Abstract
Heterokaryon incompatibility (HI) is a nonself recognition phenomenon occurring in filamentous fungi that is important for limiting resource plundering and restricting viral transfer between strains. Nonself recognition and HI occurs during hyphal fusion between strains that differ at het loci. If two strains undergo hyphal fusion, but differ in allelic specificity at a het locus, the fusion cell is compartmentalized and undergoes a rapid programmed cell death (PCD). Incompatible heterokaryons show a macroscopic phenotype of slow growth and diminished conidiation, and a microscopic phenotype of hyphal compartmentation and cell death. To understand processes associated with HI and PCD, we used whole-genome microarrays for Neurospora crassa to assess transcriptional differences associated with induction of HI mediated by differences in het-c pin-c haplotype. Our data show that HI is a dynamic and transcriptionally active process. The production of reactive oxygen species is implicated in the execution of HI and PCD in N. crassa, as are several genes involved in phosphatidylinositol and calcium signalling pathways. However, genes encoding mammalian homologues of caspases or apoptosis-inducing factor (AIF) are not required for HI or programmed cell death. These data indicate that PCD during HI occurs via a novel and possibly fungal-specific mechanism, making this pathway an attractive drug target for control of fungal infections.
INTRODUCTION
The hallmark growth habit of filamentous fungi is the interconnected mycelial network that makes up a fungal colony (Glass et al., 2004). Hyphal fusion within a single colony is required to form the mycelial network, which permits transport of cytoplasm, organelles and nutrients. Hyphal fusion events can also occur between colonies that are genetically different (nonself fusion). The stability of heterokaryons formed by nonself fusion events is dependent on allelic specificity at nonself recognition loci termed het (for heterokaryon) or vcg (for vegetative compatibility group) (Garnjobst & Wilson, 1956; Glass & Dementhon, 2006; Leslie & Zeller, 1996; Saupe, 2000). If individuals that differ in het allelic specificity undergo hyphal fusion, the fusion cell is rapidly compartmentalized by septal plugging and dies (Biella et al., 2002; Jacobson et al., 1998; Marek et al., 2003). Heterokaryon incompatibility (HI) is associated with restriction of viral transfer between fungal individuals and prevention of resource plundering (Biella et al., 2002; Debets & Griffiths, 1998; Debets et al., 1994; van Diepeningen et al., 1997). Hyphal death during HI shows some features that are similar to apoptosis in mammalian cells, including shrinkage of plasma membrane, membrane-bound vesicle formation, DNA condensation and TUNEL-positive nuclei (Biella et al., 2002; Jacobson et al., 1998; Leslie & Zeller, 1996; Marek et al., 2003). Markers associated with apoptosis have also been reported upon exposure of various filamentous fungi to treatment with H2O2, acetic acid, farnesol, sphingolipids and amphotericin B (Castro et al., 2008; Chen & Dickman, 2005; Cheng et al., 2003; Leiter et al., 2005; Mousavi & Robson, 2004; Phillips et al., 2003; Semighini et al., 2006).
One of the best-studied HI systems is the Neurospora crassa het-c pin-c (Glass & Kaneko, 2003; Glass & Dementhon, 2006). The het-c locus, which encodes a glycine-rich plasma-membrane protein (Sarkar et al., 2002), has three specificites, het-c1, het-c2 and het-c3 (Kaneko et al., 2006; Saupe & Glass, 1997; Wu et al., 1998; Wu & Glass, 2001). The pin-c locus encodes a protein with a conserved, filamentous-fungal-specific domain termed HET (Kaneko et al., 2006), which is found in many genes involved in HI (Fedorova et al., 2005; Glass & Dementhon, 2006). Similar to N. crassa het-c, three allelic variants also occur at pin-c (Kaneko et al., 2006) (Fig. 1). Alleles at het-c and pin-c show severe linkage disequilibrium, such that isolates can be classified into one of three het-c pin-c haplotypes: het-c1 pin-c1, het-c2 pin-c2 and het-c3 pin-c3. Alleles at het-c have previously been shown to be under balancing selection (Wu et al., 1998), consistent with the role of the het-c pin-c haplotypes in mediating nonself recognition. HI requires genetic interactions between alternate het-c and pin-c alleles (i.e. het-c1 and pin-c2) and interactions between alternate het-c alleles (i.e. het-c1 and het-c2) (Fig. 1) (Glass & Dementhon, 2006; Kaneko et al., 2006; Sarkar et al., 2002). Although het loci have been cloned and characterized in both N. crassa and Podospora anserina (Glass & Dementhon, 2006; Saupe, 2000), relatively little is known about how interactions between alternate incompatibility proteins translate into the phenotype of HI and cell death.
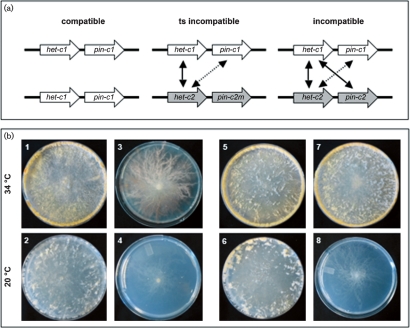
Fig. 1.
het-c and pin-c genetic interactions during HI. (a) Interactions between het-c and pin-c are required for nonself recognition (arrows), while interactions between het-c1 and het-c2 contribute to the severity of the HI phenotype (Kaneko et al., 2006). The het-c2–pin-c1 interaction is naturally temperature-sensitive (shown by dotted arrows). (b) Heterokaryons carrying identical alleles at het-c and pin-c show no genetic interactions and are fully compatible at all temperatures (plates 1 and 2; FGSC 4564+FGSC 6103). Heterokaryons of alternate het-c pin-c haplotype (het-c1 pin-c1+het-c2 pin-c2) are incompatible at all temperatures (plates 3 and 4; FGSC 4564+JH1). The temperature-sensitive incompatible (TS inc) heterokaryon (het-c1 pin-c1+het-c2 pin-c2m) is fully compatible at 34 °C, but is incompatible at 20 °C (plates 7 and 8; XK81+FGSC 456). Plates 5 and 6 contain a compatible heterokaryon (het-c2 pin-c2 +het-c2 pin-c2m; XK81+Xa-3).
In this study, we exploited the fact that the het-c2/pin-c1 interaction in laboratory strains is naturally temperature sensitive. Thus, a heterokaryon between a het-c1 pin-c1 strain and a pin-c2 loss-of-function mutant strain (het-c2 pin-c2m) is compatible and shows reduced cell death when grown at 34 °C. When transferred to 20 °C, this strain rapidly induces HI and high levels of cell death (Kaneko et al., 2006). We utilized this temperature-sensitive incompatible heterokaryon to evaluate morphological and transcriptional alterations associated with HI as a measure of how the physiological status of the colony was altered by the induction of HI, using oligonucleotide arrays representing all annotated genes for N. crassa (Kasuga et al., 2005; Tian et al., 2007). From expression profiling data, we predicted and showed that reactive oxygen species (ROS), phosphatidylinositol and calcium signalling pathways play a role early during HI and programmed cell death (PCD). However, homologues to genes involved in apoptosis in higher eukaryotic species, such as caspases (metacaspases) and apoptosis-including factor (AIF), were not required for HI in N. crassa.
METHODS
Strains and growth conditions.
All strains (see Supplementary Table S1, available with the online version of this paper) were grown on Vogel's minimal medium (Vogel, 1956) or Bird's minimal medium (BMM) (Metzenberg, 2004) with supplements as required, and crossed on Westergaard's medium (Westergaard & Mitchell, 1947). Heterokaryons with the helper strain (FGSC 4564) were used as females in crosses (Perkins, 1984). Transformations were performed as described by Margolin et al. (1997). Heterokaryons were forced by co-inoculation of conidial suspensions of each pair of isolates onto minimal medium. For profiling experiments, two BMM plates per time point were overlaid with sterilized cellophane circles and inoculated with 8 mm plugs of hyphae. Heterokaryons were grown in constant light for 16 h at 34 °C prior to transfer to 20 °C. Plates were sampled at each time point by peeling the cellophane/hyphae and freezing immediately in liquid N2. The transcriptional profiling experiment was repeated twice in its entirety.
