Abstract
Proteus mirabilis causes urinary tract infections (UTIs) in individuals requiring long-term indwelling catheterization. The pathogenesis of this uropathogen is mediated by a number of virulence factors and the formation of crystalline biofilms. In addition, micro-organisms have evolved complex systems for the acquisition of nutrients, including the phosphate-specific transport system, which has been shown to be important in biofilm formation and pathogenesis. A functional Pst system is important during UTIs caused by P. mirabilis HI4320, since transposon mutants in the PstS periplasmic binding protein and the PstA permease protein were attenuated in the CBA mouse model of UTI. These mutants displayed a defect in biofilm formation when grown in human urine. This study focuses on a comparison of the proteomes during biofilm and planktonic growth in phosphate-rich medium and human urine, and microscopic investigations of biofilms formed by the pst mutants. Our data suggest that (i) the Δpst mutants, and particularly the ΔpstS mutant, are defective in biofilm formation, and (ii) the proteomes of these mutants differ significantly from that of the wild-type. Therefore, since the Pst system of P. mirabilis HI4320 negatively regulates biofilm formation, this system is important for the pathogenesis of these organisms during complicated UTIs.
INTRODUCTION
Proteus mirabilis – a member of the family Enterobacteriaceae – is the most common aetiological agent responsible for complicated urinary tract infections (UTIs) (Mobley, 1996). Subpopulations at higher risk for infection by this pathogen include those with long-term indwelling catheterization as well as those with structural and functional abnormalities within the urinary tract (Mobley, 1996). Clinical syndromes associated with P. mirabilis include cystitis and pyelonephritis, with possible complications from stone formation and bacteraemia (Mobley, 1996). These micro-organisms survive in the urinary tract by virtue of their production of a battery of virulence factors, including urease, flagella, fimbriae, haemolysin and IgA protease (Belas, 1996; Mobley et al., 1994; Musher et al., 1975; Peerbooms et al., 1984; Walker et al., 1999), and formation of crystalline biofilm on indwelling catheters (Stickler et al., 1993).
Biofilm formation – one of the most important mechanisms of pathogenicity of this micro-organism in the urinary tract – may be defined as a community of surface-attached bacteria encased in an extracellular matrix consisting of secreted carbohydrates, proteins and DNA. Indeed, biofilms have been associated with the virulence of a number of pathogens (Castelli et al., 2006). Organisms in these communities display phenotypic differences from planktonic cells, including a slower rate of growth and increased resistance to antibiotics (Lewis, 2001; Stoodley et al., 2002). P. mirabilis biofilms in urine are unique in that the extracellular polysaccharide matrix is enmeshed with struvite and carbonate apatite crystals (Jones et al., 2006). Crystal formation occurs due to the urease-catalysed hydrolysis of urea into ammonia, leading to an increase in the local pH and the precipitation of calcium, magnesium and phosphate ions (Morris et al., 1999). The pathogenesis of P. mirabilis UTIs is not at present understood fully.
The Pst system of P. mirabilis (Table 1), a multimeric, high-affinity inorganic phosphate (Pi) transporter, is induced at limiting extracellular Pi concentrations (<1 mM, 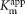 of 0.2 μM) (Rao & Torriani, 1990; Rosenberg et al., 1977). pst mutants are highly attenuated in cochallenge colonization of the CBA mouse model (Burall et al., 2004; Jacobsen et al., 2008). P. mirabilis HI4320 possesses a pst operon identical in gene organization to that observed in Escherichia coli. Previous studies have suggested that the roles of the Pst transport system in pathogenesis include regulation of invasion (Lucas et al., 2000; Mathew et al., 2001; Sinai & Bavoil, 1993), antibiotic resistance (Soualhine et al., 2005), colonization (Buckles et al., 2006; Daigle et al., 1995; Lamarche et al., 2005; Orihuela et al., 2001; Peirs et al., 2005; Runyen-Janecky et al., 2005), and biofilm formation (Monds et al., 2001).
of 0.2 μM) (Rao & Torriani, 1990; Rosenberg et al., 1977). pst mutants are highly attenuated in cochallenge colonization of the CBA mouse model (Burall et al., 2004; Jacobsen et al., 2008). P. mirabilis HI4320 possesses a pst operon identical in gene organization to that observed in Escherichia coli. Previous studies have suggested that the roles of the Pst transport system in pathogenesis include regulation of invasion (Lucas et al., 2000; Mathew et al., 2001; Sinai & Bavoil, 1993), antibiotic resistance (Soualhine et al., 2005), colonization (Buckles et al., 2006; Daigle et al., 1995; Lamarche et al., 2005; Orihuela et al., 2001; Peirs et al., 2005; Runyen-Janecky et al., 2005), and biofilm formation (Monds et al., 2001).
Table 1.
Characteristics of the P. mirabilis pst operon
| Gene | pI | MW (kDa) | Function | Cellular location |
|---|---|---|---|---|
| pstS | 8.71 | 37.0 | Phosphate binding; domain from amino acid residues 27 to 319 | Periplasmic space |
| pstC | 5.27 | 34.6 | Cytoplasmic membrane permease with six predicted transmembrane domains | Cytoplasmic membrane |
| pstA | 9.95 | 33.1 | Cytoplasmic membrane permease with six predicted transmembrane domains | Cytoplasmic membrane |
| pstB | 6.34 | 29.1 | ATPase; domain from amino acid residues 36 to 230 | Multiple |
| phoU | 5.28 | 27.9 | pho regulon negative regulator protein | Cytoplasm |
Since (i) earlier work has alluded to the importance of this system during biofilm formation (Monds et al., 2001, 2007) and (ii) the formation of crystalline biofilms is a characteristic of UTIs associated with P. mirabilis, we hypothesized that the Pst system of P. mirabilis HI4320 is involved in biofilm formation during establishment of a UTI. To assess the role of the Pst system, this study focused upon determining previously undetected in vitro phenotypes of the Δpst mutants by comparing biofilm formation between the wild-type HI4320 and ΔpstA and ΔpstS mutants via confocal laser scanning microscopy (CLSM) and quantitative analysis of surface coverage, and by comparing in vitro protein expression of these strains grown planktonically and as a biofilm.
METHODS
Bacterial strains, plasmids, reagents and culture conditions.
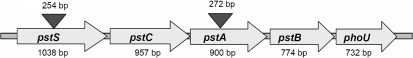
Bacterial strains and plasmids used in this study are listed in Table 2. Wild-type P. mirabilis HI4320 was isolated from the urine of a long-term-catheterized elderly woman with significant bacteriuria (≥105 c.f.u. ml−1) (Mobley & Warren, 1987; Warren et al., 1982). Δpst mutants used in this study, G1-43 (ΔpstA) and H4-34 (ΔpstS), were generated previously by signature-tagged mutagenesis and were polar mutants (Burall et al., 2004) (Fig. 1). All plasmids were initially constructed in either E. coli DH5α (Bethesda Research Laboratories) or E. coli TOP10 (Invitrogen).
Table 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source |
|---|---|---|
| P. mirabilis strains | ||
| HI4320 | Wild-type P. mirabilis isolate, Tetr | Mobley & Warren (1987) |
| G1-43 (ΔpstA) | pstA mutant of HI4320 (pstA : : Tn5 Kanr) | Burall et al. (2004) |
| H4-34 (ΔpstS) | pstS mutant of HI4320 (pstS : : Tn5 Kanr) | Burall et al. (2004) |
| Plasmids | ||
| pKHKS403 | Moderate-copy-number cloning vector used for complementation, 3159 kb, pBluescript with p15A ori from pACYC184, Ampr | H. L. T. Mobley |
| pSMJ003 | pKHKS403 with pstS-phoU (pst operon) | Jacobsen et al. (2008) |
Fig. 1.
The pst operon of P. mirabilis, showing the position and size of the ΔpstA and ΔpstS insertion sites.
