Abstract
Patulin is an acetate-derived tetraketide mycotoxin produced by several fungal species, especially Aspergillus, Penicillium and Byssochlamys species. The health risks due to patulin consumption by humans have led many countries to regulate it in human food. Previous studies have shown the involvement of cytochrome P450 monooxygenases in the hydroxylation of two precursors of patulin, m-cresol and m-hydroxybenzylalcohol. In the present study, two cytochrome P450 genes were identified in the genome sequence of Aspergillus clavatus, a patulin-producing species. Both mRNAs were strongly co-expressed during patulin production. CYP619C2, encoded by the first gene, consists of 529 aa, while the second cytochrome, CYP619C3, consists of 524 aa. The coding sequences were used to perform the heterologous expression of functional enzymes in Saccharomyces cerevisiae. The bioconversion assays showed that CYP619C3 catalysed the hydroxylation of m-cresol to yield m-hydroxybenzyl alcohol. CYP619C2 catalysed the hydroxylation of m-hydroxybenzyl alcohol and m-cresol to gentisyl alcohol and 2,5-dihydroxytoluene (toluquinol), respectively. Except for the last compound, all enzyme products are known precursors of patulin. Taken together, these data strongly suggest the involvement of CYP619C2 and CYP619C3 in the biosynthesis of patulin. CYP619C2 and CYP619C3 are located near to two other genes involved in patulin biosynthesis, namely the 6-methylsalicylic acid synthase (6msas) and isoepoxydon dehydrogenase (idh) genes. The current data associated with an analysis of the sequence of A. clavatus suggest the presence of a cluster of 15 genes involved in patulin biosynthesis.
INTRODUCTION
Patulin is a toxic fungal secondary metabolite that induces immunological, neurological and gastrointestinal effects. Assessment of the health risks due to patulin consumption by humans has led many countries to regulate its amounts in food. In Europe, a maximum level has been established of 50 μg kg−1 for apple juice and cider, while the maximum level has been reduced to 10 μg kg−1 for all products intended for infants and young children.
The genus Penicillium is the most prolific genus with respect to the ability to synthesize patulin (Frisvad et al., 2004). In this genus, Penicillium expansum is the species most commonly linked to patulin production and the one most often found on apples. In the past, a considerable number of Aspergillus and Byssochlamys species have also been reported as patulin-producing species (Moake et al., 2005). However, recent studies have limited the number of patulin-producing species, other than species of Penicillium, to Aspergillus clavatus, Aspergillus giganteus, Aspergillus longivesica and Byssochlamys nivea (Varga et al., 2007; Houbraken et al., 2006; Puel et al., 2007).
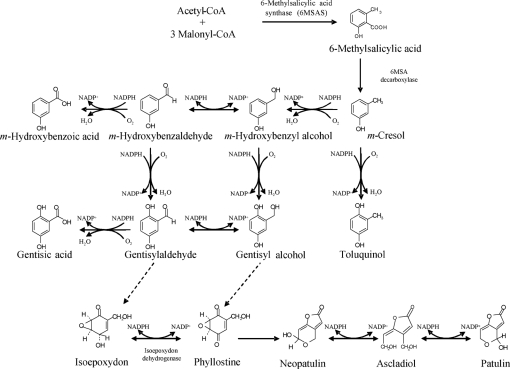
Patulin is a polyketide metabolite, like several other major mycotoxins, e.g. aflatoxins, fumonisins and ochratoxins. The biosynthetic pathway consists of about 10 steps (Fig. 1), as suggested by several biochemical studies and by the identification of several mutants which are blocked at various steps in the patulin biosynthetic pathway (Sekiguchi & Gaucher, 1977). Fungal genes encoding enzymes involved in the production of secondary metabolites are usually contained in clusters on chromosomes. The existence of a gene cluster related to mycotoxin synthesis has been demonstrated for aflatoxins, fumonisins, trichothecenes and ergot alkaloids (Yu et al., 2004; Seo et al., 2001; Proctor et al., 2003; Kimura et al., 2007; Panaccione, 2005). By contrast, a gene cluster related to patulin biosynthesis has not been described. Only a few genes implicated in patulin biosynthesis have been identified, such as 6-methylsalicylic acid synthase (6MSAS) from Penicillium griseofulvum (Beck et al., 1990; Wang et al., 1991) and the isoepoxydon dehydrogenase (IDH) gene from several Penicillium species (Fedeshko, 1992; White et al., 2006; Dombrink-Kurtzman, 2007). Recently, a gene encoding a protein with strong homology to isoamyl-alcohol oxidase has also been suggested to belong to this pathway (Dombrink-Kurtzman, 2008). Although the genes encoding cytochrome P450 monooxygenases have not been isolated, the involvement of cytochrome P450s in several steps of the patulin biosynthesis pathway has been established (Murphy et al., 1974; Murphy & Lynen, 1975).
Fig. 1.
Putative biosynthetic pathways of patulin. Adapted from Fedeshko (1992) and Priest & Light (1989).
Recently, the whole genome of the patulin-producing fungus A. clavatus was sequenced by The Institute for Genomic Research (Fedorova et al., 2008). When compared with genome sequences from A. clavatus, genes orthologous to 6msas and idh were found in the A. clavatus genome. Like all genes involved in secondary metabolite biosynthesis in filamentous fungi, these genes were located in a single cluster which contains 15 genes putatively involved in patulin biosynthesis. Among these genes, two cytochrome P450 genes were located near the 6msas gene. The average number of cytochrome P450 genes in genomes of fungal species is large, and about half of these are located in clusters of genes required for secondary metabolite synthesis.
In the current paper, we report the cloning and heterologous expression of two genes encoding cytochrome P450s and demonstrate their biochemical roles in patulin biosynthesis.
METHODS
Chemicals.
Gentisaldehyde, m-cresol, m-hydroxybenzyl alcohol, m-hydroxybenzaldehyde, toluquinol and patulin were purchased from Sigma-Aldrich. Gentisyl alcohol was kindly provided by Dr M. Blanco (Universidad Politécnica, Valencia, Spain). All solvents used in extraction, HPLC with diode-array detection (HPLC-DAD), and liquid chromatography-MS (LC-MS/MS) were analytical grade. Water for HPLC-DAD and molecular biology procedures was purified by using a Millipore MilliQ purification system. Taq DNA polymerase, SuperScript transcriptase and a GeneRacer kit were purchased from Invitrogen.