Microarray construction and hybridization.
Microarrays were constructed and performed as described by Tian et al. (2007). Briefly, RNA was extracted using Trizol (Invitrogen Life Technologies) according to the manufacturer's protocol and cleaned using the RNeasy miniprep protocol (Qiagen). cDNA was prepared using the ChipShot Indirect cDNA Synthesis and labelling protocol from Promega/Corning (Promega) according to the manufacturer's protocol with the following exceptions: dried cDNA was resuspended in 9 μl 0.06 M sodium bicarbonate, and quenching was accomplished by the addition of 4.5 μl 4 M hydroxylamine. Hybridizations were performed using Pronto kits (Promega) according to the manufacturer's protocol and as described by Tian et al. (2007). Images were acquired with an Axon GenePix 4000B scanner and GenePix Pro 6 software (Molecular Devices); each slide was also examined manually.
Data analysis.
Spots with intensities greater than the background plus 3 standard deviations and less than 0.02 % of pixels saturated were selected for further analysis. We used a closed-circuit design for microarray analysis (Townsend & Hartl, 2002; Townsend, 2003) (see Fig. 3a), and determined relative gene expression levels using Bayesian Analysis of Gene Expression Levels (BAGEL) (Townsend & Hartl, 2002; Townsend, 2004). All expression data are deposited at the Neurospora functional genomics database (http://www.yale.edu/townsend/Links/ffdatabase/introduction.html).
Fig. 3.
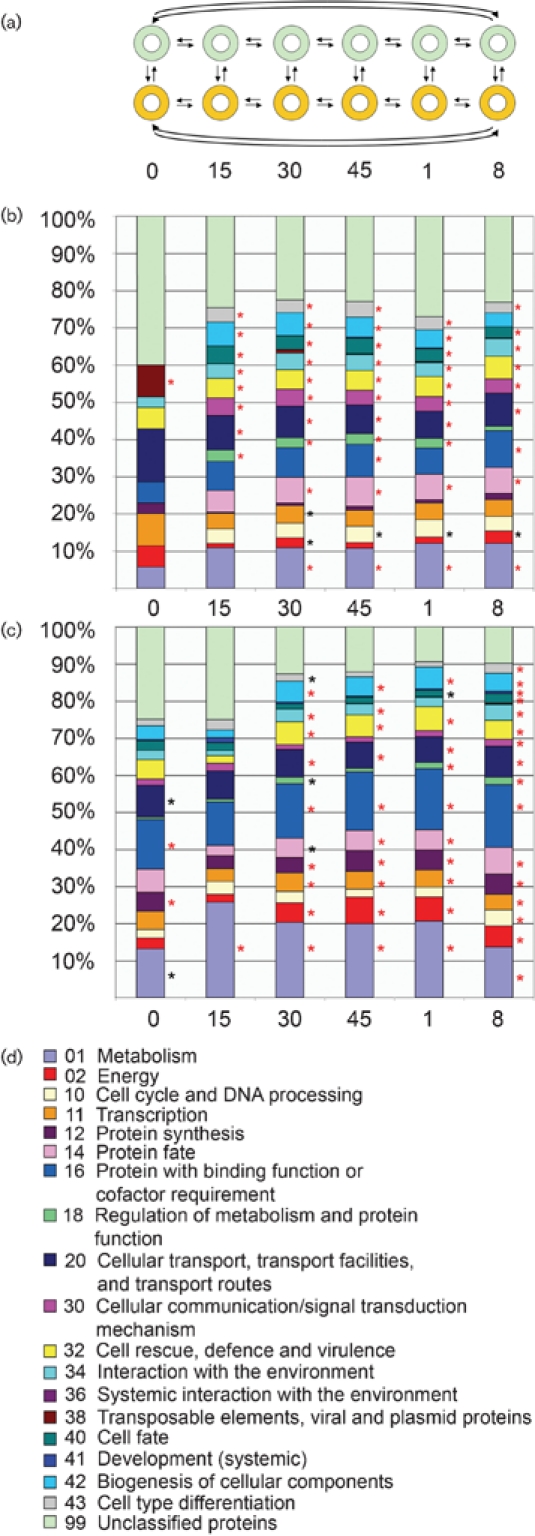
Functional categories of up- and downregulated genes during HI. (a) Closed-circuit experimental design used for microarray analysis. Doughnuts represent sampled cultures; arrows represent hybridizations, where the arrowhead points to the sample labelled with Cy5 and the tail points to the sample labelled with Cy3. Green, compatible heterokaryon (XK81+Xa-3); orange, TS inc heterokaryon (XK81+FGSC 456). Time points of analysis are indicted below. (b) Functional categories of genes that were upregulated in the TS inc heterokaryon at time points before and after transfer to 20 °C. (c) Functional categories of genes that were downregulated in the TS inc heterokaryon at time points before and after transfer to 20 °C. Asterisks indicate statistically over-represented functional categories (red asterisks indicate P<0.01, black asterisks indicate P<0.05). (d) Key for functional categories shown in (b) and (c).
Genes were clustered based on their differential expression in the incompatible heterokaryon relative to the compatible, accounting for confidence intervals (Eisen et al., 1998). Comparability of the two replicate experiments was assessed in two ways. Firstly, cDNA from the same conditions at the same time point in the two experiments was co-hybridized to a single slide and the raw data were visualized as a scatter plot. Secondly, correlations were calculated using Microsoft Excel for the normalized BAGEL data at time 0 for the two experiments, comparing compatible heterokaryons in the two experiments, incompatible heterokaryons in the two experiments, and compatible vs incompatible heterokaryons for the combined experimental data.
Methylene blue and DCF staining.
Segments of cellophane and mycelium were laid on slides and stained with a 0.003 % (w/v) solution of the vital dye methylene blue (Suzuki et al., 2000). For ROS detection, compatible and incompatible heterokaryons were grown on sterile cellophane laid over BMM plates. Sections of cellophane with hyphae from the edges of the colony were cut, laid on a microscope slide and treated with 10 μM 2′,7′-dihydrodichlorofluorescein diacetate (H2DCFDA), which is oxidized by ROS to dichlorofluorescein (DCF). Heterokaryons were incubated with the dye solution for 1–5 min prior to microscopic examination. Images were acquired under differential interference contrast (DIC) and under fluorescence with a Zeiss Axioskop II fluorescence microscope using OpenLab 4.0.3 software. Images were resized in Adobe Photoshop CS. The proportion of dead (methylene blue) or fluorescent (DCF) compartments was counted as a percentage of the total number of compartments in an image at each time point. Calculations were replicated 10–20 times per sample and per time point, and the mean taken.
Construction of strains.
The Δpp-1 and Δmak-2 strains are ascospore lethal, so we used the Diploid (Dip) mutant strain (FGSC 9537) for strain construction. In Dip crosses, about 2/3 of the ascospores are large and diploid. Streaking Dip progeny onto sorbose plates result in restoration of haploid strains. First, Dip was crossed with a his-3 strain (R15-07) to obtain a his-3; Dip strain (R23-20; Supplementary Table S1). This strain was crossed to Δpp-1 and Δmak-2 strains to construct his-3; Δpp-1 and his-3; Δmak-2 strains (Supplementary Table S1). Strain genotype was verified by PCR. Other deletion strains were crossed with het-c1 pin-c1 strains and het-c2 pin-c2 strains with auxotrophic markers to construct strains for heterokaryon tests (Supplementary Table S1).