All reagents were manufactured by Fisher Scientific, Sigma-Aldrich, American Bioanalytical, Amersham Biosciences or J. T. Baker. Multimark protein molecular mass marker was purchased from Invitrogen. PBS, urea, thiourea, Biuret solution, Folin reagent, bromophenol blue, TCA, sodium thiosulfate, iodoacetamide, formaldehyde and PMSF were obtained from Sigma Aldrich. Immobiline Dry Strips (18 cm, pH 3–10, nonlinear), Pharmalytes (pH 3–10), mineral oil, DTT, CHAPS, the Multiphor II isoelectric focuser and the Höefer DALT Vertical System were obtained from Amersham Biosciences. Most other chemicals and media, including acrylamide (40 %), silver nitrate, sodium carbonate, SDS, ammonium persulfate, yeast extract, NaCl, Tris base and agarose, were obtained from Fisher Scientific. Tetramethylethylenediamine (TEMED) was obtained from Bio-Rad. Zirconia/silica 0.1 mm beads were purchased from Biospec Products. Components of the bacterial culture medium were purchased from Becton Dickinson.
Luria broth (LB; containing, per litre, 10 g tryptone, 5 g yeast extract and 10 g NaCl) was used routinely with the appropriate antibiotics (15 μg tetracycline ml−1, 50 μg kanamycin ml−1 and/or 100 μg ampicillin ml−1) to culture P. mirabilis. Urine was collected from multiple anonymous healthy donors, pooled, filter-sterilized and stored in the dark at 4 °C until required.
Growth of planktonic P. mirabilis in vitro.
An overnight culture of either P. mirabilis HI4320 or Δpst mutants in LB and appropriate antibiotics was diluted 1 : 40 into fresh LB without antibiotics and incubated at 37 °C with shaking at 200 r.p.m. for 2 h. This was diluted 1 : 200 into 100 ml test medium and incubated at 37 °C with shaking. For the studies examining growth in LB, each strain was incubated at 37 °C until OD600 0.3–0.4 was reached. For comparisons of growth in LB against that in pooled human urine, cultures were incubated at 37 °C for 24 and 48 h to ensure growth but limit crystal formation. After the required period, cultures were pelleted and resuspended in protein preservation solution (2.8 mM PMSF, 10 mM Tris/HCl and 1 mM EDTA, pH 8.0) to prevent protein degradation and changes in protein expression.
Growth of P. mirabilis biofilm in vitro.
A variation of the in vitro flow reactor system utilized in our laboratory for Staphylococcus aureus biofilm culture (Brady et al., 2006) was used. The reactor system was constructed within a 37 °C incubator and consisted of silicone tubing through which pooled human urine (pH 5.0–6.5) flowed under the control of a peristaltic pump into a waste container. This system incorporated square-cross-section glass flow cells for use in confocal microscopy studies. Prior to inoculation, urine was pumped through the system and allowed to equilibrate to temperature for 24 h. An overnight culture of P. mirabilis HI4320 or Δpst mutants grown in LB and appropriate antibiotics was diluted 1 : 100 into fresh LB without antibiotics and allowed to grow at 37 °C with shaking for 2 h. The tubing was clamped upstream of the injection ports and 1.0 ml exponential phase bacterial culture was injected. The system was incubated without flow for 20 min at 37 °C so that bacteria could adhere to the luminal surface. The media flow was restored to a flow rate of 0.5 ml min−1 at 37 °C for 21 h. At 21 h, biofilm was harvested by (i) removal of the square glass flow cells for confocal microscopy and (ii) squeezing the biofilm from the tubing into protein preservation solution for protein expression analysis. Total biofilm protein was collected by mechanical disruption using a FastPrep instrument (Qbiogene) with 0.1 mm zirconia/silica beads and quantified using the modified method of Bradford (Bradford, 1976).
2D gel electrophoresis (2DGE).
2DGE was conducted according to the principles of O'Farrell (1975) and as outlined by Gorg et al. (2000) and Sauer & Camper (2001). Rehydration of 400 μg crude protein was accomplished by the addition of a one-tenth volume of ice-cold 1 : 10 TCA : acetone. The resulting pellet was solubilized in rehydration buffer (0.1 mM urea, 25 μM thiourea, 0.35 μM DTT, 0.5 %, w/v, CHAPS, 1.6 % Pharmalyte, pH 3–10). These samples were applied to 18 cm, pH 3–10 (nonlinear), Immobiline Dry-Strips (GE Healthcare) and allowed to swell at room temperature for at least 23 h prior to focusing. IEF was performed using a Multiphor II from Amersham as per the manufacturer's directions. Prior to the second dimension, the immobilized pH gradient (IPG) strips were equilibrated (as per the manufacturer's directions) and subsequently applied to the top edge of the 26 cm×20 cm 2D Höefer DALT Vertical System gel system (Amersham Biosciences). Crude protein extracts were separated at 10 °C on an 11 % resolving gel, which was then non-destructively silver-stained (Gharahdaghi et al., 1999).
MALDI-TOF analysis.
Protein spots excised from 2D-PAGE gels were sent for processing and analysis at the Mass Spectrometry Core of the Biomolecular Resource Facility at the University of Texas Medical Branch. For identification of trypsin-digested proteins, an Applied Biosystems Voyager-DE STR MALDI-TOF mass spectrometer or Applied Biosystems 4800 MALDI-TOF/TOF mass spectrometer was operated in the positive-ion mode with α-cyano-4-hydroxycinnamic acid matrix used for ionization. At least 100 laser shots per spectrum were averaged. Mass spectral peaks with a signal-to-noise ratio greater than 5 : 1 were deisotoped, and the resulting mono-isotopic masses were used for protein identification using mass fingerprint analysis. Proteins were identified by correlation of uninterpreted tandem mass spectra to entries in the database PAHI4320 completefirstrun.2005.01.16, utilizing the ProteinProspector MS-Fit server (IBM AIX version, University of California at San Francisco; http://prospector.ucsf.edu/) (Clauser et al., 1999). This database was generated by GLIMMER [Gene Locator and Interpolated Markov modelER; The Institute For Genomic Research (TIGR); http://www.tigr.org/software/glimmer/] (Delcher et al., 1999) from the nucleotide sequence available from the Sanger Centre, Cambridge, UK (http://www.sanger.ac.uk/Projects/P_mirabilis/). For the initial pass search, no mass or pI constraints were applied and one missed cleavage per peptide was allowed. The mass tolerance for the monoisotopic precursor ion was set at ±50 p.p.m. All matching spectra were reviewed manually, and proteins with an expectation MOWSE score of 10 or more were considered positive identities. Protein spot comparisons and analysis were accomplished using the Image-MASTER 2D Platinum software (Amersham Biosciences, version 5.00) and Melanie 3 software (GeneBio). Determination of proteins that were exclusively expressed in either the HI4320 strain or the pst mutants was accomplished through analysis of the Melanie-generated spot report and confirmation through the appearance of the protein spot in at least three separate gels. Functional and cellular localization predictions were accomplished using the KEGG GENES database (Kyoto Encyclopedia of Genes and Genomes; http://www.genome.ad.jp/kegg/genes.html) and psort-B v.2.0 (http://www.psort.org/; Gardy et al., 2003), respectively.
Biofilm formation assay.
To assess biofilm formation during static conditions, an adaptation of the 96-well microtitre dish biofilm assay developed by O'Toole and co-workers (O'Toole & Kolter, 1998; O'Toole et al., 1999) based on viable counts was performed. An overnight culture of P. mirabilis HI4320 and the Δpst mutants was diluted 1 : 40 into fresh LB and allowed to grow at 37 °C with shaking for 2 h. Pooled human urine was inoculated with 25 μl of each 2 h culture into polystyrene six-well plates (Becton Dickinson) in triplicate. Samples were incubated at 37 °C statically and after 24 h, growth medium was replaced with fresh urine. After 48 h, excess urine was discarded from each well and 1.0 ml PBS was added to remove any planktonic culture that remained. Well bottoms were scraped to remove biofilms; these were resuspended in PBS. Biofilm suspension was diluted 1 : 10 in PBS and homogenized with the Polytron Pt 1200 CL homogenizer (Kinematica), probe set 5 for 30 s. Bacterial counts (c.f.u. ml−1) for the 48 h samples were determined by diluting samples 10-fold in PBS and plating each dilution on Luria agar plates.