Organisms and growth conditions.
Escherichia coli Top10F′ (Invitrogen) was used for cloning experiments. This strain was grown at 37 °C on solid Luria–Bertani (LB) medium or in liquid LB medium. Saccharomyces cerevisiae InvSC1 (Invitrogen) was used for heterologous expression experiments. The patulin-producing A. clavatus strain NRRL 1980 was chosen instead of strain NRRL 1, whose complete genome has been sequenced, because Varga et al. (2003) have shown that strain IMI 015949 (NRRL 1) is not able to produce patulin. This strain was provided by S. Peterson, ARS Culture Collection, Northern Regional Research Laboratory, Peoria, IL, USA. The strain was cultured on Potato Dextrose Agar (PDA) medium (Sigma-Aldrich) at 25 °C for 7 days in the dark. To determine the transcription kinetics and patulin production of CYP619C2 and CYP619C3, 5×105 conidia from a spore suspension from a late culture were used to inoculate, in triplicate, 1 l Roux flasks containing 100 ml sterile Czapek-glucose broth [35 g Czapek medium (Difco), 2 g yeast extract (Sigma-Aldrich), 8 g glucose, 1 l distilled water]. This culture was placed in an incubator in the dark at 25 °C for 1–5 days, without shaking. Then, the supernatants were extracted and analysed by HPLC-DAD, and total RNAs were extracted from harvested mycelia. The fungus was grown on Eagle's minimal essential medium (MEM; Sigma-Aldrich) supplemented with 5 % sterile fetal calf serum, with shaking (180 r.p.m.) for 5 days, for restrictive growth, i.e. the production of only traces of patulin.
cDNA cloning.
The procedures used in this study are standard molecular biology techniques. Total RNA was isolated from a 4-day-old A. clavatus culture as previously described (Puel et al., 2007). The specific primers were designed from preliminary sequence data obtained from The Institute for Genomic Research website at http://www.tigr.org and deposited in GenBank (XM_001273089 and XM_001273090). These primers are listed in Table 1. Total RNA was used as the template to generate first-strand cDNAs in 20 μl reaction mixtures containing 5 μg total RNA, 820 ng GeneRacer Oligo dT primer (Invitrogen), 1× RT buffer, 40 U RNAsin RNase inhibitor (Promega), 0.1 M DTT and 200 U SuperScript reverse transcriptase (Invitrogen). By using the GeneRacer kit as specified by the manufacturer, the 5′ and 3′ untranslated regions (UTRs) of cytochrome P450 genes were amplified by the rapid amplification of cDNA ends (RACE)-PCR method, as described elsewhere (Shen et al., 2004). The obtained products were ligated in the pCR2.1-TOPO vector for amplification in a TOP10F′ host. Nucleotide sequences of both DNA strands were determined via ABI Prism dye (Applied Biosystems) terminator cycle sequencing of templates consisting of maxiprep-purified (Qiagen) plasmids.
Table 1.
Primers used in this study
| Primer* | Sequence (5′–3′) | Annealing temperature |
|---|---|---|
| P450Iext5′1 | GGGTAGCTCAGGTGGCCGATATCCTGAGCGCA | 60 °C |
| P450Iext5′2 | TCAGCAGCACCCCGAGGATGTACGATGGCGG | 60 °C |
| P450IIext5′1 | CCAGTCGGCGATCCGTGACGACGGCC | 60 °C |
| P450IIext5′2 | GCCATTCCCAACTTTGAGCATGTAGAGGCCGCCG | 60 °C |
| P450Iext3′1 | CGTCTGGGGCATGCACCACGACCCCGACC | 60 °C |
| P450Iext3′2 | CGCTGGTCCGAACCGGAGCACTTCCAGCCC | 60 °C |
| P450IIext3′1 | GTGGGACGGCGCATCTGCCCGGGG | 60 °C |
| P450IIext3′2 | GGCGTTCGAGTTCCAGCGCGGGGAGG | 60 °C |
| P450IexpF | AAGATGGATTTCACACAAGTACCGCCATCG | 55 °C |
| P450IexpR | ATCAAACCGGCTAAACACCCCCTGG | 55 °C |
| P450IIexpF | ACCATGGAACCAATGCTGCTCCTGATATTG | 55 °C |
| P450IIexpR | GCCCTCCGTAAACTGCGAAAACACC | 55 °C |
| CYP619C2F3 | ACCTGAGCTACCCAGCGGA | 60 °C |
| CYP619C2R3 | GCCCATTTGGTGAACTGAAAA | 60 °C |
| CYP619C3F2 | TCAGTAACAGTATCACCAACTCAATCGTA | 60 °C |
| CYP619C3R2 | GGCGCCGGTTTCCATAAT | 60 °C |
| TubACF3 | GCCCTACAACGCCACTCTCTC | 60 °C |
| TubACR3 | GCATGCAGATATCGTACAGAGCC | 60 °C |
*P450I (CYP619C2) and P450II (CYP619C3) are equivalent, respectively, to sequences submitted under GenBank accession numbers XM_001273090 and XM_001273089.
Kinetics of CYP619C2 and CYP619C3 expression in A. clavatus.
After assessment of concentration, purity and quality, 5 μg total RNA was used as the template to generate first-strand cDNA in 20 μl reaction mixture, as described above. The amount of RNA transcripts from genes encoding cytochrome P450s was measured in triplicate by real-time quantitative RT-PCR (qRT-PCR). Briefly, 8 ng cDNA was mixed with the Power SYBR Green PCR Master Mix (Applied Biosystems), specific A. clavatus primers for a housekeeping gene (β-tubulin), and genes encoding cytochrome P450s CYP619C2 and CYP619C3. The optimal primers used in the reaction were identified by Primer Express 1.0 software (Applied Biosystems) and are listed in Table 1. Special care was taken to design or choose sets of primers that spanned exon–exon conjunctions to exclude amplification of genomic DNA. The reactions were denatured at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min, in an ABI Prism 7000 Sequence Detection System (Applied Biosystems). A dissociation curve analysis was performed to confirm the specificity of the amplified target nucleic acid sequence. Data were analysed using the method of Peirson et al. (2003), and individual values for cytochrome P450 expression were obtained after normalization to β-tubulin (relative quantification).
Heterologous expression of cytochrome P450 genes in yeasts.