RESULTS
Transfer of the temperature-sensitive incompatible (het-c1 pin-c1+het-c2 pin-c2m) heterokaryon to permissive temperature results in rapid induction of cell death
Under laboratory conditions, strains of alternate het specificity can be forced to grow as heterokaryons by plating individuals with complementary nutritional requirements onto minimal medium; growth is extremely slow, conidiation is suppressed (Fig. 1) and hyphal compartmentation and death occurs in ∼30–35 % of the hyphal compartments (Kaneko et al., 2006; Perkins, 1988b; Xiang & Glass, 2002). Forced heterokaryons of identical het specificity are indistinguishable from a wild-type strain (Fig. 1). When grown at 34 °C, the macroscopic phenotype of the temperature-sensitive (het-c1 pin-c1+het-c2 pin-c2m) (TS inc) heterokaryon is also indistinguishable from a wild-type heterokaryon (Kaneko et al., 2006), but shows a typical HI phenotype upon transfer to 20 °C (Kaneko et al., 2006) (Fig. 1). To evaluate the rapidity of induction of HI and PCD in the TS inc heterokaryon, we assessed both the macroscopic and microscopic events associated with the induction of HI over an 8 h time-course. As a control, we used a heterokaryon between the het-c2 pin-c2m mutant strain and a strain of identical het-c pin-c haplotype (het-c2 pin-c2 arg-5; pan-2 A; Supplementary Table S1). The macroscopic phenotype of this heterokaryon was identical to that of a wild-type strain at all temperatures.
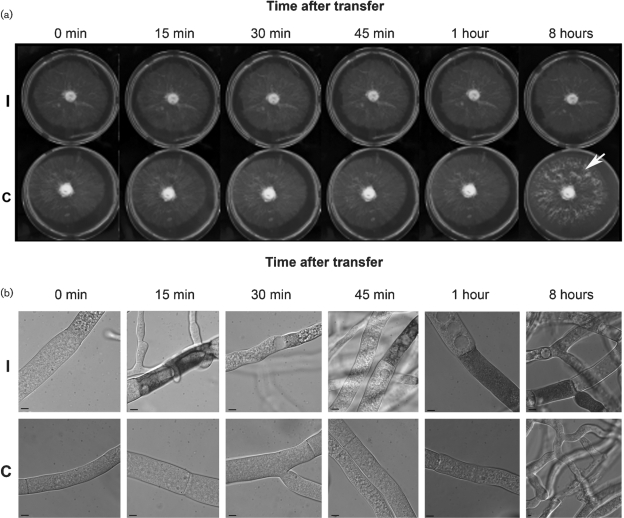
The het-c2 pin-c2 compatible and TS inc heterokaryons were grown for 16 h at 34 °C and then transferred to 20 °C. Prior to transfer, the compatible and the TS inc heterokaryons showed a similar macroscopic and microscopic phenotype (Fig. 2, time 0; Table 1). Although few macroscopic differences are apparent between the compatible and the TS inc heterokaryons following transfer to 20 °C (Fig. 2a), many differences are evident at a microscopic level (Fig. 2b). Hyphal compartments in the TS inc heterokaryon became highly vacuolated and exhibited a granular appearance (Fig. 2b, row I). Using the vital dye methylene blue, many compartmentalized and dead hyphal segments were observed in the TS inc heterokaryon (Fig. 2b, row I), which were evident as early as 15 min after transfer. The percentage of dead hyphal compartments greatly increased in the TS inc heterokaryon, reaching a peak of ∼37 % at 1 h after transfer (Table 1). Macroscopic morphological differences between the compatible and the TS inc heterokaryons were apparent after ∼8 h of incubation at permissive temperature. As shown in Fig. 2(a), the compatible heterokaryon grew to the edge of the plate and produced aerial hyphae and conidia (row C), while the TS inc heterokaryon showed little growth and remained aconidial (row I).
Fig. 2.
Macroscopic and microscopic characteristics of HI. (a) Growth of a heterokaryon between XK81 and FGSC 456 (temperature-sensitive incompatible, TS inc, row I) compared to growth of a heterokaryon between XK81 and Xa-3 (compatible; row C) grown at permissive temperature for HI (20 °C). Both heterokaryons were grown at 34 °C for 16 h (0 min) and then transferred to 20 °C, and photographs of heterokaryons growing on Petri dishes were taken at the time points indicated following shift to 20 °C. The arrow shows conidiation observed in the compatible heterokaryon after 8 h, while the TS inc heterokaryon shows no growth or conidiation during the same time period. (b) DIC micrographs of hyphae from the TS inc (row I) and compatible (row C) heterokaryons stained with the vital dye methylene blue and photographed prior to transfer (0 min) and at the indicated time points after transfer to 20 °C.
Table 1.
Percentage of dead hyphal compartments in the compatible (Xa-3+XK81) and TS incompatible (XK81+FGSC 456) heterokaryons
| Time point | Compatible heterokaryon | TS incompatible heterokaryon |
|---|---|---|
| 0 min | 8.4 % | 11.6 % |
| 15 min | 15.4 % | 20.7 % |
| 30 min | 17.1 % | 32.9 % |
| 45 min | 14.0 % | 31.5 % |
| 1 h | 10.0 % | 37.1 % |
| 8 h | 7.8 % | 22.4 % |
Transcriptional profiling reveals that the expression of a large number of genes is elevated during induction of HI
The rapid induction of HI and PCD upon transfer of the TS inc heterokaryon to permissive temperature makes it an attractive model for assessing physiological responses associated with HI. Although growth rate is severely reduced (from 7 cm day−1 to 1–2 cm day−1), only ∼30 % of the hyphal compartments are dead, even when all nuclei carry an incompatible het-c pin-c haplotype (Glass & Kaneko, 2003; Mylyk, 1975; Perkins, 1988a). Thus, we anticipated that transcriptional profiling would reveal physiological processes associated both with cell death and with cell survival.
We sampled RNA from the het-c2 pin-c2 compatible and the TS inc heterokaryons prior to transfer (0 min) and at 15 min, 30 min, 45 min, 60 min and 8 h post-transfer to permissive temperature. Expression levels of a number of candidate genes were verified using quantitative RT-PCR (Supplementary Fig. S1). Strong statistical support was obtained for relative expression levels of 3447 genes (Supplementary Table S2). To identify variation in expression profiles at each time point between the compatible heterokaryon and the TS inc heterokaryon, the data were grouped into clusters of genes whose expression levels significantly increased or decreased in the TS inc heterokaryon relative to the compatible heterokaryon at each time point. Thus, for each time point, two clusters (‘UP’ and ‘DOWN’) for a total of 12 clusters were generated (Supplementary Table S2). Genes that showed a statistically significant difference in expression levels for each cluster were evaluated by functional category analysis using FunCat (http://mips.gsf.de/projects/funcat) (Ruepp et al., 2004) (Fig. 3b–d). Fig. 3(b) shows the fraction of genes in each category in each ‘UP’ cluster, while Fig. 3(c) shows the fraction of genes in each category in the ‘DOWN’ clusters. Asterisks indicate functional categories that were statistically over-represented in each cluster.
Prior to transfer, the compatible and TS inc heterokaryons were similar in expression patterns, although a number of genes in specific pathways showed transcriptional differences even at this time point (Fig. 3b, c; time 0). The functional category ‘Transposable elements, viral and plasmid proteins’ was enriched in the UP category (27 genes) in the TS inc heterokaryon at time 0, the only time point during the entire time-course that showed enrichment for this category. There were more genes in the DOWN category at time 0 (218 genes), showing enrichment of functional categories ‘Protein synthesis’ and ‘Metabolism’. These differences could possibly be due to the fact that the TS inc and compatible heterokaryons are not completely isogenic. However, upon transfer, expression profiling revealed a rapid and diverse transcriptional response to HI, with the numbers of both down- and upregulated genes increasing to ∼500–700.
At all time points after transfer (15 min to 8 h), genes in the functional categories ‘Cellular transport, transport facilities and transport routes’ (P<0.001), ‘Cellular communication/signal transduction mechanism’ (P<0.0001), ‘Cell rescue, defence and virulence’ (P<0.01), ‘Interaction with the environment’ (P<0.05), ‘Cell fate’ (P<0.0001) and ‘Cell type differentiation’ (P<0.001) increased in relative expression level in the TS inc heterokaryon (Fig. 3b). Within the ‘Cellular communication/signal transduction mechanism’ functional category, genes involved in enzyme-mediated signal transduction (e.g. kinases, GTPases, heterotrimeric G proteins), second messenger signal transduction and transmembrane signal transduction pathways showed a significant increase in expression level (Supplementary Table S2). In addition, a number of genes involved in phosphatidylinositol, Ca2+ and cAMP signalling pathways also showed increased expression levels at the early time points, including the metacaspase-2 homologue NCU09882.