CLSM.
To determine whether mutations in the Pst system of P. mirabilis HI4320 affect biofilm formation under flow conditions, visualization of the wild-type and mutant strains was accomplished through differential viability staining and CLSM. Biofilms were grown under flow in pooled human urine as described above. Prior to staining, flow cells were washed with 0.85 % NaCl to remove excess urine. Bacterial cells were stained in the dark for at least 15 min at room temperature with 3 ml BacLight Live/Dead stain (3.34 mM SYTO 9, 20 mM propidium iodide in 0.85 %, w/v, NaCl; Molecular Probes, Invitrogen). Samples were washed in fresh 0.85 % (w/v) NaCl and then visualized using an LSM510 Meta laser scanning confocal microscope (Carl Zeiss). The microscope was configured with two lasers (argon 488 nm/514 nm/543 nm and HeNe 633 nm) and micrographs were taken at random with the Plan-Apochromat ×20/0.75 lens or, if otherwise noted, with the Plan Neofluar ×10/0.3 lens. Green fluorescence was used as an indicator of the presence of living cells in the biofilm, as SYTO 9 (480 nm excitation) is a freely diffusible nucleic acid intercalator that labels all cells in the microbial population regardless of viability. To distinguish viable cells from dead organisms, the membrane-impermeant DNA intercalator propidium iodide (536 nm excitation) was used as a counterstain to stain cells with compromised membrane integrity. A series of horizontal (xy) optical sections were taken throughout the full-length of the biofilm. Image capture and 2D projections of z-stacks were performed using the LSM 510 Meta image acquisition software (Zeiss). The subsequent micrographs were analysed quantitatively for biomass, mean and maximum thickness, and roughness coefficient using the Community Statistics (COMSTAT) image analysis software (Heydorn et al., 2000).
Statistical analyses.
The Student's t test was used to determine significant differences in biofilm growth throughout all of the biofilm plate assays; a P value <0.05 was considered statistically significant. All determinations were performed in triplicate.
RESULTS
Biofilm formation by P. mirabilis HI4320 and its pst mutants
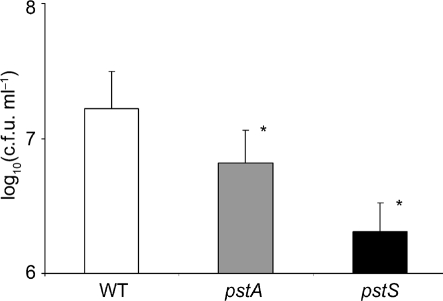
Since it has been shown that biofilm formation plays a role in early P. mirabilis UTIs and that gene products of the pst operon negatively regulate biofilm formation (Monds et al., 2001), quantitative biofilm microtitre assays were performed. After 48 h of static growth in pooled human urine, a 2.5- and eightfold reduction in biofilm formation between the HI4320 wild-type and the ΔpstA and ΔpstS mutants, respectively, was observed [wild-type, 1.65×107 c.f.u. ml−1; ΔpstA, 6.6×106 c.f.u. ml−1 (P=0.03); ΔpstS, 2.1×106 c.f.u. ml−1 (P=0.02); Fig. 2]. These data suggest that biofilm formation is impaired in the ΔpstA and ΔpstS mutants.
Fig. 2.
Viable cell count biofilm assay comparing 48 h biofilm formation during growth in pooled human urine between wild-type P. mirabilis HI4320 and the ΔpstA and ΔpstS mutants. Bacterial counts [in log10(c.f.u. cm−1)] were determined by serial dilution and spread plating. Data are presented as mean±sd of triplicate assays; asterisks denote statistically significant differences (P<0.05).
Comparison of biofilm formation by CLSM
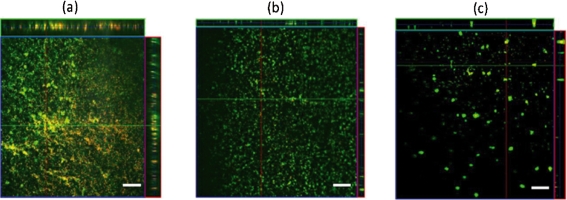
Visualization of 21 h flow-grown biofilms by CLSM and BacLight staining revealed differences in surface coverage and biofilm architecture between the wild-type strain and the Δpst mutants (Fig. 3). Wild-type HI4320 biofilms, grown in human pooled urine, possess a dense 3D organization consisting of towering mushroom structures with water channels flowing throughout. Conversely, the biofilms formed by the Δpst mutants lacked this distinct 3D architecture and were composed only of large, irregularly shaped microcolonies/aggregates (Fig. 3). These images revealed that the Δpst mutants are defective in biofilm formation as compared with the wild-type strain and apparently cannot form mature biofilm structures under flow conditions during growth in pooled human urine.
Fig. 3.
CLSM images of wild-type P. mirabilis HI4320 and Δpst mutant biofilms grown under flow in pooled human urine. Biofilms on glass flow cells were stained with Live/Dead stain and images were captured using a Plan-Apochromat ×20/0.75 lens, except for the side views (Plan-Neofluar ×10/0.3). (a) Wild-type P. mirabilis, stack size 460.7×460.7×48.0 μm; (b) ΔpstA mutant P. mirabilis, stack size 651.5×651.5×24.5 μm; (c) ΔpstS mutant P. mirabilis, stack size 651.5×651.5×42.0 μm. Lines in the xy plane depict the location of z projections, 97, 49 and 42 μm deep, respectively, shown at the top and right of the images. Bars, 50 μm.
To quantify the data obtained from the CLSM images, COMSTAT analysis software was used to assess the quantity (biomass, and mean and maximum thickness) and quality of the biofilms (roughness coefficient) grown under flow in pooled human urine. CLSM images of a single biofilm (total images; HI4320, 97; ΔpstA, 49; ΔpstS, 42) were taken randomly and converted into black and white for analysis by this software. Quantitative computer analysis of biofilm architecture using COMSTAT verified visual observations, as biofilms formed by Δpst mutants grown in pooled human urine differed from wild-type biofilm with respect to mean and maximum thickness (μm) as well as total biomass (Table 3). Under flow conditions, biofilms formed by the ΔpstS mutant during growth in pooled human urine demonstrated a reduction in biomass as compared with the HI4320 wild-type strain (Table 3; ΔpstS, 0.04 μm3 μm−2; ΔpstA, 1.40 μm3 μm−2; HI4320, 1.21 μm3 μm−2). These findings, in conjunction with the other biofilm analyses, indicate that mutants in the Pst system of P. mirabilis HI4320 are defective in biofilm formation. As stated previously, the Δpst mutants are defective in biofilm formation as compared with the wild-type strain as visualized by CLSM, and apparently these mutants cannot form mature biofilm structures.
Table 3.
COMSTAT analysis of the wild-type HI4320 and pst mutant strains after 21 h biofilm growth in pooled human urine
| Strain | Biomass (μm3 μm−2)* | Mean thickness (μm)† | Maximum thickness (μm)‡ | Roughness coefficient§ |
|---|---|---|---|---|
| Wild-type | 1.21 | 2.70 | 25.0 | 1.83 |
| pstA | 1.40 | 2.46 | 17.33 | 1.72 |
| pstS | 0.04|| | 0.07|| | 5.63|| | 1.97 |
*A measurement of the overall volume of the biofilm.
†A measurement of the spatial area occupied by the biofilm.
‡A measurement of the maximum thickness at a given location in the biofilm.
§A measurement of variation in biofilm thickness and an indicator of biofilm heterogeneity.
||Statistically significant difference (P<0.05).
Proteomic comparison of P. mirabilis HI4320 and the pst mutants
Proteomic analysis using 2DGE was used to compare the wild-type HI4320 strain with the Δpst mutants grown as a biofilm in pooled human urine (Fig. 4, Table 4) and planktonically in LB in phosphate-excess conditions (Figs 5 and 6, Table 5).