The entire coding sequences of CYP genes were amplified by RT-PCR using, respectively, P450IexpF/R and P450IIexpF/R primer pairs. The cDNA devoid of PCR errors was inserted into a yeast expression vector, pYES2.1/V5/His-Topo (Invitrogen). The expression vectors were purified from overnight culture and were introduced into strain InvSc1 of S. cerevisiae by the lithium acetate procedure.
Preparation of yeast microsomes and Western blotting.
First, yeast were grown at 29 °C in 500 ml YPGE (yeast extract, peptone, glucose, ethanol) for 24 h, as described by Pompon et al. (1996). Cells were induced by addition of 10 % (v/v) of a sterile aqueous solution of 20 g galactose l−1 and grown for 16 h. Cells were harvested by centrifugation, resuspended in TEK buffer (50 mM Tris/HCl, pH 7.4, 1 mM EDTA, 0.1 M KCl) and left at room temperature for 5 min. Cells were recovered by centrifugation and resuspended in 0.5 ml TE buffer (50 mM Tris/HCl, 1 mM EDTA, pH 7.4) containing 0.6 M sorbitol. Cells were then broken by mechanical disruption using glass beads and a BeadBeater apparatus, according to the method described by Chung et al. (2004). Microsomal protein concentration was determined using the method of Schaffner & Weissmann (1973). The proteins (15 μg per lane) were separated by 12 % SDS-PAGE (Laemmli, 1970) and transferred to a nitrocellulose membrane. Detection of overexpressed proteins was performed by Western blotting with mouse monoclonal anti-V5–HRP antibody (Invitrogen) and developed using ECL chemiluminescence technology (Amersham).
Bioconversion assays.
For induction of candidate CYP genes, the transgenic yeasts were inoculated into SC minimal medium broth plus 2 % glucose and incubated for 3 days at 30 °C. Then, the yeast cells were harvested and transferred to SC minimal medium containing 2 % galactose as the sole carbon source. The OD600 was adjusted to 0.4. After 24 h of induction, 1 mM of substrate (m-cresol, m-hydroxybenzyl alcohol, m-hydroxybenzaldehyde, toluquinol) was added to the cultures. After 6 and 24 h, the medium was analysed by HPLC-DAD and LC-MS/MS.
HPLC and MS analysis.
After centrifugation, 100 ml medium was extracted with 70 ml ethyl acetate. The organic phase was evaporated in vacuo at 50 °C on a rotary evaporator. The residue was taken up in 200 μl methanol, and this suspension was filtered through a 0.45 μm pore-size filter before injection (20 μl) into the chromatograph apparatus. Mycotoxin analysis was performed by HPLC-DAD (Kontron Instruments) with a 250 mm×4.6 mm Kromasil 5 μm C18 column (Interchim). For the determination of patulin production, HPLC analysis was performed according to the method previously described (Puel et al., 2005). For bioconversion assays, HPLC analysis was performed with a linear elution gradient using 33 mM acetic acid (solvent A) and methanol (solvent B) with a flow rate of 0.6 ml min−1. The compounds were eluted by starting from 18 % of solvent B for 15 min, followed by a step to 30 % solvent B, then a linear gradient up to 75 % within 10 min. After an isocratic elution for 20 min, the gradient was decreased to the initial value within 10 min, and remained at this value for the last 10 min.
The HPLC-MS/MS instrument used was an LCQ Deca XP Ion Trap coupled with an HPLC system from Thermo Finnigan. HPLC was performed on a reverse-phase 125×2.0 mm LiChrospher C18 column (Phenomenex). Methanol suspension (20 μl) was injected directly into the HPLC system. Gradient chromatography (run of 33 min) was performed with 33 mM acetic acid in water (eluent A)/methanol (eluent B) as the mobile phases at a flow rate of 0.2 ml min−1. The program initiated with 95/5 % eluent A/B for 5 min, then eluent B was increased to 10 % within 5 min, increased to 60 % in 10 min, held constant for 10 min, and then increased to 70 % in 10 min. The mobile phases remained constant for 6 min and finally returned to 90/10 % eluent A/B within 1 min and were allowed to reequilibrate for 5 min. HPLC-MS measurements were performed using the electrospray ionization mode (ESI). ESI was performed at room temperature in a negative mode; the tension was maintained at 4.5 kV with the capillary temperature at 300 °C. MS and MS-MS adjustments were made using m-hydroxybenzyl alcohol as chemical standard. The collision energy was set at 35 %.
Computer-assisted sequence analysis.
Sequence similarities were obtained by alignments of amino acid sequences using the blast 2 sequence program (http://www.ncbi.nih.gov/blast/bl2seq/wblast2.cgi).
The different domains of the P450 conserved core were identified from alignments performed using MultAlin (http://bioinfo.genopole-toulouse.prd.fr/multalin). The membrane-targeting hydrophobic segment was located in both cytochrome P450s using the TMpred program (http://www.ch.embnet.org/software/TMPRED_form.html).
RESULTS
Sequence analysis of the putative cytochrome P450s from A. clavatus
First, patulin production by A. clavatus NRRL 1980 strain was determined by HPLC-DAD after 2, 3, 4 and 5 days of culture. Patulin appeared on chromatograms from the third day, and RNA was extracted from mycelium harvested from this 3-day-old culture.