The functional category ‘Metabolism’ was enriched at all time points in the DOWN categories (Fig. 3c). In the remaining time points, genes within functional categories ‘Energy’ (P<1.5×10−13), ‘Transcription’ (P<0.01), ‘Protein synthesis’ (P<0.0005), ‘Protein fate’ (P<0.05), ‘Protein with binding function or cofactor requirement’ (P<0.0005), ‘Regulation of metabolism and protein function’ (P<0.05), ‘Cellular transport, transport facilities and transport routes’ (P<0.05), ‘Cell rescue, defence and virulence’ (P<5×10−9) and ‘Biogenesis of cellular components’ (P<5×10−8) were all enriched. The reduction in the expression in genes within these functional categories is consistent with the severe reduction in growth rate associated with HI.
It is possible that the induction of HI is associated with stress responses, ultimately resulting in cell death. We therefore evaluated whether the transcriptional changes in the TS inc heterokaryon were similar to other stress-response profiles reported in N. crassa (Tian et al., 2007; Videira et al., 2009). We first compared the HI transcriptional profile (30 min time point) to the transcriptional profile of N. crassa treated with the cell-death-inducing reagent phytospingosine (PHS) (Videira et al., 2009); treatment of germinated conidia with 10 μg PHS ml−1 resulted in ∼30 % cell death as assayed via vital dye staining. Of the 386 genes induced during HI, 147 overlapped with the 659 genes induced during PHS treatment, a significant enrichment (P<0.005) (Supplementary Table S3). Functional categories enriched in the overlapping dataset included ‘Cellular communication/signal transduction’ (P<6×10−7), ‘Cell fate’ (P<2×10−6), ‘Cell rescue, defence and virulence’ (P<1×10−4) and ‘Protein fate’ (P<2×10−4). Enriched subcategories included many signal transduction categories such as protein kinases, modification by phosphorylation, calcium binding, second messenger signalling and regulation of protein activity. In particular, three genes that showed very high expression levels during HI – NCU04554 (putative endochitinase), NCU05309 (putative RNase) and NCU05693 (putative GTPase) (Table 2; asterisks) – were also upregulated in the PHS dataset.
Table 2.
Genes tested for function in HI
| Gene name/ID† | MIPS-defined function | Fold induction at 30 min |
|---|---|---|
| Phosphatidylinositol-related genes | ||
| NCU01047 | Related to inositol polyphosphate 5-phosphatase OCRL-1 | 3.23 |
| NCU04379* | Probable regulator of phosphatidylinositol-4-OH kinase protein | 6.92 |
| NCU06245* | Related to 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase (phospholipase C) | 2.67 |
| NCU06877 | Related to phosphatidylinositol transfer protein | 2.22 |
| NCU10397 | Related to phosphatidylinositol 4-kinase | - |
| Calcium-related genes | ||
| NCU06177 | Related to calcium/calmodulin-dependent protein kinase C | 3.33 |
| NCU06366* | Related to vacuolar Ca2+/H+ antiporter | 2.03 |
| Highly upregulated genes | ||
| NCU04554* | Probable endochitinase class V precursor | 34 |
| NCU05309* | Related to Cut9 interacting protein Scn1 | 62 |
| NCU05693* | Related to RBTMx2 protein | 36 |
*Genes upregulated following exposure to PHS (Videira et al., 2009).
†Gene name/ID from Borkovich et al. (2004) and the Broad Institute (http://www.broad.mit.edu/annotation/genome/neurospora/Home.html).
To determine whether the overlap dataset between HI- and PHS-induced genes represented a general stress response, we also compared the transcriptional profile of N. crassa undergoing HI to the profile of N. crassa undergoing amino acid starvation (Tian et al., 2007). Unlike the HI/PHS comparison, only 15 genes in the HI dataset (336 genes) overlapped with the amino acid starvation response (334 genes), which was not significant. However, of the 491 genes that showed reduced expression level in the TS inc heterokaryon, 35 genes overlapped the set of genes that showed reduced expression during amino acid starvation (P<0.005). Sixteen of these genes in the HI dataset also overlapped with the 103 genes downregulated during PHS treatment. Within this overlap dataset, almost half of the genes are predicted to be involved in metabolism or protein synthesis. Thus, downregulation of genes involved in both metabolism and protein synthesis may be a part of a general stress response in N. crassa.
Screening mutants in HI-induced genes for effects on cell death
We constructed homozygous deletion strains (Dunlap et al., 2007) (Supplementary Table S1) of alternate het-c pin-c haplotype (het-c* pin-c*) via crosses to determine whether deletions of selected genes affected HI and/or cell death. We first evaluated strains containing deletions of three genes that showed a significant increase in expression level in both the HI and PHS datasets (Table 2). The NCU04554 gene encodes an endochitinase (gh18-5), NCU05309 encodes a hypothetical protein similar to the Schizosaccharomyces pombe gene scn3, which has an RNase domain (Kimata & Yanagida, 2004), and NCU05693 contains a dynamin domain and is homologous to an Mx (myxovirus resistance) GTPase in mouse and human (Sadler & Williams, 2008). NCU05693 was also recently identified as a dsRNA-activated gene in N. crassa (Choudhary et al., 2007).
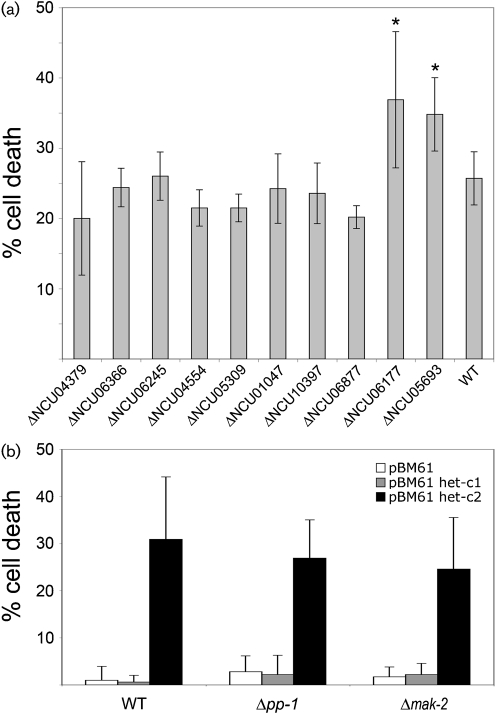
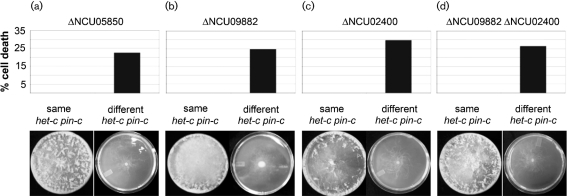
Heterokaryons of alternate het-c pin-c haplotype (het-c* pin-c*) with homozygous deletions of gh18-5 (NCU04554) or NCU05309 showed an identical incompatibility and cell death phenotype to that of a wild-type incompatible heterokaryon (Fig. 4a). A heterokaryon of het-c* pin-c* with homozygous deletions in NCU05693 had a similar macroscopic phenotype to a wild-type incompatible heterokaryon, but showed significantly more cell death (∼35 %; Fig. 4a). These data suggest that NCU05693 may function to promote cell survival in an incompatible heterokaryon.
Fig. 4.
Effect of deletion mutations on the induction and phenotype of HI. (a) Cell death percentages in heterokaryons carrying homozygous deletions of the indicated genes, but of alternate het-c pin-c haplotype compared to cell death in a wild-type incompatible heterokaryon. (b) Cell death percentages in wild-type (FGSC 6103), Δpp-1 and Δmak-2 transformants bearing either het-c1, het-c2 or pBM61 integrated at the his-3 locus. Numbers are means±sem of 10 plates, and for each plate 50–100 cells were counted.