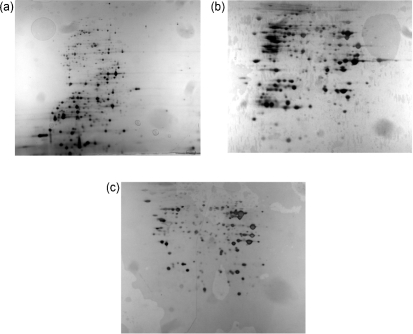
Fig. 4.
Proteomic comparison of the wild-type HI4320 strain and the Δpst mutant strains after biofilm growth in pooled human urine, as visualized by 2D PAGE. Overnight cultures were utilized to inoculate pooled human urine and grown in a biofilm flow reactor system for 21 h. After standardization, whole-cell lysates were prepared and 400 μg protein was loaded. pH range 3–10 was utilized, with pH 3 on the left. Protein spots were excised and examined by MALDI-TOF or MALDI-TOF/TOF. (a) HI4320, (b) ΔpstA, (c) ΔpstS.
Table 4.
Proteins of the wild-type and pst mutant strains expressed exclusively during biofilm growth in pooled human urine
| PMI number* | Protein | Function | Cellular location |
|---|---|---|---|
| Proteins upregulated in pst mutants only | |||
| 242 | Pgk | Phosphoglycerate kinase | Cytoplasm |
| 454 | NagB | Glucosamine-6-phosphate isomerase | Unknown |
| 700 | SerS | Glycine, serine, threonine metabolism | Cytoplasm |
| 708† | AnsB | l-Asparaginase II | Periplasm |
| 729† | AphA | Class B acid phosphatase precursor | Unknown |
| 1043 | ArnB | PbgP1 protein | Cytoplasm |
| 1094 | YcdW | 2-Hydroxyacid dehydrogenase | Cytoplasm |
| 1966 | – | NADPH-dependent FMN reductase protein | Unknown |
| 2024 | PepP | Proline aminopeptidase | Cytoplasm |
| 2092 | BudC | Acetoin phosphatase | Cytoplasm |
| 2416 | DeoC | Deoxyribose-phosphate aldolase | Cytoplasm |
| 2500 | PhoA | Alkaline phosphatase | Periplasm |
| 2893 | PstS | Phosphate-binding periplasmic protein precursor | Periplasm |
| 3545† | PepE | α-Aspartyl dipeptidase | Cytoplasm |
| 3617 | UgpB | Glycerol 3-phosphate-binding periplasmic protein | Periplasm |
| 3674 | AbgA | Aminobenzoyl-glutamate utilization protein | Unknown |
| Proteins upregulated in the wild-type only | |||
| 9 | DnaK | Heat-shock protein 70 chaperone protein DnaK | Cytoplasm |
| 3464 | PepA | Cytosol aminopeptidase A/I | Cytoplasm |
*PMI – Proteus mirabilis index; a P. mirabilis genome databank hosted at the Sanger Institute, Cambridge, UK; http://www.sanger.ac.uk/Projects/P_mirabilis/.
†Expressed only in the pstA mutant. Proteins with no designation were left blank.
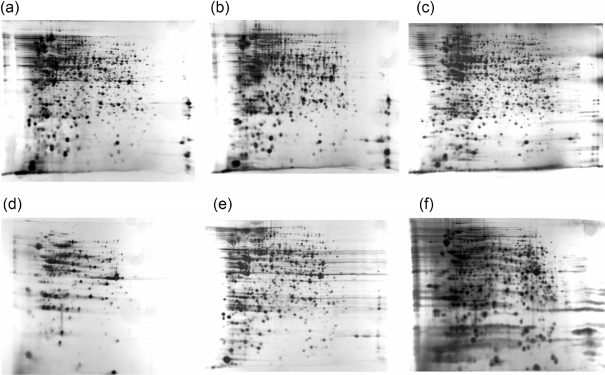
Fig. 5.
Proteomic comparison of the wild-type HI4320 strain and the pst mutant strains during planktonic growth in LB, as visualized by 2D PAGE. pH range 3–10 was utilized, with pH 3 on the left. Six hundred and sixty-nine spots were excised and examined by MALDI-TOF or MALDI-TOF/TOF. Samples were run at least in triplicate. (a) HI4320, (b) ΔpstS, (c) ΔpstA, (d) HI4320(pKHKS403), (e) ΔpstS(pKHKS403), (f) ΔpstA(pKHKS403).
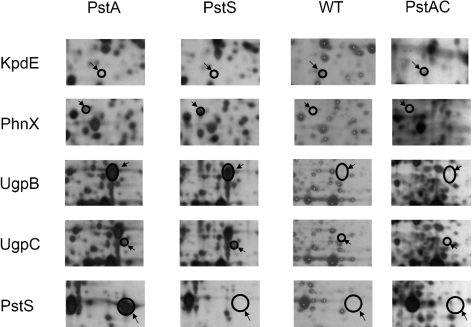
Fig. 6.
Unique protein expression of the wild-type HI4320 strain and the pst mutant strains during planktonic growth in LB, as visualized by 2D PAGE. pH range 3–10 was utilized and the gels were silver-stained. Proteins were designated as being exclusively expressed if the spot was present in at least three separate gels. The transcription regulator KpdE was only expressed in the HI4320 wild-type strain. The phosphonoacetaldehyde phosphonohydrolase PhnX, periplasmic glycerol 3-phosphate binding protein UgpB, and sn-glycerol 3-phosphate transport ATP-binding protein UgpC were expressed in both pst mutants. The PstS protein was expressed exclusively by the pstA mutant. WT, P. mirabilis HI4320 strain; PstA, P. mirabilis HI4320 pstA strain; PstS, P. mirabilis HI4320 pstS strain; PstAC, P. mirabilis HI4320 pstA strain with pSMJ003 (pKHKS403 possessing the entire pst operon).
Table 5.
Proteins expressed exclusively by the wild-type HI4320 or pst mutant strains during planktonic growth in LB
| PMI number* | Protein | Function | Cellular location |
|---|---|---|---|
| Proteins expressed only in the wild-type strain | |||
| 1476 | OppA1 | ABC transporter | Periplasm |
| 2021 | GcvT | Glycine, serine, threonine metabolism | Cytoplasm |
| 3299 | IlvG | Valine, leucine, isoleucine biosynthesis | Cytoplasmic membrane |
| 3551 | PepQ | Degradation | Cytoplasm |
| 601 | YbhE | Hypothetical protein, 3-carboxymuconate cyclase | Unknown |
| 2673 | GntK | Putative glucokinase | Cytoplasm |
| 420 | LipA | Lipoic acid synthetase | Cytoplasm |
| 1810 | FadL | Long-chain fatty acid transporter | Outer membrane |
| 2386 | TerB | Tellurite resistance | Cytoplasm |
| 3424 | DeaD | Cold-shock RNA helicase | Cytoplasm |
| 3455 | PyrB | Aspartate carbamoyltransferase | Miscellaneous |
| 2240 | TruD | tRNA pseudouridine synthase D | Cytoplasm |
| 2351 | YgiC | Hypothetical protein, glutathionylspermidine synthase | Cytoplasm |
| 3270 | RplF | 50S ribosomal subunit protein L6 | Cytoplasm |
| 3420 | TruB | tRNA pseudouridine synthase B | Cytoplasm |
| 868 | YcfH | Highly similar to DNase YcfH | Unknown |
| 2631 | – | Undefined product, lipoprotein, putative | Unknown |
| 2759 | IclR | Putative IclR family transcriptional regulator | Unknown |
| 2798 | – | Hypothetical protein | Unknown |
| 3673 | AlbA | Putative MerR-family regulator | Periplasm |
| 1226 | KdpE | Transcriptional regulator of kdp operon | Cytoplasm |
| 2883 | NtrB | Nitrogen regulation protein NR(II) | Cytoplasm |
| Proteins expressed only in pst mutants | |||
| 361 | – | Unknown | Unknown |
| 2282 | Frr | Ribosomal recycling factor | Cytoplasm |
| 2893† | PstS | Phosphate-binding periplasmic protein precursor | Periplasm |
| 3079 | PhnX | Phosphonoacetaldehyde phosphonohydrolase | Cytoplasm |
| 3296 | – | Hypothetical protein | Cytoplasm |
| 3357† | Orn | OligoRNase | Cytoplasm |
| 3614 | UgpC | sn-Glycerol 3-phosphate transport ATP-binding protein | Miscellaneous |
| 3617 | UgpB | Periplasmic glycerol 3-phosphate binding protein | Periplasm |
| 3618 | UgpQ | Glycerophosphoryl diester esterase | Cytoplasm |
| 3715 | – | Hypothetical protein | Unknown |
*PMI – Proteus mirabilis index; a P. mirabilis genome databank hosted at the Sanger Institute, Cambridge, UK; http://www.sanger.ac.uk/Projects/P_mirabilis/.
