer's instructions. The resulting cDNA (equivalent to 25 ng of total RNA) was amplified using the SYBR Green real-time PCR kit and detected using the Prism 7300 system (Applied Biosystems). Real-time qPCR was performed with forward and reverse primers (Table S3) designed using Primer Express version 1.0 (Applied Biosystems). On completion of the PCR amplification, a DNA melting curve analysis was carried out to confirm the presence of a single amplicon. β-actin was used as an internal reference gene to normalize the transcript levels. Relative mRNA levels (2-ΔΔCt) were determined by comparing (i) the PCR cycle thresholds (Ct) for the gene of interest and Actb (ΔCt) and (ii) ΔCt values for the treated and control groups (ΔΔCt).
Immunohistochemistry.
DEFB1 immunostaining was performed using a rabbit polyclonal antibody as described previously (15). The specific anti-DEFB1 antibody was kindly provided by Dr. T. Ganz (UCLA, Los Angeles, CA). Colonic biopsy specimens were fixed in 4% paraformaldehyde/phosphate-buffered formalin and embedded in paraffin. In brief, sections were first deparaffinized and rehydrated. Human colonic sections were preincubated in 3% H2O2 methanol for 20 min to quench the endogenous peroxidase activity and with a blocking solution containing avidin D and biotin (Blocking Kit SP2001; Vector Laboratories). Then sections were blocked for 15 min with 5% milk and 1% BSA in PBS and exposed for 30 min to the primary rabbit polyclonal antibody directed against DEFB1 (1:300 dilution) at room temperature. Sections were incubated for 30 min at room temperature with goat anti-rabbit IgG (Dako), and then under the same conditions with an avidin–biotinylated peroxidase complex that was prepared at least 30 min before use.
For mDefB10 immunostaining, we generated an immune affinity–purified rabbit polyclonal F(ab′)2 fragment against the mDefB10-derived synthetic peptide Ser-Arg-Phe-Met-Ser-Asn-Cys-His-Pro-Glu-Asn-Leu-Arg. Sections were processed for peroxidase immunostaining using the Dako system following the manufacturer's recommendations. Immunohistochemistry was performed on formalin-fixed, paraffin-embedded tissue sections using the streptavidin-biotin-peroxydase method in a DakoCytomation AutoStainer. Sections were first deparaffinized and rehydrated. Antigen retrieval was performed by incubating the slides in Tris-citrate buffer (pH 6.0) for 20 min at 97 °C (PT Link; DakoCytomation). Endogenous peroxydase activity was blocked by incubation in 3% hydrogen peroxide for 10 min. The newly generated polyclonal rabbit anti-mDefB10 (2 mg/L) was incubated on slides for 12 h at 4 °C. The biotinylated secondary antibody was a polyclonal swine anti-rabbit (DakoCytomation).
Sections were incubated with 3,3′-diaminobenzidine substrate (Dako) for 1 min, after which the reaction was stopped in distilled water and the sections were counterstained with hematoxylin. Withdrawal of the primary antibody and replacement with a nonspecific antibody were used as negative controls.
Flow Cytometric Antimicrobial Assay.