The functional category of cellular communication and signal transduction was enriched in the UP category at almost all of the time points in the TS inc heterokaryon (Fig. 3b), including many genes in the phosphatidylinositol signalling pathways (Supplementary Table S4; FunCat 30.01.09.11, P<0.05). Phosphatidylinositol is found in the membranes of all eukaryotic species, and derivatives of this molecule function in diverse cellular processes, including cell signalling, cell motility, GPI protein anchoring, vesicular trafficking, cytoskeleton remodelling and cytoprotection (Michell, 2008; Strahl & Thorner, 2007). Deletion strains were available for predicted phophatidylinositol signalling genes including NCU01047 (inositol polyphosphate 5-phosphatase), NCU04379 (regulator of phosphatidylinositol-4-OH kinase), NCU06245 (plc-1; phospholipase C), NCU06877 (phosphatidylinositol transfer protein) and NCU10397 (phosphatidylinositol 4-kinase) (Table 2). NCU04379 and NCU06245 also showed high relative expression levels during PHS-induced cell death (Videira et al., 2009). However, heterokaryons of alternate het-c pin-c haplotype and carrying homozygous deletions of NCU01047, NCU04379, NCU06245, NCU06877 or NCU10397 (Supplementary Table S1) showed a typical HI phenotype, with cell death percentages comparable to a wild-type incompatible heterokaryon (Fig. 4).
In addition to the phosphatidylinositol signalling pathway, enrichment for genes involved in calcium signalling, homeostasis, transport and binding was also observed in the TS inc heterokaryon (Supplementary Table S4; FunCat 30.01.09.03, P<0.05). Calcium is involved in multiple aspects of growth and development in filamentous fungi, including circadian rhythms and pathogenesis (Borkovich et al., 2004). One of the canonical Ca2+ signalling pathways involves the hydrolysis of phosphatidylinositol 4,5-diphosphate by phospholipase C into 1,2-diacylglycerol and inositol 1,4,5-triphosphate (IP3), resulting in protein kinase C activation and Ca2+ release (Strahl & Thorner, 2007). We therefore constructed strains of alternate het-c* pin-c* haplotype carrying deletions of NCU06177, encoding calcium/calmodulin-dependent protein kinase C (PKC) and NCU06366, which encodes a protein related to vacuolar Ca2+/H+ antiporter; NCU06366 also showed increased expression levels during PHS-induced cell death (Videira et al., 2009). The phenotype of the homozygous ΔNCU06366 het-c* pin-c* heterokaryons was indistinguishable from that of a wild-type incompatible heterokaryon. Although the ΔNCU06177 het-c* pin-c* heterokaryon was macroscopically similar to an incompatible heterokaryon, it exhibited a statistically significant increase in cell death (Fig. 4a). N. crassa contains two PKC homologues; NCU06544 (pkc) has been shown to be involved in regulating the N. crassa circadian clock via modification of the transcription factor White Collar-1 (Arpaia et al., 1999), while the function of NCU06177 has not been characterized.
Among the enriched subcategories within the ‘Cellular communication/signal transduction mechanism’ functional category were genes encoding protein kinases. Two well-characterized members of a MAPK pathway in N. crassa are pp-1 and mak-2. The pp-1 gene (NCU00340) encodes a transcription factor homologous to Saccharomyces cerevisiae STE12, while mak-2 encodes a homologue of FUS3 (Li et al., 2005); STE12 and FUS3 constitute part of the pheromone response signal transduction pathway. In S. cerevisiae, inappropriate stimulation of this pathway results in cell death that exhibits apoptotic features (Severin & Hyman, 2002; Zhang et al., 2006). The Δpp-1 and Δmak-2 mutants are also defective in hyphal fusion (Fleissner et al., 2008), suggesting the possibility of an interaction between the cell death and cell fusion pathways.
Because both Δpp-1 and Δmak-2 mutants fail to form heterokaryons, we evaluated the effect of these mutations on HI by a transformation protocol (Sarkar et al., 2002; Saupe et al., 1996; Saupe & Glass, 1997; Wu & Glass, 2001). We constructed his-3; het-c1 pin-c1; Δpp-1 and his-3; het-c1 pin-c1; Δmak-2 strains using a Dip (Diploid) strain because mutations in both Δpp-1 and Δmak-2 result in ascospore lethality (see Methods; Supplementary Table S1). Wild-type, Δpp-1 and Δmak-2 strains were transformed with pBM61-het-c1 or pBM61-het-c2 (Margolin et al., 1997). The het-c1 pin-c1; Δpp-1 and het-c1 pin-c1; Δmak-2 transformants bearing the het-c1 allele showed an identical phenotype to the untransformed and pBM61 vector control strains (growth rate ∼1.8 cm day−1 and ∼1.4 cm day−1, respectively). The het-c1 pin-c1; Δpp-1 and het-c1 pin-c1; Δmak-2 transformants bearing the het-c2 allele showed a reduced growth rate (0.8 cm day−1 and 0.7 cm day−1, respectively) and wild-type levels of cell death (Fig. 4b).
ROS are induced during HI
Many proteins play a role in the reaction to oxidative stress, either in the reduction of oxygen radicals or in the induction of other genes that respond to oxidative stress. A subset of these genes showed increased expression levels in the TS inc heterokaryon, including genes for generation of ROS (NADPH oxidase nox-1; NCU02110) and ROS scavengers (glutaredoxin, NCU01219; cyc-1 cytochrome c, NCU01808) (Supplementary Table S2). ROS and oxidative stress have been implicated in apoptosis in mammals, S. cerevisiae and other filamentous fungi (Chandra et al., 2000; Chen & Dickman, 2005; Ludovico et al., 2005; Madeo et al., 1999; Semighini et al., 2006).
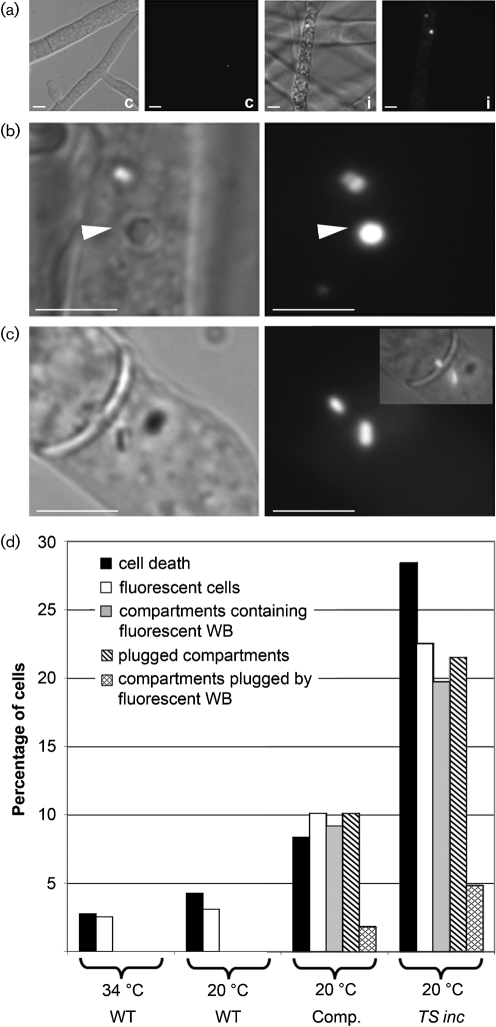
The compatible and TS inc heterokaryons were evaluated for the oxidation of 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) to dichlorofluorescein (DCF), an indicator dye for ROS (LeBel et al., 1992). As shown in Fig. 5, ROS were generated in the cytoplasm of TS inc heterokaryon within 5–15 min post-transfer to permissive conditions, but not in the compatible heterokaryon. The percentage of ROS-producing compartments in the TS inc heterokaryon was similar to the percentage of dead compartments (Fig. 5d). The compatible heterokaryon (Xa-3+XK81) showed a slight elevation in the percentage of dead/fluorescent cells as compared to parental strains (Fig. 5d), most likely due to strain differences.
Fig. 5.