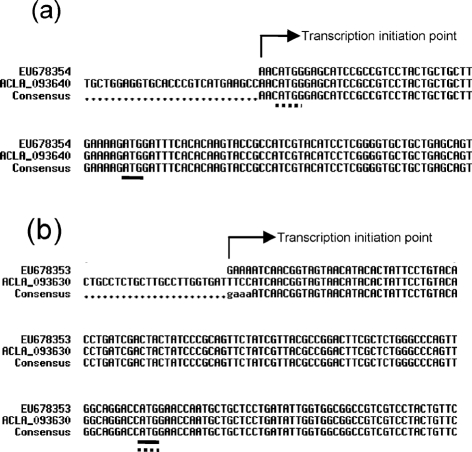
In order to determine the location of the start and stop codons of the two putative cytochrome P450 genes involved in patulin biosynthesis, we used RACE-PCR to isolate, clone and sequence the 5′ and 3′ UTR extremities from A. clavatus strain NRRL 1980. Then, the whole coding sequences of these genes were sequenced. For the first putative cytochrome P450 gene, AcP450I (ACLA_093640), the transcript sequence began only 3 bp upstream of a start codon predicted by TIGR bioinformatics tools (http://www.tigr.org/tdb/e2k1/acla1/). It is surprising that the 5′ UTR sequence is so short, but another ATG codon is present at the +40 position (Fig. 2). The sequences upstream and downstream of the second ATG were searched with the Kozak sequence consensus in Neurospora crassa; these results suggested that this new ATG could be the real start codon. The sequencing of the whole cDNA of AcP450I confirmed the presence of four introns of 55, 73, 81 and 91 bp in ACP450I. The sequence obtained from A. clavatus NRRL 1980 (GenBank accession no. EU678354) and those from A. clavatus NRRL 1 differed in only two transition mutations. However, after translation, these differences did not change the amino acid sequence. With the shift of the start codon, the predicted gene product comprised only 529 aa instead of the 541 aa initially predicted. Concerning the second putative cytochrome P450 gene, AcP450II (ACLA_093630), the whole transcript sequence began 107 bp upstream of the predicted start codon and ended 130 bp downstream of the predicted stop codon. The gene consists of five exons of 206, 64, 380, 573 and 588 bp interrupted by four introns. The sequence of A. clavatus strain NRRL 1980 (GenBank accession no. EU678353) differs from that of strain NRRL 1 by only six modifications (five transition and one tranversion mutations). Among these, only the transition mutation (A changed to G) located at position 1791 modifies the 524 aa amino acid sequence, by a change of glycine to glutamic acid at position 468. This sequence displays, respectively, 54 % identity with AcP450I, 55 % identity with CYP619C1 (EAU32821) of Aspergillus terreus, and 53 % identity with a putative cytochrome P450 (CAP71842) of Podospora anserina. On other hand, the AcP450I sequence is 65 % identical to CYP619C1 and 51 % to CAP71842 of Podospora anserina. Two sequences are close enough to both be in the CYP619C subfamily. On the basis of homology (a minimum 40 % amino acid sequence identity for family and 55 % for subfamily) (Nelson et al., 1996), the P450 Nomenclature Committee assigned AcP450I to CYP619C2 and AcP450II to CYP619C3.
Fig. 2.
Sequences of 3′ UTR extremities of genes encoding cytochrome P450s. (a) Alignment of the gene encoding CYP619C2 (GenBank accession no. EU678354) with gene ACLA_093640. (b) Alignment of the gene encoding CYP619C3 (EU678353) with gene ACLA_093630. Arrows show the first nucleotide of the respective mRNAs determined by RACE-PCR. Underlined are the start codons determined experimentally. Underlined with dashed lines are the start codons determined by an ORF finder automatic algorithm.
For both cytochrome P450s, the transmembrane prediction method (TMpred) detected in the N-terminal region a 35 aa hydrophobic segment that forms a membrane anchor indispensable to most eukaryotic P450s associated with microsomal membranes. This segment is followed by a P450-characteristic cluster of basic residues and prolines. Also, certain sequence motifs corresponding to the conserved tertiary structures were identifiable, and the sequences displayed several domains considered to be P450 signatures (Werck-Reichhart & Feyereisen, 2000): the central part of helix I containing the consensus sequence (Ala/Gly-Gly-X-Asp/Glu-Thr-Thr/Ser); the absolutely conserved (Glu-X-X-Arg) motif important for locking the haem pocket and stabilizing the core structure; and the haem-binding loop (Phe-X-X-Gly-X-Arg-X-Cys-X-Gly), although Phe was replaced by Tyr.
Kinetics of AcP450I and AcP450II expression under permissive and restrictive conditions
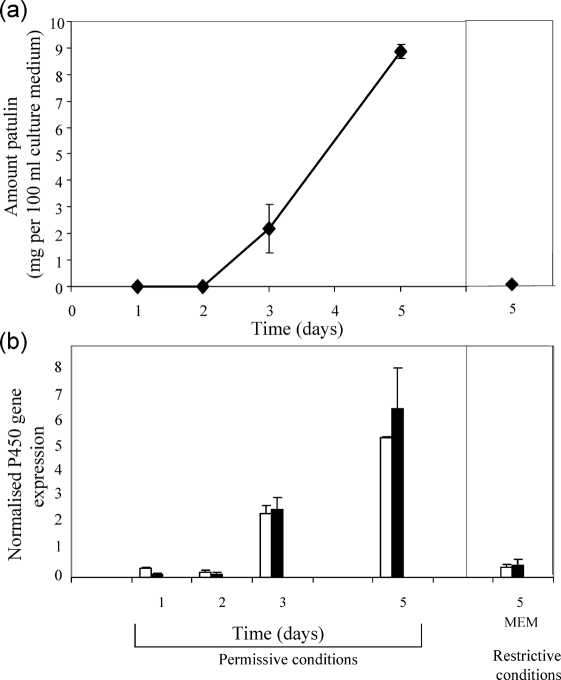
A. clavatus was inoculated in Czapek-glucose medium, without shaking. This medium supports patulin production. At certain time intervals (1, 2, 3 and 5 days), the expression of AcP450I and AcP450II encoding, respectively, CYP619C2 and CYP619C3 was quantified by qRT-PCR, whereas the patulin produced was quantified by HPLC. The results are shown in Fig. 3. The expression of AcP450I and AcP450II genes increased with a pattern similar to the increase in patulin concentration in the medium. In contrast, when A. clavatus was grown under restrictive conditions (MEM medium+5 % fetal calf serum, with shaking), which do not support patulin production, the expression of AcP450I and AcP450II genes remained at a basal level, and only traces of patulin were detected in the medium (0.7 μg 100 ml−1). After 5 days incubation, the expression levels of both genes were higher under patulin-permissive conditions, indicating a putative link between these genes and patulin biosynthesis in the fungus.
Fig. 3.
(a) Amount of patulin (mg 100 ml−1) in the culture medium. (b) Relative quantification expressed as P450 gene starting fluorescence R0 normalized to β-tubulin R0 (Peirson et al., 2003). Black bars, AcP450I gene; white bars, AcP450II gene. Patulin-permissive conditions were culture on Czapek-glucose without shaking; patulin-restrictive conditions were MEM supplemented with 5 % fetal calf serum, with shaking.
Heterologous expression of CYP619C2 and CYP619C3
The coding regions of these genes were amplified by PCR from cDNA of A. clavatus strain NRRL 1980. For CYP619C2, the forward primer was designed to consider the ATG located at the +40 position as the new start codon. For expression, the coding sequences were cloned into vector pYES2.1/V5/His-Topo. After galactose induction, the proteins were extracted and analysed by SDS-PAGE. The overexpressed proteins were detected by Western blotting using anti-V5 antibody. For each P450 protein, the observed molecular mass was 60 kDa, in good accordance with the calculated value of 64 kDa for V5 His6-tagged proteins (Fig. 4).