†Expressed only in the pstA mutant. Proteins with no designation are left blank.
Biofilm growth in pooled human urine
Comparisons of the proteomes of the HI4320 and the pst mutant biofilms cultured under flow in pooled human urine were performed (Fig. 4, Table 4). A total of 18 proteins were tentatively identified as being exclusively expressed (16 proteins in the Δpst mutants, two proteins in the HI4320 strain) (Table 4). Four proteins (AnsB, AphA, PstS and PepE; Table 4) were expressed in the ΔpstA mutant but not in ΔpstS.
Sixteen proteins were present only in the Δpst mutant strains and not in the wild-type (Table 4). Three were known members of the pho regulon (alkaline phosphatase PhoA, PstS and glycerol 3-phosphate periplasmic binding protein UgpB) and would be expected to be upregulated in these Δpst mutant strains. The other proteins were identified as performing a variety of functions (Table 4). Four were found to be involved in nutrient scavenging, including the proline aminopeptidase PepP, the α-aspartyl dipeptidase PepE, the aminobenzoyl-glutamate utilization protein AbgA, and the deoxyribose-phosphate aldolase DeoC. One protein, ArnB, is an amino transferase known to be involved in LPS biosynthesis. Two proteins, the class B acid phosphatase AphA and the acetoin reductase BudC, are known to be involved in the prevention of intracellular acidification. Two were identified only as putative proteins (2-hydroxyacid dehydrogenase YcdW and PMI1966), and three other proteins are known to be involved in general metabolism (phosphoglycerate kinase Pgk, seryl-tRNA synthetase SerS and l-asparaginase II AnsB). Some proteins expressed in the mutants have been demonstrated in other studies to be indirectly associated with the downregulation of biofilm formation (NagB, AphA). These analyses provide some insight as to which systems are affected by mutations in the pst operon.
Two proteins, the 70 kDa heat-shock protein DnaK and the cytosol aminopeptidase PepA, were expressed exclusively in the HI4320 strain. Currently, there are no known roles for these proteins during biofilm formation. Thus, they may assist the organism in adapting to growth in human urine.
Planktonic growth in LB
Proteomes of the HI4320 strain, the Δpst mutants, and the latter strains transformed with the pKHKS403 vector possessing the complete pst operon, were compared after planktonic growth in phosphate-rich LB (Figs 5 and 6, Supplementary Figs S1 and S2). A total of 32 proteins were exclusively expressed in either the wild-type (22 proteins) or mutant (10 proteins) strains (Table 5). Of the 22 proteins that were identified as being exclusively upregulated in the wild-type, several are associated with amino acid metabolism and translation, such as the amino acid transporter protein OppA1, the glycine cleavage enzyme GcvT, the enzyme IlvG, the peptidase PepQ, the 50S ribosomal subunit protein L6 RplF, the tRNA pseudouridine synthases TruB and TruD, and the putative glutathionylspermidine synthase YgiC. A number of transcriptional regulators were expressed by this strain, including a putative MerR-family regulator, an IclR transcriptional regulator, KdpE, and NtrB. Other metabolic proteins that were exclusively expressed included the long-chain fatty acid transporter FadL, the aspartate carbamoyltransferase PyrB, the cold-shock RNA helicase DeaD (nucleotide metabolism), the tellurite-resistance protein TerB, and the coenzyme lipoic acid synthesis protein LipA. Three putative proteins were expressed in the HI4320 strain (putative lipoprotein 2631, hypothetical protein 2798, DNase YcfH). Proteins involved in carbohydrate metabolism were identified and included the 3-carboxymuconate cyclase YbhE and the putative gluconokinase GntK.
A variety of proteins were upregulated exclusively in the pst mutants, including phosphate assimilation and transport proteins (UgpB, UgpC, UgpQ, PhnX and PstS), the ribosomal recycling factor Frr, the oligoRNase Orn and two hypothetical proteins (PMI3296 and PMI3715). As expected, the PstS protein was expressed only in the ΔpstA mutant (Fig. 6, Table 5). Introduction of the complete pst operon into the Δpst mutants demonstrated that complementation occurred: known members of the pho regulon (UgpB, UgpC, PstS and PhnX) were downregulated in these strains (Fig. 6, Table 5).
These data suggest that mutations in the pst system of P. mirabilis HI4230 have an effect on global protein expression; several proteins were present or absent in these mutants, as evaluated under planktonic and biofilm growth conditions and visualized by 2D PAGE. Several of the proteins exclusively expressed by these mutants were associated with phosphate assimilation and transport. However, our analyses have revealed proteins that may be involved in biofilm formation by P. mirabilis and thus also have a role in the attenuation of virulence of the pst mutant strains.
DISCUSSION
Mutagenesis studies have elucidated several potential roles for the Pst transport system in pathogenesis, the most intensively studied being the changes in cell surface properties that occur under low-phosphate conditions. Such changes include alterations in the polysaccharide content of the membrane (Mendrygal & Gonzalez, 2000; Ruberg et al., 1999) and synthesis of phosphate-free membrane lipids [sulfolipids, ornithine-containing lipids, diacylglyceryl-N,N,N-trimethylhomoserine (DGTS) and teichoic acid], thought to conserve Pi during new membrane synthesis (Benning et al., 1995; Danhorn et al., 2004; Minnikin et al., 1974; Soldo et al., 1999). Other studies have postulated that the Pst system is involved in intracellular invasion: Tn5 insertion mutants of the ΔpstS gene demonstrate a reduction in hilA and/or invasin gene expression in Salmonella enterica serotype Typhimurium (Lucas et al., 2000). A ΔpstS insertional mutant in Shigella flexneri (Runyen-Janecky et al., 2005) forms smaller plaques on Henle cell monolayers as compared with the wild-type; this correlates with the ability of the organism to cause disease, suggesting that the Pst system plays a role as an internal sensor of phosphate during cellular internalization. In E. coli, insertional mutants of the pst operon result in hyperinvasiveness to HEp-2 cells (Sinai & Bavoil, 1993). Some studies have suggested that P. mirabilis invades epithelial cells (Chippendale et al., 1994; Oelschlaeger & Tall, 1996); thus, it may be that the Pst system has a role in invasion during UTIs.
Finally, there exists evidence – in other micro-organisms – that the Pst transport system plays a role in biofilm formation, critical for the establishment of chronic, catheter-associated UTIs. Studies using null mutants in pstC and pstA genes have revealed that the Pst system negatively regulates biofilm formation in Pseudomonas aureofaciens PA147-2 (Monds et al., 2001). Since biofilm formation is critical for the establishment of P. mirabilis in the human host, mutations in the Pst system may explain the observed attenuation of the mutant strains. This was the hypothesis under examination in our study.