ROS are produced during HI and are associated with Woronin bodies. (a) As assessed by DCF fluorescence, the presence of ROS was infrequent in a compatible heterokaryon (c; XK81+Xa-3). DCF fluorescence was observed when HI was induced in the TS inc heterokaryon (i; XK81+FGSC 456). DCF fluorescence was within discrete hyphal compartments, indicating that the septal pores were plugged in these compartments. (b) Bright dots of fluorescence co-localized with Woronin bodies (WBs) (arrowheads). WB are visible in the DIC images (distinguished by their hexagonal shape) and fluoresced brightly when stained with DCF. (c) Fluorescent WBs were only occasionally associated with plugged septa (merged image in inset). (d) Cell death and DCF fluorescence was quantified in strains Xa-3 (WT) and XK-81 (data not shown) at 34 °C and 20 °C. Data for XK-81 did not differ statistically from those for Xa-3. No fluorescent WBs were observed. Cell death and DCF fluorescence were quantified in the compatible heterokaryon (Xa-3+XK81) and the TS inc heterokaryon (XK81+FGSC 456) at 20 °C. The number of dead hyphal compartments with fluorescent WB and how often fluorescent WB were associated with septa was quantified in the compatible (Xa-3+XK81) and TS inc (XK81+FGSC 456) heterokaryons. DCF staining indicated an almost complete overlap between compartments that were dead, fluorescent, contained fluorescent WB, and were plugged. However, less than 5 % of the plugged compartments showed fluorescent WB at the septa.
In the TS inc heterokaryon, DCF fluorescence was concentrated at organelles that resembled the peroxisome-like Woronin body (Fig. 5b). Woronin bodies are membrane-bound modified peroxisomes found in the cytoplasm of filamentous ascomycete species and which plug the septal pore following catastrophic injury (Jedd & Chua, 2000; Markham & Collinge, 1987; Tenney et al., 2000); previous studies have associated an unknown peroxidase/catalase activity with Woronin bodies (Schliebs et al., 2006; Tenney et al., 2000). In HI, septal plugging is a rapid response during het-c pin-c incompatibility that occurs less than 5 min post-fusion (Glass & Kaneko, 2003). DCF fluorescence in hyphal compartments in the TS inc heterokaryon was always associated with Woronin bodies, although these were only occasionally observed at septa (Fig. 5c, d). These data indicate that although ROS were associated with Woronin bodies during HI, septal plugging during HI is independent of the Woronin body.
Homologues of caspases or apoptosis-inducing factor (AIF1) are not required for HI or PCD in N. crassa
The induction of PCD in a number of systems includes activation of a MAPK cascade, accumulation of ROS, release of cytochrome c from mitochondria and activation of caspases (Chandra et al., 2000; Stridh et al., 1998) (reviewed by Adams, 2003). A survey of mammalian and fungal proteins known to play a role in PCD revealed that ∼42 genes are conserved in filamentous fungal genomes (Fedorova et al., 2005). The ‘Cell death and apoptosis’ functional category of genes was enriched at time points 15 min to 1 h (FunCat 40.10.02, P<0.05). Of the 42 N. crassa cell death homologues, 11 increased in expression level in the TS inc heterokaryon upon induction of HI (Table 3), including genes encoding metacaspase (NCU09882 and NCU02400), cytochrome c (NCU01808) and heat-shock protein HSP70 (NCU09602). We chose a subset of these genes to evaluate for HI phenotypes.
Table 3.
Predicted homologues of apoptotic genes that increase in relative expression level during HI
| Protein† | Name/ID‡ | MIPS-defined function | Largest fold induction (time) |
|---|---|---|---|
| Cyc1p | NCU01808 | Cytochrome c | 2.4 (8 h) |
| Yca1p | NCU09882; NCU02400 | Related to metacaspase 2; probable caspase | 6.3 (30 min); 4.0 (45 min) |
| Hel10p | NCU04928 | Conserved hypothetical protein | 2.9 (1 h) |
| Uth1p | NCU02668 | Related to cell cycle regulation and ageing protein | 2.1 (45 min) |
| FadA/GpaA | NCU06493 | GNA-1 G protein alpha chain | 4.4 (1 h) |
| Ste4p/CGB1 | NCU00440 | Gnb-1 G protein beta subunit | 2.3 (45 min) |
| Lag1p | NCU00008 | Probable longevity-assurance protein LAG1 | 2.9 (1 h) |
| Ppa1p | NCU09747 | Probable vacuolar ATP synthase 22 kDa proteolipid subunit | 2.2 (1 h) |
| TEL1 | NCU00247 | Hypothetical protein | 3.1 (45 min) |
| HSP70 | NCU09602 | HSP70 heat-shock protein 70 (hsp70) | 2.1 (45 min) |
†Predicted apoptotic and PCD homologues according to Fedorova et al. (2005).
‡Gene name/ID from Borkovich et al. (2004) and the Broad Institute (http://www.broad.mit.edu/annotation/genome/neurospora/Home.html).
Caspases are important executors of the apoptotic pathway in metazoan animals and are present in the cell as inactive zymogens. Upon exposure to an apoptotic signal they are activated via cleavage (executioner caspases) or oligomerization (initiator caspases) (Adams, 2003). In S. cerevisiae, the single metacaspase gene, YCA1, has been implicated in cell death pathways in response to a variety of chemicals and stresses (Guaragnella et al., 2006; Herker et al., 2004; Madeo et al., 2002; Mazzoni & Falcone, 2008). We constructed strains of alternate het-c* pin-c* haplotype containing deletions of predicted metacaspase genes (ΔNCU02400 and ΔNCU09882) and strains of alternate het-c* pin-c* haplotype containing deletions of both predicted metacaspase genes (ΔNCU02400 ΔNCU09882) (Supplementary Table S1). However, all the het-c* pin-c* ΔNCU09882, ΔNCU02400 and ΔNCU09882 ΔNCU02400 heterokaryons showed a typical HI phenotype, with cell death percentages similar to a wild-type incompatible heterokaryon (Fig. 6). These data indicate that metacaspases are not required for PCD during HI in N. crassa.
Fig. 6.
Effect of the deletion of predicted cell death genes on HI. (a) Heterokaryons carrying homozygous deletions of Δaif-1 (NCU05850) but of alternate het-c pin-c haplotype showed a typical macroscopic HI phenotype and WT levels of cell death. (b, c) Heterokaryons carrying homozygous deletions of predicted metacaspase genes (NCU09882 or NCU02400) but of alternate het-c pin-c haplotype show a macroscopic and microscopic phenotype indistinguishable from a WT incompatible heterokaryon. (d) A heterokaryon between two strains carrying homozygous deletions of both predicted metacaspase genes (NCU09882 and NCU02400), but of alternate het-c pin-c haplotype, show a typical HI phenotype and cell death percentages.
We also tested whether deletions of the apoptosis-inducing factor (AIF) homologue in N. crassa (NCU05850) affected HI or cell death. In metazoans, AIF1 is released from mitochondria during apoptosis, and is predicted to act in the caspase-mediated apoptotic pathway (Eisenberg et al., 2007; Modjtahedi et al., 2006). Mutations in NCU05850 (Δaif-1) result in strains that show resistance to PHS-induced cell death (Castro et al., 2008), while aif-1 deletion strains in Aspergillus nidulans are more sensitive to farnesol-induced cell death (Savoldi et al., 2008). Heterokaryons carrying homozygous Δaif-1 deletions, but of alternate het-c pin-c haplotype, showed an identical phenotype to a wild-type incompatible heterokaryon (Fig. 6). These data indicate that major components of the apoptotic pathway are not required for cell death during HI, implying that a novel and perhaps fungal-specific PCD pathway is functioning during nonself recognition and HI in filamentous ascomycete fungi.