Fig. 4.
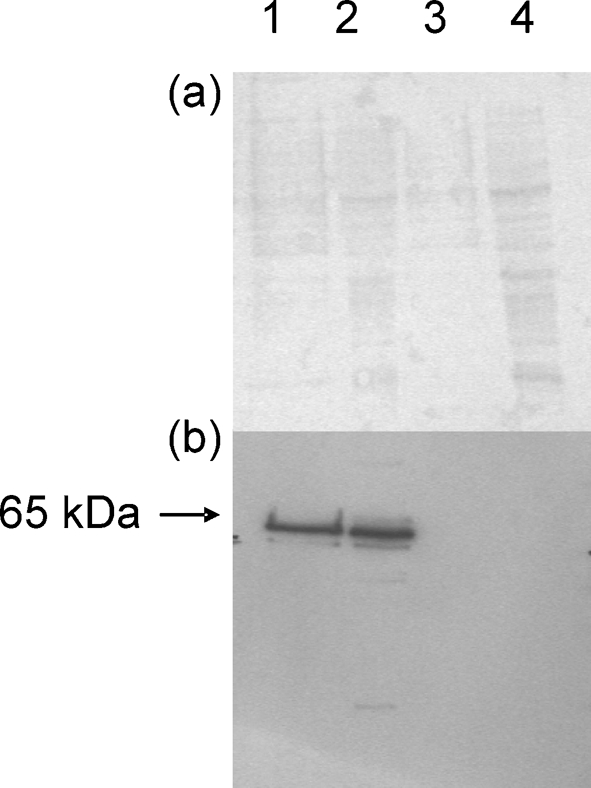
Analysis of the transformants after induction with galactose. (a) Electrophoresis on a 12 % SDS-polyacrylamide gel stained with Ponceau red. (b) Western immunoblots performed with anti-V5 antibody. Lanes: 1, total proteins from yeast cells expressing CYP619C2; 2, from yeast cells expressing CYP619C3; 3, from CYP619C2 control; 4, from CYP619C3 control. For both controls, each gene was inserted in the non-coding direction into the pYES vector with which the yeast cells were transformed.
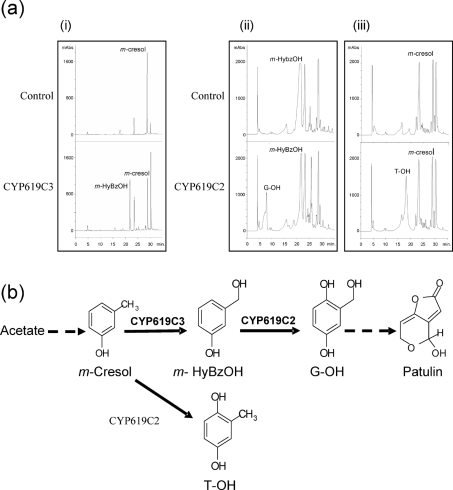
The galactose-induced yeast cells transformed with CYP619C2 were incubated with candidate phenolic compounds, and the supernatant was analysed by reverse-phase HPLC. When m-hydroxybenzyl alcohol was the substrate, a new peak of the reaction product appeared (Fig. 5). This product was characterized as gentisyl alcohol according to its retention time, UV spectrum and mass spectrum compared with an authentic standard. When m-cresol was the substrate, this compound was converted to a product which shared chromatographic features with toluquinol. The negative control for the transformed yeast was obtained using the same plasmid vector, including the reverse gene sequence. No conversion was observed in the negative controls. Another patulin precursor, m-hydroxybenzaldehyde, was tested for the reaction. Only m-hydroxybenzyl alcohol appeared on the chromatogram of yeasts overexpressing CYP619C2 and of control yeasts. For CYP619C2 yeast, gentisyl alcohol was detected by HPLC, but we did not observe any new peaks on the chromatogram. These last results show that S. cerevisiae Sc1 cells transformed with CYP619C2 were able to reduce m-hydroxybenzaldehyde to m-hydroxybenzyl alcohol but were unable to convert m-hydroxybenzaldehyde to gentisaldehyde.
Fig. 5.
(a) HPLC analysis of biotransformation assays. Chromatograms at 270 nm of extracts from yeast cultures, after 24 h substrate introduction: (i) biotransformation of cresol by yeasts overexpressing CYP619C3; (ii) biotransformation of m-hydroxybenzyl alcohol by yeasts overexpressing CYP619C2; and (iii) biotransformation of cresol by yeasts overexpressing CYP619C2. For both controls, genes were inserted in the non-coding direction into the pYES vector and transformed yeast cells were reacted with each substrate for 24 h. M-HyBzOH, m-hydroxybenzyl alcohol; G-OH, gentisyl alcohol; T-OH, toluquinol; mABs, milliabsorbance units. (b) Enzymic reactions catalysed by CYP619C2 and CYP619C3, and their involvement in the hydroxylation steps of the patulin biosynthesis pathway. Dashed arrows represent the steps which occur upstream and downstream of steps catalysed by CYP619C2 and CYP619C3.
For CYP619C3, m-cresol, m-hydroxybenzyl alcohol and toluquinol were tested. We did not observe any new peaks when m-hydroxybenzyl alcohol and toluquinol were used as substrates. On the other hand, m-cresol yielded a new product which displayed retention times and a UV spectrum similar to those of m-hydroxybenzyl alcohol. This last metabolite was identified as m-hydroxybenzyl alcohol on the basis of MS and MS2 spectra. Indeed, it showed a base peak at m/z 123, and the MS2 peaks were observed at m/z 105 and 93. Control yeasts transformed with vector containing reverse-inserted AcP450II failed to convert m-cresol to m-hydroxybenzyl alcohol.
DISCUSSION
In this study, we cloned and overexpressed two cytochrome P450 genes from the patulin-producing species A. clavatus in a heterologous host. Our aim was to determine their functions and to demonstrate that these genes are involved in the patulin biosynthesis pathway. Several enzymes involved in patulin biosynthesis have previously been characterized from P. griseofulvum, such as 6-methylsalicylic acid carboxylase, IDH, m-cresol hydroxylase and m-hydroxybenzyl alcohol hydroxylase (Light, 1969; Murphy et al., 1974; Murphy & Lynen, 1975; Sekiguchi & Gaucher, 1979), but among these enzymes, only the gene encoding IDH has been isolated (Fedeshko, 1992).