Extraction of cationic proteins from colonic and ileal tissue of Pparγ+/− (n = 6) and Pparγ+/+ (n = 5) mice was performed as described previously (24). C. albicans (clinical isolate 526; Institute of Laboratory Medicine, Klinik am Eichert), E. faecalis (clinical isolate 404), and E. coli (clinical isolate 304446) were grown aerobically at 37 °C, whereas B. fragilis (ATCC 25285) was cultured anaerobically (Anaero Gen; Oxoid). All clinical isolates were kindly provided by the Institute of Laboratory Medicine, Klinik am Eichert. Then cell suspensions in Schaedler broth bouillion (1:6 dilution) were incubated at a concentration of 1.5 × 106 cells/mL with cationic proteins isolated from 10 μg total extract at 37 °C. After 90 min, 1 μg/mL of the membrane potential–sensitive dye bis-(1,3-dibutylbarbituric acid) trimethine oxonol [DiBAC4(3)] (Invitrogen) was added. After 10 min, the suspensions were centrifuged for 10 min at 4,500× g, and then the bacterial or fungal pellets were resuspended in 300 μL of PBS (pH 7.4). For blocking experiments with anti-mDefB10 antibody, cationic extracts of colonic tissue from WT and PPARγ+/− mice (35 μg/mL) were incubated for 1 h at 37 °C with or without anti-mDefB10 antibody resuspended in 0.1 M K phosphate buffer at a concentration of 10 mg/L. Subsequently, E. coli (clinical isolate 304446) was added in a concentration of 1.5 × 106 cells/mL in Schaedler broth bouillon (1:6) with aqua dest. A total of 10,000 events were analyzed with a FACSCalibur flow cytometer and Cell Quest software (BD) for light scattering and green fluorescence. Antimicrobial activity was determined as percentage of depolarized fluorescent cells with respect to the bacterial control incubated with solvent.
Genotyping.
Genotyping for the DEFB1 promoter polymorphisms rs11362 and rs1800972 was performed using TaqMan SNP Genotyping Assays (assay c_11636793_20 [rs11362] and assay c_11636794_10 [rs1800972]) on a Prism 7900 System (Applied Biosystems), according to the supplier's instructions. Initial and postassay analyses were performed using the Sequence Detection System version 2.3 (Applied Biosystems). One-third of the samples were analyzed in duplicates as an internal control, and DNase-free water was used as a nontemplate control.
Statistics.
Data were analyzed using Prism 4.0 (GraphPad Software). The unpaired Student t test was used to test for significant differences between activities of different promoter constructs. Statistical analysis was performed using (i) the Spearman test for nonparametric correlation analysis and (ii) the Mann-Whitney test for normalized gene expression in mice and antimicrobial assays, and (iii) Kruskall-Wallis test for normalized gene expression in humans. Differences were considered significant at P < 0.05. Values represent the mean of normalized data ± SEM.
Supplementary Material
Acknowledgments
We thank C. Winkler and A. Coutts for their excellent technical assistance; T. Ganz and P. Chavatte for kindly providing the anti-DEFB1 antibody and rosiglitazone, respectively; and Professor M-A. Bigard and L. Dubuquoy for helpful discussion. This work was supported by the Association des chefs de service du CHU de Nancy and by grants from the Fondation pour la Recherche Médicale, UCB Pharma, and Sanofi-Aventis. The study also was supported by the Robert Bosch Foundation, the Deutsche Forschungsgemeinschaft Emmy Noether program (WE 436/1-1), and Sonderforschungsbereich 685 (immunotherapy; Project A9).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0905745107/-/DCSupplemental.
References
- 1.Loftus EV., Jr. Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences. Gastroenterology. 2004;126:1504–1517. doi: 10.1053/j.gastro.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 2.Koslowski MJ, et al. Genetic variants of Wnt transcription factor TCF-4 (TCF7L2) putative promoter region are associated with small intestinal Crohn's disease. PLoS One. 2009;4:e4496. doi: 10.1371/journal.pone.0004496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wehkamp J, Koslowski M, Wang G, Stange EF. Barrier dysfunction due to distinct defensin deficiencies in small intestinal and colonic Crohn's disease. Mucosal Immunol. 2008;1(Suppl 1):S67–S74. doi: 10.1038/mi.2008.48. [DOI] [PubMed] [Google Scholar]