DISCUSSION
In this study, we assessed transcriptional alterations associated with HI and PCD in N. crassa and from these data evaluated the role of various genes/processes implicated in HI and PCD. Analysis of the induction of HI in N. crassa has a number of advantages over studies of PCD in other organisms. First of all, the induction of HI and PCD is a very rapid response (Biella et al., 2002; Garnjobst & Wilson, 1956; Glass & Kaneko, 2003; Sbrana et al., 2007). For both metazoan apoptosis and plant-induced PCD, a significant time lag occurs between induction and cell death (Adams, 2003; Reape et al., 2008; Reed, 2000). Second, PCD induced during HI does not require the application of exogenous substances. Thus, studies on HI are in contrast to many studies of apoptosis in S. cerevisiae and other filamentous fungi where responses to treatment with toxic metabolites are evaluated (for reviews, see Jin & Reed, 2002; Lu, 2006; Ramsdale, 2008). In the literature, controversy exists over whether cell death in S. cerevisiae is a process with real similarities to apoptosis in metazoan cells (Modjtahedi et al., 2006; Vachova & Palkova, 2007; Vercammen et al., 2007), primarily because of the difficulty of assessing mechanisms of cell death following exposure to toxic compounds and because of secondary effects of mutations in genes that affect the physiology of cells responding to exposure to death-inducing agents.
Mutations in the S. cerevisiae metacaspase gene YCA1 result in strains that are resistant to cell death induced by a variety of stresses/chemicals (Guaragnella et al., 2006; Herker et al., 2004; Khan et al., 2005; Madeo et al., 2002; Mazzoni & Falcone, 2008). However, strains containing mutations in the two predicted metacaspase genes (ΔNCU02400, ΔNCU09882 and ΔNCU02400 ΔNCU09882) and an AIF1 (ΔNCU05850) homologue did not affect any phenotypic aspects associated with HI in N. crassa. Although HI displays some downstream characteristics of apoptosis observed in metazoans (Jacobson et al., 1998; Leslie & Zeller, 1996; Marek et al., 2003), the induction of death via nonself recognition and HI may occur by a filamentous-fungal-specific process. Many genes required for HI in filamentous fungi encode proteins containing a HET domain (PF06985), including PIN-C (Kaneko et al., 2006), and which are ubiquitous in the genomes of filamentous ascomycete species, but are notably absent from all other eukaryotic and prokaryotic species. Importantly, the overexpression of just the HET domain is sufficient to induce an HI-like phenotype in P. anserina (Paoletti & Clave, 2007). Thus, the filamentous-ascomycete-specific HI pathway is an attractive system to consider for development of novel fungal-specific drugs for plant and human filamentous ascomycete pathogens.
Expression profiling provided insight not only into the physiological processes occurring during the induction of cell death, but also into the cellular response in hyphal compartments of the colony that survive. The transcriptional response to HI induction overlapped significantly with the transcriptional profile for PHS-induced cell death for both up- and downregulated gene sets, suggesting a common cellular response to the induction of cell death/cell survival. However, both the HI and PHS datasets overlapped significantly with the downregulated gene set identified during amino acid starvation, which likely represents a general response to a broad range of stresses in N. crassa. The upregulation of genes for the synthesis and signalling pathways of phosphatidylinositol as well as the Ca2+ signalling pathways indicate that these two processes may be coordinately regulated during HI. Little is known about phosphatidylinositol signalling in N. crassa, although there is evidence for IP3-activated Ca2+ release from vacuoles (Cornelius et al., 1989; Silverman-Gavrila & Lew, 2002). We hypothesize that N. crassa responds to HI by increasing phosphatidylinositol synthesis and modification that eventually promotes Ca2+ release from the vacuole, which further stimulates the activity of Ca2+-binding proteins and other Ca2+-dependent processes involved in incompatibility. In other systems, IP3-dependent Ca2+ signalling has been implicated in both cell survival and cell death pathways (Rong & Distelhorst, 2008).
Septal plugging is associated with HI, occurring within 5 min of fusion between incompatible strains (Biella et al., 2002; Garnjobst & Wilson, 1956; Glass & Kaneko, 2003; Newhouse & MacDonald, 1991; Sbrana et al., 2007). Catastrophic injury of hyphae in filamentous ascomycete species results in the rapid plugging of septa by the Woronin body; strains containing mutations in the gene, hex-1, encoding the crystalline body core of the Woronin body, show extensive cytoplasmic bleeding (Jedd & Chua, 2000; Markham & Collinge, 1987; Tenney et al., 2000). In this study, we observed DCF fluorescence associated with Woronin bodies during HI. Woronin bodies are a modified peroxisome (Jedd & Chua, 2000; Liu et al., 2008; Tenney et al., 2000) and ROS production has been associated with peroxisomes in a wide variety of organisms (Schrader & Fahimi, 2006). In this study, genes encoding NADPH oxidase, glutaredoxin and cytochrome c also increased in relative expression during HI induction. These enzymes are typically involved in ROS responses within the cell (Eisenberg et al., 2007; Takemoto et al., 2007). These results suggest that ROS are generated during induction of HI and may be a downstream effector of PCD.
It is likely that upon nonself recognition and HI, activation of multiple cell death and cell survival pathways is induced. In this case, it is unlikely that most single gene deletions would have a large effect on the HI phenotype. However, mutations in two genes, NCU06177 (encoding a PKC homologue) and NCU05693 (encoding a GTPase), significantly increased cell death in het-c* pin-c* heterokaryons. It is likely that these genes may be involved in promoting cell survival in cells adjacent to the dying compartments during HI. In P. anserina, genes encoding proteins involved in autophagy were identified as being preferentially expressed during HI (Pinan-Lucarre et al., 2003). Similar to the ΔNCU06177 and ΔNCU05693 mutants, mutations in genes essential for autophagy in P. anserina resulted in strains showing accelerated cell death during HI (Pinan-Lucarre et al., 2005). Future experiments will delineate the roles of these pathways on cell death and cell survival during HI in N. crassa.
Acknowledgments
We thank Drs Takao Kasuga and Betty Gilbert for help with technical aspects of transcriptional profiling with the Neurospora oligonucleotide arrays. We thank Drs Natalie Catlett and Andre Fleissner for Dip strain construction. We thank Dr Jeff Townsend for assistance with BAGEL analysis of the profiling data. We thank Anna Simonin, David Kowbel, and Drs Jianping Sun, Charles Hall and Jingyi Li for critical reading of this manuscript. This work was supported by a National Institutes of Health grant to N. L. G. (GM60468).
Abbreviations
BAGEL, Bayesian analysis of gene expression levels
DCF, dichlorofluorescein
DIC, differential interference contrast
GPI, glycosylphosphatidylinositol
H2DCFDA, 2′,7′-dichlorodihydrofluorescein diacetate
HI, heterokaryon incompatibility
IP3, inositol 1,4,5-triphosphate
MAPK, mitogen-activated protein kinase
PCD, programmed cell death
ROS, reactive oxygen species
PHS, phytosphingosine
TS inc, temperature-sensitive incompatible
Footnotes
The expression data associated with this paper have been deposited at the Neurospora functional genomics database (http://www.yale.edu/townsend/Links/ffdatabase/introduction.html) (Experiment ID# 50).
Four supplementary tables and a supplementary figure are available with the online version of this paper.