The early studies showed that all hydroxylase reactions are inhibited in a reversible manner by carbon monoxide, and require molecular oxygen and NADPH, suggesting cytochrome P450 activities. The use of radiolabelled m-cresol showed that methyl hydroxylation of m-cresol constitutes a key step in patulin biosynthetic pathways (Scott & Yalpani, 1967). Here, we report that CYP619C3 catalyses the hydroxylation of m-cresol to yield m-hydroxybenzyl alcohol, while CYP619C2 transforms m-hydroxybenzyl alcohol to gentisyl alcohol and m-cresol to 2,5-dihydroxytoluene (toluquinol). The importance of gentisyl alcohol as a direct precursor of patulin was controversial for a long time, although this compound has been isolated from several patulin-producing fungal species: P. expansum, P. griseofulvum and Penicillium novae-zeelandiae (Abo-Dahab et al., 1996; Alfaro et al., 2003; Birkinshaw et al., 1943). Iijima et al. (1986) have determined that patulin is formed after incubation with microsomal preparations only when gentisyl alcohol is added and not from gentisaldehyde. The importance of m-hydroxybenzaldehyde as a patulin precursor remains uncertain, since CYP619C2 did not display in vivo m-hydroxybenzaldehyde-2-hydroxylase activity. This result is consistent with earlier studies, in which ring-hydroxylation of m-hydroxybenzyl alcohol to gentisyl alcohol in cell-free preparations of P. griseofulvum could be demonstrated, but ring-hydroxylation of m-hydroxybenzaldehyde to gentisaldehyde could not (Murphy & Lynen, 1975). This questions whether m-hydroxybenzaldehyde and gentisaldehyde are direct patulin precursors. One hypothesis is that these last compounds are the products of an alcohol dehydrogenase unrelated to the patulin pathway.
In earlier studies, the role of toluquinol, a metabolite isolated from patulin-producing species, was discussed. Initially, based on a demonstration that P. griseofulvum could convert deuterio-labelled m-cresol into toluquinol, gentisyl alcohol, gentisaldehyde and patulin, a major route for patulin biosynthesis via m-cresol, toluquinol and gentisyl alcohol was proposed (Scott & Yalpani, 1967). Use of a pulse–chase radiolabelling technique or a culture force-fed deuterated toluquinol has shown that toluquinol is not incorporated into either gentisyl alcohol or patulin, but is metabolized to deoxyepoxidon (Forrester & Gaucher, 1972; Scott et al., 1973). In the present experiments, we demonstrated that CYP619C2 catalyses the hydroxylation of m-cresol to toluquinol, but CYP619C3 is not able to convert toluquinol into gentisyl alcohol. In addition, the conversion of m-cresol to both the hydroxylated products previously reported, toluquinol and m-hydroxybenzyl alcohol (Murphy et al., 1974), has been confirmed.
Partial sequences of two putative cytochrome P450 monooxygenase genes, p450-1 and p450-2, were recently described for the patulin-producing species P. expansum (White et al., 2006). These authors show that the genes are expressed only in patulin production-permissive conditions, not in restrictive conditions, and conclude that they have a potential role in patulin biosynthesis. Except for a weak identity (28 %) between CYP619C3 and P450-2, blast analysis failed to find a significant similarity among CYP619C2, CYP619C3 and proteins predicted from these genes, indicating that these genes are not orthologous.
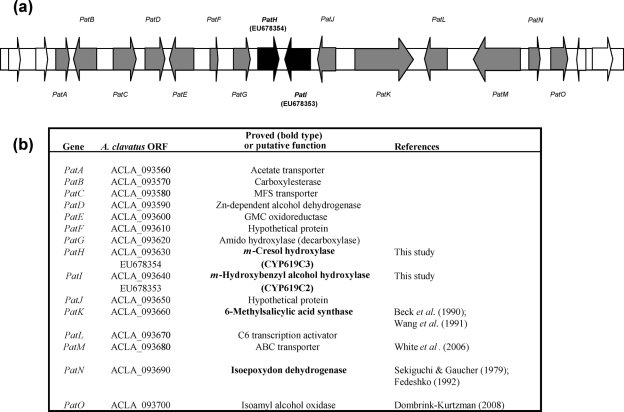
In A. clavatus, the genes encoding both cytochrome P450s are located in a 40 kb cluster (Fig. 6) consisting of approximately 15 genes: one putative transcription factor gene (PatL); three transporter (one ABC transporter, one MFS transporter and one acetate transporter) genes (PatM, PatC and PatA); two genes with unknown function (PatF and PatJ); and nine biosynthetic genes, i.e. the 6MSAS gene (PatK) isolated from P. griseofulvum (Beck et al., 1990), the genes encoding cytochrome P450s CYP619C2 and CYP619C3 described in this study (PatI and PatH), the IDH gene isolated from P. griseofulvum (PatN) (Fedeshko, 1992), a putative isoamyl alcohol oxidase described from P. griseofulvum (PatO) (Dombrink-Kurtzman, 2008), a gene encoding a putative decarboxylase displaying an amido hydrolase conserved domain (PatG), a putative carboxylesterase gene (PatB), and two genes encoding, respectively, a putative Zn-dependent alcohol dehydrogenase and a glucose-methanol choline oxidoreductase (PatD and PatE). Among these genes, two encode proteins known to be involved in patulin biosynthesis: 6MSAS, responsible for synthesis of 6-methylsalicylic acid from one acetate unit and three malonate units; and IDH, which converts isoepoxydon to phyllostine. Therefore, the proximity of four genes involved in patulin biosynthesis strongly suggests that the organization of patulin genes follows the general fungal law of secondary metabolite biosynthetic gene clustering. The distal extremity of this cluster has been described elsewhere for P. griseofulvum and P. expansum (Dombrink-Kurtzman, 2008; White et al., 2006). This part of the cluster is highly conserved, since the direction of transcription and the order of the last three genes (PatM, PatN and PatO) remain unchanged for the three species A. clavatus, P. griseofulvum and P. expansum.
Fig. 6.
(a) Putative patulin gene cluster of A. clavatus. Grey arrows represent genes of the cluster, white arrows the genes adjacent to the patulin cluster. Both arrow types show the direction of transcription. The genes encoding cytochrome P450s are in bold type, PatH for CYP619C3 and PatI for CYP619C2. (b) Characteristics of genes identified as belonging to the patulin gene cluster.