The data presented in Fig. 2 support our hypothesis: c.f.u. recovered from biofilms of the ΔpstA and ΔpstS mutant strains were significantly lower than in the wild-type. Furthermore, these data were reinforced by CLSM imaging and COMSTAT analysis (Fig. 3, Table 3). The ΔpstS mutant strain showed marked reductions in biomass, mean thickness and maximum thickness, while the ΔpstA mutant showed a less marked reduction in mean thickness and maximum thickness when compared with the wild-type (Table 3). Qualitative visual analysis of P. mirabilis biofilm by CLSM lent support to these data (Fig. 3). Therefore, not surprisingly, mutations in both the pstA and the pstS genes caused a reduction in biofilm growth by P. mirabilis HI4320.
A comparison of protein expression during P. mirabilis biofilm growth in pooled human urine under flow by MALDI-TOF/TOF was performed to provide some insight into the biofilm growth defect displayed by the ΔpstA and ΔpstS mutants. The data tentatively identified 18 proteins as being expressed exclusively under these conditions: 16 in the pst mutants and two in wild-type HI4320 (Table 4). Of the 16 proteins present exclusively in the pst mutants, three were known members of the pho regulon, including the alkaline phosphatase PhoA, the periplasmic phosphate binding protein PstS, and the glycerol 3-phosphate periplasmic binding protein UgpB. These would be expected to be upregulated in the pst mutant strains both because of the function of the pho regulon in phosphate uptake and because of the polarity of the mutants affecting expression of the genes downstream of the transposon insertion site, including phoU, a negative regulator of the pho regulon.
Other proteins expressed exclusively in the pst mutants may or may not be linked to biofilm formation. However, the functions of the upregulated proteins may aid our understanding of the cellular processes affected by the Pst system. Of interest are two proteins that have been shown in previous studies to be indirectly associated with the downregulation of biofilm formation (NagB and AphA). The glucosamine-6-phosphate isomerase NagB is part of the nag operon, whose genes are involved in the metabolism of N-acetylglucosamine 6-phosphate (GlcNAc6P), an important precursor of peptidoglycan and LPS synthesis. In E. coli, increases in the intracellular level of this precursor lead to the upregulation of the nag operon (Barnhart et al., 2006) and downregulation of curli and type 1 pili (Barnhart et al., 2006; Sohanpal et al., 2004), both known to be involved in biofilm formation (Kikuchi et al., 2005; Pratt & Kolter, 1998). Curli have been shown to assist in uroepithelial cell attachment in E. coli (Kikuchi et al., 2005). It may be that expression of NagB was induced in the pst mutants as a method of acquiring additional nutrients. Since the expression of NagB – encoded by the first gene in the nag operon – is upregulated in the Δpst mutants, type 1 pili may be downregulated, as occurs in E. coli. A homologue of the major type 1 pilin subunit FimA (PMI3435) is present in P. mirabilis HI4320. It is likely that downregulation of type 1 pili would negatively affect the ability of pst mutants to form biofilms.
The quorum-sensing-regulated activator AphA was also present in the proteomes of the Δpst mutant biofilms cultured under flow in pooled human urine. This protein is known to regulate virulence determinants in Vibrio cholerae (Kovacikova et al., 2005), in which it acts to prevent intracellular acidification during growth in glucose and at low pH as the organism is approaching stationary phase (Kovacikova et al., 2005). Previous studies in V. cholerae have determined that AphA is indirectly involved in biofilm formation, as this protein reduces the intracellular c-di-GMP concentration, which in turn decreases biofilm formation (Kovacikova et al., 2005). The acetoin reductase BudC, also expressed exclusively by the pst mutants, is involved in the 2,3-butanediol pathway, which protects the cell from acidification. These findings suggest that factors which may be important in biofilm formation are downregulated in the Δpst mutants, thus providing an explanation for the observed defect in biofilm formation.
Four proteins exclusively expressed in the pst mutants were related to nutrient scavenging. The mutants expressed the aminopeptidases PepE and PepP. The aminobenzoyl-glutamate utilization protein AbgA is part of the abg operon involved in the catabolism of folate, a compound essential for the synthesis of DNA, RNA and amino acids (Carter et al., 2007). The deoxyribose-phosphate aldolase DeoC is known to be important for the catabolism of deoxyribonucleosides (Han et al., 2004). Lastly, the protein ArnB is known to be involved in LPS biosynthesis by modifying lipid A through the addition of 4-deoxy-l-arabinose (l-Ara4N) on phosphate residues (Breazeale et al., 2005). Due to these modifications, Proteus spp. are not susceptible to cationic antimicrobial peptides. It is possible that although ArnB is being expressed, the addition of l-Ara4N is not possible due to the absence of phosphate residues on lipid A. If such modifications in LPS do indeed occur in P. mirabilis during times of phosphate limitation, they may affect the ability of these mutants to evade the immune response. Thus, these modifications in LPS structure may be partly or wholly responsible for the attenuation observed in vivo.
It has been suggested that the Δpst mutants are defective in biofilm formation due to the constitutive induction of the genes of the pho regulon by PhoB. Currently, it is not known whether this defect is the direct result of the loss of the Pst system or is the indirect effect of the constitutive expression of the pho regulon. Numerous pho regulon genes have been identified (Torriani-Gorini, 1994). One gene that has been demonstrated to be involved in biofilm formation and is found to be upregulated during exponential phase in pst mutants is the general stress stationary-phase regulator RpoS (Ruiz & Silhavy, 2003).
RpoS regulates a number of genes involved in protecting the bacterial cell against various stresses (starvation, osmotic shock, pH, oxidative) (Hengge-Aronis, 2002). There have been conflicting reports about the function of RpoS during biofilm formation. Some have reported that RpoS expression is enhanced during biofilm formation (Adams & McLean, 1999; Xu et al., 2001), while others have shown that mutations in the rpoS gene enhance biofilm formation (Corona-Izquierdo & Membrillo-Hernandez, 2002; Heydorn et al., 2002; Yun et al., 2007). It seems possible that RpoS is expressed transiently during some phase of biofilm development, as due to limited access to nutrients, bacterial growth rates within the centre of the biofilm – and especially deep inside microcolonies – are lower than those of bacteria located in the microcolony periphery (Moller et al., 1996). A homologue of rpoS is present in the HI4320 genome (PMI2236) (data not shown). Thus, it seems possible that mutations in pst affect biofilm development through an rpoS-dependent mechanism, the precise nature of which is as yet unknown.
Thus, our data suggest that the pst operon has a role in the formation of biofilm by P. mirabilis and furthermore that this effect may be at least in part responsible for the attenuated virulence of the pst mutant strains. Proteomic analysis suggests that this is due to altered expression of one or more of a number of proteins differentially expressed in the mutant strains. Future studies will focus upon elucidating precisely which of these is involved in biofilm formation and in the attenuated virulence of P. mirabilis.
Acknowledgments
We thank Anthony Haag at the Mass Spectrometry Core of the Biomolecular Resource Facility at the University of Texas Medical Branch for conducting MALDI-TOF MS and database analyses. This research was funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health grant R01-AI69568.
Abbreviations
CLSM, confocal laser scanning microscopy
2DGE, 2D gel electrophoresis
UTI, urinary tract infection
Footnotes
Two supplementary figures, showing 2D gel electrophoresis images of the proteome of P. mirabilis HI4320 cultured planktonically in LB, with and without annotation, are available with the online version of this paper.
References
- Adams, J. L. & McLean, R. J. (1999). Impact of rpoS deletion on Escherichia coli biofilms. Appl Environ Microbiol 65, 4285–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart, M. M., Lynem, J. & Chapman, M. R. (2006). GlcNAc-6P levels modulate the expression of Curli fibers by Escherichia coli. J Bacteriol 188, 5212–5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belas, R. (1996). Proteus mirabilis swarmer cell differentiation and urinary tract infection. In Urinary Tract Infections: Molecular Pathogenesis and Clinical Management, pp. 271–298. Edited by H. L. Mobley & J. W. Warren. Washington, DC: American Society for Microbiology.