- 4.Nuding S, Fellermann K, Wehkamp J, Stange EF. Reduced mucosal antimicrobial activity in Crohn's disease of the colon. Gut. 2007;56:1240–1247. doi: 10.1136/gut.2006.118646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubuquoy L, et al. PPARgamma as a new therapeutic target in inflammatory bowel diseases. Gut. 2006;55:1341–1349. doi: 10.1136/gut.2006.093484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis JD, et al. Rosiglitazone for Ulcerative Colitis Study Group Rosiglitazone for active ulcerative colitis: A randomized placebo-controlled trial. Gastroenterology. 2008;134:688–695. doi: 10.1053/j.gastro.2007.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peyrin-Biroulet L, et al. NODs in defence: From vulnerable antimicrobial peptides to chronic inflammation. Trends Microbiol. 2006;14:432–438. doi: 10.1016/j.tim.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 8.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 9.Bouskra D, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- 10.Varnat F, et al. PPARbeta/delta regulates Paneth cell differentiation via controlling the hedgehog signaling pathway. Gastroenterology. 2006;131:538–553. doi: 10.1053/j.gastro.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Swidsinski A, et al. Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002;122:44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- 12.Lakatos PL, et al. Hungarian IBD Study Group Interaction between seroreactivity to microbial antigens and genetics in Crohn's disease: Is there a role for defensins? Tissue Antigens. 2008;71:552–559. doi: 10.1111/j.1399-0039.2008.01049.x. [DOI] [PubMed] [Google Scholar]
- 13.Kocsis AK, et al. Association of beta-defensin 1 single nucleotide polymorphisms with Crohn's disease. Scand J Gastroenterol. 2008;43:299–307. doi: 10.1080/00365520701682615. [DOI] [PubMed] [Google Scholar]
- 14.Peyrin-Biroulet L, Standaert-Vitse A, Branche J, Chamaillard M. IBD serological panels: Facts and perspectives. Inflamm Bowel Dis. 2007;13:1561–1566. doi: 10.1002/ibd.20226. [DOI] [PubMed] [Google Scholar]
- 15.Valore EV, et al. Human beta-defensin-1: An antimicrobial peptide of urogenital tissues. J Clin Invest. 1998;101:1633–1642. doi: 10.1172/JCI1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi J, et al. Porcine epithelial beta-defensin 1 is expressed in the dorsal tongue at antimicrobial concentrations. Infect Immun. 1999;67:3121–3127. doi: 10.1128/iai.67.6.3121-3127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng Z, et al. Human beta-defensins: Differential activity against candidal species and regulation by Candida albicans. J Dent Res. 2005;84:445–450. doi: 10.1177/154405910508400509. [DOI] [PubMed] [Google Scholar]
- 18.Jurevic RJ, Bai M, Chadwick RB, White TC, Dale BA. Single-nucleotide polymorphisms (SNPs) in human beta-defensin 1: High-throughput SNP assays and association with Candida carriage in type I diabetics and nondiabetic controls. J Clin Microbiol. 2003;41:90–96. doi: 10.1128/JCM.41.1.90-96.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Standaert-Vitse A, et al. Candida albicans is an immunogen for anti–Saccharomyces cerevisiae antibody markers of Crohn's disease. Gastroenterology. 2006;130:1764–1775. doi: 10.1053/j.gastro.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Kalus AA, et al. Association of a genetic polymorphism (-44 C/G SNP) in the human DEFB1 gene with expression and inducibility of multiple beta-defensins in gingival keratinocytes. BMC Oral Health. 2009;9:21. doi: 10.1186/1472-6831-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogino S, et al. Colorectal cancer expression of peroxisome proliferator–activated receptor gamma (PPARG, PPARgamma) is associated with good prognosis. Gastroenterology. 2009;136:1242–1250. doi: 10.1053/j.gastro.2008.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jansson EA, et al. The Wnt/beta-catenin signaling pathway targets PPARgamma activity in colon cancer cells. Proc Natl Acad Sci USA. 2005;102:1460–1465. doi: 10.1073/pnas.0405928102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braissant O, Foufelle F, Scotto C, Dauça M, Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): Tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology. 1996;137:354–366. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- 24.Wehkamp J, et al. The Paneth cell alpha-defensin deficiency of ileal Crohn's disease is linked to Wnt/Tcf-4. J Immunol. 2007;179:3109–3118. doi: 10.4049/jimmunol.179.5.3109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.