The role of the genes included in the cluster can be discussed with regard to the steps identified biochemically in the synthesis of patulin (Fig. 1). PatG possesses the signature sequence motifs of the amidohydroxylase superfamily, which contains γ-resorcylate decarboxylase involved in the biodegradation of hydroxybenzoates, and 5-carboxyvanillic acid decarboxylase involved in the metabolism of lignin (Liu & Zhang, 2006). PatG is most likely responsible for the decarboxylation of 6-methylsalicylic acid, producing m-cresol. PatE shares 59 % similarity with versicolorin B synthase (VBS), which catalyses a step of the aflatoxin biosynthetic pathway: the closing of the fifth ring of versiconal to yield versicolorin B. On the basis of similarity of chemical structures, it can be speculated that PatE is responsible for the final step of the pathway, the transformation of ascladiol into patulin. The presence of other enzymes remains obscure, and their involvement in the opening of the final aromatic intermediate ring, which has been a matter of speculation, cannot be determined without further research. Disruption analysis of PatE and PatG is under way in order to confirm the hypotheses proposed above.
In conclusion, the results clarify two steps of the patulin biosynthesis pathway. The identification of the patulin cluster will help to define strategies to reduce patulin contamination in apples and other food/feed products.
Acknowledgments
This work was supported in part by a Midi-Pyrénées Regional grant (no. 01002728), and by Agence Française de Sécurité Sanitaire de l'Environnement et du Travail (AFSSET) grant ES-2007-063. Preliminary sequence data were obtained from The Institute for Genomic Research website; http://www.tigr.org. Sequencing of A. clavatus was funded by National Institute of Allergy and Infectious Disease (NIAID) funding to William C. Nierman. We thank S. Peterson, a curator of the ARS Culture Collection, for providing A. clavatus strain NRRL 1980, and M. Blanco for the generous gift of a gentisyl alcohol standard. We thank D. Nelson, correspondent of the Cytochrome P450 Nomenclature Committee, for assignments of names for each cytochrome P450. We would like to thank N. Shaw for her editorial assistance, A. Polizzi for his help in development of qRT-PCR, and P. Galtier for his constant support.
Abbreviations
DAD, diode-array detection
IDH, isoepoxydon dehydrogenase
6MSAS, 6-methylsalicylic acid synthase
qRT-PCR, real-time quantitative RT-PCR
RACE, rapid amplification of cDNA ends
UTR, untranslated region
Footnotes
The GenBank/EMBL/DDBJ accession numbers for the nucleotide sequences reported in this paper are EU678353–EU678354.
References
- Abo-Dahab, N. F., Paterson, R. R. & Razak, A. A. (1996). Effect of fungistatic agent 2-deoxy-d-glucose on mycotoxins from Penicillium expansum. Lett Appl Microbiol 23, 171–173. [DOI] [PubMed] [Google Scholar]
- Alfaro, C., Urios, A., González, M. C., Moya, P. & Blanco, M. (2003). Screening for metabolites from Penicillium novae-zeelandiae displaying radical-scavenging activity and oxidative mutagenicity: isolation of gentisyl alcohol. Mutat Res 539, 187–194. [DOI] [PubMed] [Google Scholar]
- Beck, J., Ripka, S., Siegner, A., Schiltz, E. & Schweizer, E. (1990). The multifunctional 6-methylsalicylic acid synthase gene of Penicillium patulum. Its gene structure relative to that of other polyketide synthases. Eur J Biochem 192, 487–498. [DOI] [PubMed] [Google Scholar]
- Birkinshaw, J. H., Bracken, A. & Raistrick, H. (1943). Studies in the biochemistry of micro-organisms: 72. Gentisyl alcohol (2:5-dihydroxybenzyl alcohol), a metabolic product of Penicillium patulum Bainier. Biochem J 37, 726–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, W. G., Sen, A., Wang-Buhler, J. L., Yang, Y. H., Lopez, N., Merrill, G. F., Miranda, C. L., Hu, C. H. & Buhler, D. R. (2004). cDNA-directed expression of a functional zebrafish CYP1A in yeast. Aquat Toxicol 70, 111–121. [DOI] [PubMed] [Google Scholar]
- Dombrink-Kurtzman, M. A. (2007). The sequence of the isoepoxydon dehydrogenase gene of the patulin biosynthetic pathway in Penicillium species. Antonie Van Leeuwenhoek 91, 179–189. [DOI] [PubMed] [Google Scholar]
- Dombrink-Kurtzman, M. A. (2008). A gene having sequence homology to isoamyl alcohol oxidase is transcribed during patulin production in Penicillium griseofulvum. Curr Microbiol 56, 224–228. [DOI] [PubMed] [Google Scholar]
- Fedeshko, R. W. (1992). Polyketide enzymes and genes in Penicillium urticae. PhD thesis, University of Calgary, Calgary, AB, Canada.