- Benning, C., Huang, Z. H. & Gage, D. A. (1995). Accumulation of a novel glycolipid and a betaine lipid in cells of Rhodobacter sphaeroides grown under phosphate limitation. Arch Biochem Biophys 317, 103–111. [DOI] [PubMed] [Google Scholar]
- Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Brady, R. A., Leid, J. G., Camper, A. K., Costerton, J. W. & Shirtliff, M. E. (2006). Identification of Staphylococcus aureus proteins recognized by the antibody-mediated immune response to a biofilm infection. Infect Immun 74, 3415–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breazeale, S. D., Ribeiro, A. A., McClerren, A. L. & Raetz, C. R. (2005). A formyltransferase required for polymyxin resistance in Escherichia coli and the modification of lipid A with 4-amino-4-deoxy-l-arabinose. Identification and function of UDP-4-deoxy-4-formamido-l-arabinose. J Biol Chem 280, 14154–14167. [DOI] [PubMed] [Google Scholar]
- Buckles, E. L., Wang, X., Lockatell, C. V., Johnson, D. E. & Donnenberg, M. S. (2006). PhoU enhances the ability of extraintestinal pathogenic Escherichia coli strain CFT073 to colonize the murine urinary tract. Microbiology 152, 153–160. [DOI] [PubMed] [Google Scholar]
- Burall, L. S., Harro, J. M., Li, X., Lockatell, C. V., Himpsl, S. D., Hebel, J. R., Johnson, D. E. & Mobley, H. L. (2004). Proteus mirabilis genes that contribute to pathogenesis of urinary tract infection: identification of 25 signature-tagged mutants attenuated at least 100-fold. Infect Immun 72, 2922–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, E. L., Jager, L., Gardner, L., Hall, C. C., Willis, S. & Green, J. M. (2007). Escherichia coli abg genes enable uptake and cleavage of the folate catabolite p-aminobenzoyl-glutamate. J Bacteriol 189, 3329–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelli, P., Caronno, R., Ferrarese, S., Mantovani, V., Piffaretti, G., Tozzi, M., Lomazzi, C., Rivolta, N. & Sala, A. (2006). New trends in prosthesis infection in cardiovascular surgery. Surg Infect (Larchmt) 7 (suppl. 2), S45–S47. [DOI] [PubMed] [Google Scholar]
- Chippendale, G. R., Warren, J. W., Trifillis, A. L. & Mobley, H. L. (1994). Internalization of Proteus mirabilis by human renal epithelial cells. Infect Immun 62, 3115–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauser, K. R., Baker, P. & Burlingame, A. L. (1999). Role of accurate mass measurement (± 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal Chem 71, 2871–2882. [DOI] [PubMed] [Google Scholar]
- Corona-Izquierdo, F. P. & Membrillo-Hernandez, J. (2002). A mutation in rpoS enhances biofilm formation in Escherichia coli during exponential phase of growth. FEMS Microbiol Lett 211, 105–110. [DOI] [PubMed] [Google Scholar]
- Daigle, F., Fairbrother, J. M. & Harel, J. (1995). Identification of a mutation in the pst-phoU operon that reduces pathogenicity of an Escherichia coli strain causing septicemia in pigs. Infect Immun 63, 4924–4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danhorn, T., Hentzer, M., Givskov, M., Parsek, M. R. & Fuqua, C. (2004). Phosphorus limitation enhances biofilm formation of the plant pathogen Agrobacterium tumefaciens through the PhoR–PhoB regulatory system. J Bacteriol 186, 4492–4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher, A. L., Harmon, D., Kasif, S., White, O. & Salzberg, S. L. (1999). Improved microbial gene identification with GLIMMER. Nucleic Acids Res 27, 4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardy, J. L., Spencer, C., Wang, K., Ester, M., Tusnády, G. E., Simon, I., Hua, S., deFays, K., Lambert, C. & other authors (2003). psort-B: improving protein subcellular localization prediction for Gram-negative bacteria. Nucleic Acids Res 31, 3613–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharahdaghi, F., Weinberg, C. R., Meagher, D. A., Imai, B. S. & Mische, S. M. (1999). Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis 20, 601–605. [DOI] [PubMed] [Google Scholar]
- Gorg, A., Obermaier, C., Boguth, G., Harder, A., Scheibe, B., Wildgruber, R. & Weiss, W. (2000). The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 21, 1037–1053. [DOI] [PubMed] [Google Scholar]
- Han, T. K., Zhu, Z. & Dao, M. L. (2004). Identification, molecular cloning, and sequence analysis of a deoxyribose aldolase in Streptococcus mutans GS-5. Curr Microbiol 48, 230–236. [DOI] [PubMed] [Google Scholar]
- Hengge-Aronis, R. (2002). Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol Mol Biol Rev 66, 373–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydorn, A., Nielsen, A. T., Hentzer, M., Sternberg, C., Givskov, M., Ersboll, B. K. & Molin, S. (2000). Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146, 2395–2407. [DOI] [PubMed] [Google Scholar]
- Heydorn, A., Ersboll, B., Kato, J., Hentzer, M., Parsek, M. R., Tolker-Nielsen, T., Givskov, M. & Molin, S. (2002). Statistical analysis of Pseudomonas aeruginosa biofilm development: impact of mutations in genes involved in twitching motility, cell-to-cell signaling, and stationary-phase sigma factor expression. Appl Environ Microbiol 68, 2008–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, S. M., Lane, M. C., Harro, J. M., Shirtliff, M. E. & Mobley, H. L. (2008). The high-affinity phosphate transporter Pst is a virulence factor for Proteus mirabilis during complicated urinary tract infection. FEMS Immunol Med Microbiol 52, 180–193. [DOI] [PubMed] [Google Scholar]
- Jones, G. L., Muller, C. T., O'Reilly, M. & Stickler, D. J. (2006). Effect of triclosan on the development of bacterial biofilms by urinary tract pathogens on urinary catheters. J Antimicrob Chemother 57, 266–272. [DOI] [PubMed] [Google Scholar]
- Kikuchi, T., Mizunoe, Y., Takade, A., Naito, S. & Yoshida, S. (2005). Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol Immunol 49, 875–884. [DOI] [PubMed] [Google Scholar]
- Kovacikova, G., Lin, W. & Skorupski, K. (2005). Dual regulation of genes involved in acetoin biosynthesis and motility/biofilm formation by the virulence activator AphA and the acetate-responsive LysR-type regulator AlsR in Vibrio cholerae. Mol Microbiol 57, 420–433. [DOI] [PubMed] [Google Scholar]
- Lamarche, M. G., Dozois, C. M., Daigle, F., Caza, M., Curtiss, R., III, Dubreuil, J. D. & Harel, J. (2005). Inactivation of the pst system reduces the virulence of an avian pathogenic Escherichia coli O78 strain. Infect Immun 73, 4138–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, K. (2001). Riddle of biofilm resistance. Antimicrob Agents Chemother 45, 999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas, R. L., Lostroh, C. P., DiRusso, C. C., Spector, M. P., Wanner, B. L. & Lee, C. A. (2000). Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J Bacteriol 182, 1872–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew, J. A., Tan, Y. P., Srinivasa Rao, P. S., Lim, T. M. & Leung, K. Y. (2001). Edwardsiella tarda mutants defective in siderophore production, motility, serum resistance and catalase activity. Microbiology 147, 449–457. [DOI] [PubMed] [Google Scholar]
- Mendrygal, K. E. & Gonzalez, J. E. (2000). Environmental regulation of exopolysaccharide production in Sinorhizobium meliloti. J Bacteriol 182, 599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnikin, D. E., Abdolrahimzadeh, H. & Baddiley, J. (1974). Replacement of acidic phosphates by acidic glycolipids in Pseudomonas diminuta. Nature 249, 268–269. [DOI] [PubMed] [Google Scholar]
- Mobley, H. L. (1996). Virulence of Proteus mirabilis. In Urinary Tract Infections: Molecular Pathogenesis and Clinical Management, pp. 245–269. Edited by H. L. Mobley & J. W. Warren. Washington, DC: American Society for Microbiology.