- Fedorova, N. D., Khaldi, N., Joardar, V. S., Maiti, R., Amedeo, P., Anderson, M. J., Crabtree, J., Silva, J. C., Badger, J. H. & other authors (2008). Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genet 4, e1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester, P. I. & Gaucher, G. M. (1972). Conversion of 6-methylsalicylic acid into patulin by Penicillium urticae. Biochemistry 11, 1102–1107. [DOI] [PubMed] [Google Scholar]
- Frisvad, J. C., Smedsgaard, J., Larsen, T. O. & Samson, R. A. (2004). Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Stud Mycol 49, 201–241. [Google Scholar]
- Houbraken, J., Samson, R. A. & Frisvad, J. C. (2006). Byssochlamys: significance of heat resistance and mycotoxin production. Adv Exp Med Biol 571, 211–224. [DOI] [PubMed] [Google Scholar]
- Iijima, H., Ebizuka, Y. & Sankawa, U. (1986). Biosynthesis of patulin; in vitro conversion of gentisyl alcohol into patulin by microsomal enzyme(s) and retention of one of the carbinol protons in this reaction. Chem Pharm Bull (Tokyo) 34, 3534–3537. [DOI] [PubMed] [Google Scholar]
- Kimura, M., Tokai, T., Takahashi-Ando, N., Ohsato, S. & Fujimura, M. (2007). Molecular and genetic studies of Fusarium trichothecene biosynthesis: pathways, genes, and evolution. Biosci Biotechnol Biochem 71, 2105–2123. [DOI] [PubMed] [Google Scholar]
- Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Light, R. J. (1969). 6-Methylsalicylic acid decarboxylase from Penicillium patulum. Biochim Biophys Acta 191, 430–438. [DOI] [PubMed] [Google Scholar]
- Liu, A. & Zhang, H. (2006). Transition metal-catalyzed nonoxidative decarboxylation reactions. Biochemistry 45, 10407–10411. [DOI] [PubMed] [Google Scholar]
- Moake, M. M., Padilla-Zakour, O. I. & Worobo, R. W. (2005). Comprehensive review of patulin control methods in foods. Comp Rev Food Sci Food Safety 4, 8–21. [DOI] [PubMed] [Google Scholar]
- Murphy, G. & Lynen, F. (1975). Patulin biosynthesis: the metabolism of m-hydroxybenzyl alcohol and m-hydroxybenzaldehyde by particulate preparations from Penicillium patulum. Eur J Biochem 58, 467–475. [DOI] [PubMed] [Google Scholar]
- Murphy, G., Vogel, G., Krippahl, G. & Lynen, F. (1974). Patulin biosynthesis: the role of mixed-function oxidases in the hydroxylation of m-cresol. Eur J Biochem 49, 443–455. [DOI] [PubMed] [Google Scholar]
- Nelson, D. R., Koymans, L., Kamataki, T., Stegeman, J. J., Feyereisen, R., Waxman, D. J., Waterman, M. R., Gotoh, O., Coon, M. J. & other authors (1996). P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics 6, 1–42. [DOI] [PubMed] [Google Scholar]
- Panaccione, D. G. (2005). Origins and significance of ergot alkaloid diversity in fungi. FEMS Microbiol Lett 251, 9–17. [DOI] [PubMed] [Google Scholar]
- Peirson, S. N., Butler, J. N. & Foster, R. (2003). Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res 31, e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompon, D., Louerat, B., Bronine, A. & Urban, P. (1996). Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol 272, 51–64. [DOI] [PubMed] [Google Scholar]
- Priest, J. W. & Light, R. J. (1989). Patulin biosynthesis: epoxidation of toluquinol and gentisyl alcohol by particulate preparations from Penicillium patulum. Biochemistry 28, 9192–9200. [DOI] [PubMed] [Google Scholar]
- Proctor, R. H., Brown, D. W., Plattner, R. D. & Desjardins, A. E. (2003). Co-expression of 15 contiguous genes delineates a fumonisin biosynthetic gene cluster in Gibberella moniliformis. Fungal Genet Biol 38, 237–249. [DOI] [PubMed] [Google Scholar]
- Puel, O., Tadrist, S., Galtier, P., Oswald, I. P. & Delaforge, M. (2005). Byssochlamys nivea as a source of mycophenolic acid. Appl Environ Microbiol 71, 550–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puel, O., Tadrist, S., Delaforge, M., Oswald, I. P. & Lebrihi, A. (2007). The inability of Byssochlamys fulva to produce patulin is related to absence of 6-methylsalicylic acid synthase and isoepoxydon dehydrogenase genes. Int J Food Microbiol 115, 131–139. [DOI] [PubMed] [Google Scholar]
- Schaffner, W. & Weissmann, C. (1973). A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal Biochem 56, 502–514. [DOI] [PubMed] [Google Scholar]
- Scott, A. I. & Yalpani, M. (1967). A mass-spectrometric study of biosynthesis: conversion of deuteron-m-cresol into patulin. Chem Commun (Camb) 945–946.
- Scott, A. I., Zamir, L., Phillips, G. T. & Yalpani, M. (1973). The biosynthesis of patulin. Bioorg Chem 2, 124–139. [Google Scholar]
- Sekiguchi, J. & Gaucher, G. M. (1977). Conidiogenesis and secondary metabolism in Penicillium urticae. Appl Environ Microbiol 33, 147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi, J. & Gaucher, G. M. (1979). Patulin biosynthesis: the metabolism of phyllostine and isoepoxydon by cell-free preparations from Pencillium urticae. Can J Microbiol 25, 881–887. [DOI] [PubMed] [Google Scholar]
- Seo, J. A., Proctor, R. H. & Plattner, R. D. (2001). Characterization of four clustered and coregulated genes associated with fumonisin biosynthesis in Fusarium verticillioides. Fungal Genet Biol 34, 155–165. [DOI] [PubMed] [Google Scholar]
- Shen, D. K., Noodeh, A. D., Kazemi, A., Grillot, R., Robson, G. & Brugère, J. F. (2004). Characterisation and expression of phospholipases B from the opportunistic fungus Aspergillus fumigatus. FEMS Microbiol Lett 239, 87–93. [DOI] [PubMed] [Google Scholar]
- Varga, J., Rigó, K., Molnár, J., Tóth, B., Szencz, S., Téren, J. & Kozakiewicz, Z. (2003). Mycotoxin production and evolutionary relationships among species of Aspergillus section Clavati. Antonie Van Leeuwenhoek 83, 191–200. [DOI] [PubMed] [Google Scholar]
- Varga, J., Due, M., Frisvad, J. C. & Samson, R. A. (2007). Taxonomic revision of Aspergillus section Clavati based on molecular, morphological and physiological data. Stud Mycol 59, 89–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, I. K., Reeves, C. & Gaucher, G. M. (1991). Isolation and sequencing of a genomic DNA clone containing the 3′ terminus of the 6-methylsalicylic acid polyketide synthetase gene of Penicillium urticae. Can J Microbiol 37, 86–95. [DOI] [PubMed] [Google Scholar]
- Werck-Reichhart, D. & Feyereisen, R. (2000). Cytochromes P450: a success story. Genome Biol 1, REVIEWS3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, S., O'Callaghan, J. & Dobson, A. D. (2006). Cloning and molecular characterization of Penicillium expansum genes upregulated under conditions permissive for patulin biosynthesis. FEMS Microbiol Lett 255, 17–26. [DOI] [PubMed] [Google Scholar]
- Yu, J., Chang, P. K., Ehrlich, K. C., Cary, J. W., Bhatnagar, D., Cleveland, T. E., Payne, G. A., Linz, J. E., Woloshuk, C. P. & Bennett, J. W. (2004). Clustered pathway genes in aflatoxin biosynthesis. Appl Environ Microbiol 70, 1253–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]