- Mobley, H. L. & Warren, J. W. (1987). Urease-positive bacteriuria and obstruction of long-term urinary catheters. J Clin Microbiol 25, 2216–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley, H. L., Island, M. D. & Massad, G. (1994). Virulence determinants of uropathogenic Escherichia coli and Proteus mirabilis. Kidney Int Suppl 47, S129–S136. [PubMed] [Google Scholar]
- Moller, S., Pedersen, A. R., Poulsen, L. K., Arvin, E. & Molin, S. (1996). Activity and three-dimensional distribution of toluene-degrading Pseudomonas putida in a multispecies biofilm assessed by quantitative in situ hybridization and scanning confocal laser microscopy. Appl Environ Microbiol 62, 4632–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monds, R. D., Silby, M. W. & Mahanty, H. K. (2001). Expression of the Pho regulon negatively regulates biofilm formation by Pseudomonas aureofaciens PA147–2. Mol Microbiol 42, 415–426. [DOI] [PubMed] [Google Scholar]
- Monds, R. D., Newell, P. D., Gross, R. H. & O'Toole, G. A. (2007). Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0–1 biofilm formation by controlling secretion of the adhesin LapA. Mol Microbiol 63, 656–679. [DOI] [PubMed] [Google Scholar]
- Morris, N. S., Stickler, D. J. & McLean, R. J. (1999). The development of bacterial biofilms on indwelling urethral catheters. World J Urol 17, 345–350. [DOI] [PubMed] [Google Scholar]
- Musher, D. M., Griffith, D. P., Yawn, D. & Rossen, R. D. (1975). Role of urease in pyelonephritis resulting from urinary tract infection with Proteus. J Infect Dis 131, 177–181. [DOI] [PubMed] [Google Scholar]
- Oelschlaeger, T. A. & Tall, B. D. (1996). Uptake pathways of clinical isolates of Proteus mirabilis into human epithelial cell lines. Microb Pathog 21, 1–16. [DOI] [PubMed] [Google Scholar]
- O'Farrell, P. H. (1975). High resolution two-dimensional electrophoresis of proteins. J Biol Chem 250, 4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Orihuela, C. J., Mills, J., Robb, C. W., Wilson, C. J., Watson, D. A. & Niesel, D. W. (2001). Streptococcus pneumoniae PstS production is phosphate responsive and enhanced during growth in the murine peritoneal cavity. Infect Immun 69, 7565–7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole, G. A. & Kolter, R. (1998). Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28, 449–461. [DOI] [PubMed] [Google Scholar]
- O'Toole, G. A., Pratt, L. A., Watnick, P. I., Newman, D. K., Weaver, V. B. & Kolter, R. (1999). Genetic approaches to study of biofilms. Methods Enzymol 310, 91–109. [DOI] [PubMed] [Google Scholar]
- Peerbooms, P. G., Verweij, A. M. & MacLaren, D. M. (1984). Vero cell invasiveness of Proteus mirabilis. Infect Immun 43, 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirs, P., Lefèvre, P., Boarbi, S., Wang, X. M., Denis, O., Braibant, M., Pethe, K., Locht, C., Huygen, K. & Content, J. (2005). Mycobacterium tuberculosis with disruption in genes encoding the phosphate binding proteins PstS1 and PstS2 is deficient in phosphate uptake and demonstrates reduced in vivo virulence. Infect Immun 73, 1898–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt, L. A. & Kolter, R. (1998). Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 30, 285–293. [DOI] [PubMed] [Google Scholar]
- Rao, N. N. & Torriani, A. (1990). Molecular aspects of phosphate transport in Escherichia coli. Mol Microbiol 4, 1083–1090. [DOI] [PubMed] [Google Scholar]
- Rosenberg, H., Gerdes, R. G. & Chegwidden, K. (1977). Two systems for the uptake of phosphate in Escherichia coli. J Bacteriol 131, 505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruberg, S., Puhler, A. & Becker, A. (1999). Biosynthesis of the exopolysaccharide galactoglucan in Sinorhizobium meliloti is subject to a complex control by the phosphate-dependent regulator PhoB and the proteins ExpG and MucR. Microbiology 145, 603–611. [DOI] [PubMed] [Google Scholar]
- Ruiz, N. & Silhavy, T. J. (2003). Constitutive activation of the Escherichia coli Pho regulon upregulates rpoS translation in an Hfq-dependent fashion. J Bacteriol 185, 5984–5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runyen-Janecky, L. J., Boyle, A. M., Kizzee, A., Liefer, L. & Payne, S. M. (2005). Role of the Pst system in plaque formation by the intracellular pathogen Shigella flexneri. Infect Immun 73, 1404–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, K. & Camper, A. K. (2001). Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J Bacteriol 183, 6579–6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinai, A. P. & Bavoil, P. M. (1993). Hyper-invasive mutants define a novel Pho-regulated invasion pathway in Escherichia coli. Mol Microbiol 10, 1125–1137. [DOI] [PubMed] [Google Scholar]
- Sohanpal, B. K., El-Labany, S., Lahooti, M., Plumbridge, J. A. & Blomfield, I. C. (2004). Integrated regulatory responses of fimB to N-acetylneuraminic (sialic) acid and GlcNAc in Escherichia coli K-12. Proc Natl Acad Sci U S A 101, 16322–16327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldo, B., Lazarevic, V., Pagni, M. & Karamata, D. (1999). Teichuronic acid operon of Bacillus subtilis 168. Mol Microbiol 31, 795–805. [DOI] [PubMed] [Google Scholar]
- Soualhine, H., Brochu, V., Menard, F., Papadopoulou, B., Weiss, K., Bergeron, M. G., Legare, D., Drummelsmith, J. & Ouellette, M. (2005). A proteomic analysis of penicillin resistance in Streptococcus pneumoniae reveals a novel role for PstS, a subunit of the phosphate ABC transporter. Mol Microbiol 58, 1430–1440. [DOI] [PubMed] [Google Scholar]
- Stickler, D. J., King, J. B., Winters, C. & Morris, S. L. (1993). Blockage of urethral catheters by bacterial biofilms. J Infect 27, 133–135. [DOI] [PubMed] [Google Scholar]
- Stoodley, P., Sauer, K., Davies, D. G. & Costerton, J. W. (2002). Biofilms as complex differentiated communities. Annu Rev Microbiol 56, 187–209. [DOI] [PubMed] [Google Scholar]
- Torriani-Gorini, A. (1994). Introduction: the pho regulon of Escherichia coli. In Phosphate in Microbiology: Cellular and Molecular Biology, pp. 1–4. Edited by S. Silver, A. Torriani-Gorini & E. Yagil. Washington, DC: American Society for Microbiology.
- Walker, K. E., Moghaddame-Jafari, S., Lockatell, C. V., Johnson, D. & Belas, R. (1999). ZapA, the IgA-degrading metalloprotease of Proteus mirabilis, is a virulence factor expressed specifically in swarmer cells. Mol Microbiol 32, 825–836. [DOI] [PubMed] [Google Scholar]
- Warren, J. W., Tenney, J. H., Hoopes, J. M., Muncie, H. L. & Anthony, W. C. (1982). A prospective microbiologic study of bacteriuria in patients with chronic indwelling urethral catheters. J Infect Dis 146, 719–723. [DOI] [PubMed] [Google Scholar]
- Xu, K. D., Franklin, M. J., Park, C. H., McFeters, G. A. & Stewart, P. S. (2001). Gene expression and protein levels of the stationary phase sigma factor, RpoS, in continuously-fed Pseudomonas aeruginosa biofilms. FEMS Microbiol Lett 199, 67–71. [DOI] [PubMed] [Google Scholar]
- Yun, J. I., Cho, K. M., Kim, J. K., Lee, S. O., Cho, K. & Lee, K. (2007). Mutation of rpoS enhances Pseudomonas sp. KL28 growth at higher concentrations of m-cresol and changes its surface-related phenotypes. FEMS Microbiol Lett 269, 97–103. [DOI] [PubMed] [Google Scholar]