References
- Adams, J. M. (2003). Ways of dying: multiple pathways to apoptosis. Genes Dev 17, 2481–2495. [DOI] [PubMed] [Google Scholar]
- Arpaia, G., Cerri, F., Baima, S. & Macino, G. (1999). Involvement of protein kinase C in the response of Neurospora crassa to blue light. Mol Gen Genet 262, 314–322. [DOI] [PubMed] [Google Scholar]
- Biella, S., Smith, M. L., Aist, J. R., Cortesi, P. & Milgroom, M. G. (2002). Programmed cell death correlates with virus transmission in a filamentous fungus. Proc Biol Sci 269, 2269–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich, K. A., Alex, L. A., Yarden, O., Freitag, M., Turner, G. E., Read, N. D., Seiler, S., Bell-Pedersen, D., Paietta, J. & other authors (2004). Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol Mol Biol Rev 68, 1–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro, A., Lemos, C., Falcao, A., Glass, N. L. & Videira, A. (2008). Increased resistance of complex I mutants to phytosphingosine-induced programmed cell death. J Biol Chem 283, 19314–19321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra, J., Samali, A. & Orrenius, S. (2000). Triggering and modulation of apoptosis by oxidative stress. Free Radic Biol Med 29, 323–333. [DOI] [PubMed] [Google Scholar]
- Chen, C. & Dickman, M. B. (2005). Proline suppresses apoptosis in the fungal pathogen Colletotrichum trifolii. Proc Natl Acad Sci U S A 102, 3459–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, J., Park, T. S., Chio, L. C., Fischl, A. S. & Ye, X. S. (2003). Induction of apoptosis by sphingoid long-chain bases in Aspergillus nidulans. Mol Cell Biol 23, 163–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary, S., Lee, H. C., Maiti, M., He, Q., Cheng, P., Liu, Q. & Liu, Y. (2007). A double-stranded-RNA response program important for RNA interference efficiency. Mol Cell Biol 27, 3995–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelius, G., Gebauer, G. & Techel, D. (1989). Inositol trisphosphate induces calcium release from Neurospora crassa vacuoles. Biochem Biophys Res Commun 162, 852–856. [DOI] [PubMed] [Google Scholar]
- Debets, A. J. M. & Griffiths, A. J. F. (1998). Polymorphism of het-genes prevents resource plundering in Neurospora crassa. Mycol Res 102, 1343–1349. [Google Scholar]
- Debets, F., Yang, X. & Griffiths, A. J. F. (1994). Vegetative incompatibility in Neurospora – its effect on horizontal transfer of mitochondrial plasmids and senescence in natural populations. Curr Genet 26, 113–119. [DOI] [PubMed] [Google Scholar]
- Dunlap, J. C., Borkovich, K. A., Henn, M. R., Turner, G. E., Sachs, M. S., Glass, N. L., McCluskey, K., Plamann, M., Galagan, J. E. & other authors (2007). Enabling a community to dissect an organism: overview of the Neurospora functional genomics project. Adv Genet 57, 49–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg, T., Büttner, S., Kroemer, G. & Madeo, F. (2007). The mitochondrial pathway in yeast apoptosis. Apoptosis 12, 1011–1023. [DOI] [PubMed] [Google Scholar]
- Fedorova, N. D., Badger, J. H., Robson, G. D., Wortman, J. R. & Nierman, W. C. (2005). Comparative analysis of programmed cell death pathways in filamentous fungi. BMC Genomics 6, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleissner, A., Simonin, A. R. & Glass, N. L. (2008). Cell fusion in the filamentous fungus, Neurospora crassa. Methods Mol Biol 475, 21–38. [DOI] [PubMed] [Google Scholar]
- Garnjobst, L. & Wilson, J. F. (1956). Heterocaryosis and protoplasmic incompatibility in Neurospora crassa. Proc Natl Acad Sci U S A 42, 613–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass, N. L. & Dementhon, K. (2006). Non-self recognition and programmed cell death in filamentous fungi. Curr Opin Microbiol 9, 553–558. [DOI] [PubMed] [Google Scholar]
- Glass, N. L. & Kaneko, I. (2003). Fatal attraction: nonself recognition and heterokaryon incompatibility in filamentous fungi. Eukaryot Cell 2, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass, N. L., Rasmussen, C., Roca, M. G. & Read, N. D. (2004). Hyphal homing, fusion and mycelial interconnectedness. Trends Microbiol 12, 135–141. [DOI] [PubMed] [Google Scholar]
- Guaragnella, N., Pereira, C., Sousa, M. J., Antonacci, L., Passarella, S., Corte-Real, M., Marra, E. & Giannattasio, S. (2006). YCA1 participates in the acetic acid induced yeast programmed cell death also in a manner unrelated to its caspase-like activity. FEBS Lett 580, 6880–6884. [DOI] [PubMed] [Google Scholar]
- Herker, E., Jungwirth, H., Lehmann, K. A., Maldener, C., Fröhlich, K. U., Wissing, S., Büttner, S., Fehr, M., Sigrist, S. & Madeo, F. (2004). Chronological aging leads to apoptosis in yeast. J Cell Biol 164, 501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson, D. J., Beurkens, K. & Klomparens, K. L. (1998). Microscopic and ultrastructural examination of vegetative incompatibility in partial diploids heterozygous at het loci in Neurospora crassa. Fungal Genet Biol 23, 45–56. [DOI] [PubMed] [Google Scholar]
- Jedd, G. & Chua, N. H. (2000). A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat Cell Biol 2, 226–231. [DOI] [PubMed] [Google Scholar]
- Jin, C. & Reed, J. C. (2002). Yeast and apoptosis. Nat Rev Mol Cell Biol 3, 453–459. [DOI] [PubMed] [Google Scholar]
- Kaneko, I., Dementhon, K., Xiang, Q. & Glass, N. L. (2006). Nonallelic interactions between het-c and a polymorphic locus, pin-c, are essential for nonself recognition and programmed cell death in Neurospora crassa. Genetics 172, 1545–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga, T., Townsend, J. P., Tian, C., Gilbert, L. B., Mannhaupt, G., Taylor, J. W. & Glass, N. L. (2005). Long-oligomer microarray profiling in Neurospora crassa reveals the transcriptional program underlying biochemical and physiological events of conidial germination. Nucleic Acids Res 33, 6469–6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, M. A. S., Chock, P. B. & Stadtman, E. R. (2005). Knockout of caspase-like gene, YCA1, abrogates apoptosis and elevates oxidized proteins in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 102, 17326–17331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata, Y. & Yanagida, M. (2004). Suppression of a mitotic mutant by tRNA-Ala anticodon mutations that produce a dominant defect in late mitosis. J Cell Sci 117, 2283–2293. [DOI] [PubMed] [Google Scholar]
- LeBel, C. P., Ischiropoulos, H. & Bondy, S. C. (1992). Evaluation of the probe 2′,7′-dichlorofluorescin as an indicator of reactive oxygen species formation and oxidative stress. Chem Res Toxicol 5, 227–231. [DOI] [PubMed] [Google Scholar]
- Leiter, E., Szappanos, H., Oberparleiter, C., Kaiserer, L., Csernoch, L., Pusztahelyi, T., Emri, T., Pócsi, I., Salvenmoser, W. & Marx, F. (2005). Antifungal protein PAF severely affects the integrity of the plasma membrane of Aspergillus nidulans and induces an apoptosis-like phenotype. Antimicrob Agents Chemother 49, 2445–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie, J. F. & Zeller, K. A. (1996). Heterokaryon incompatibility in fungi – more than just another way to die. J Genet 75, 415–424. [Google Scholar]
- Li, D., Bobrowicz, P., Wilkinson, H. H. & Ebbole, D. J. (2005). A mitogen-activated protein kinase pathway essential for mating and contributing to vegetative growth in Neurospora crassa. Genetics 170, 1091–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F., Ng, S. K., Lu, Y., Low, W., Lai, J. & Jedd, G. (2008). Making two organelles from one: Woronin body biogenesis by peroxisomal protein sorting. J Cell Biol 180, 325–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, B. C. K. (2006). Programmed cell death in fungi. In The Mycota: Growth, Differentiation and Sexuality, pp. 167–187. Edited by K. Esser. Berlin, Heidelberg, New York: Springer.
- Ludovico, P., Madeo, F. & Silva, M. (2005). Yeast programmed cell death: an intricate puzzle. IUBMB Life 57, 129–135. [DOI] [PubMed] [Google Scholar]
- Madeo, F., Froehlich, E., Ligr, M., Grey, M., Sigrist, S. J., Wolf, D. H. & Froehlich, K.-U. (1999). Oxygen stress: a regulator of apoptosis in yeast. J Cell Biol 145, 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo, F., Herker, E., Maldener, C., Wissing, S., Lächelt, S., Herlan, M., Fehr, M., Lauber, K. & Sigrist, S. J. (2002). A caspase-related protease regulates apoptosis in yeast. Mol Cell 9, 911–917. [DOI] [PubMed] [Google Scholar]
- Marek, S. M., Wu, J., Glass, N. L., Gilchrist, D. G. & Bostock, R. M. (2003). Nuclear DNA degradation during heterokaryon incompatibility in Neurospora crassa. Fungal Genet Biol 40, 126–137. [DOI] [PubMed] [Google Scholar]
- Margolin, B. S., Freitag, M. & Selker, E. U. (1997). Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet Newsl 44, 34–36. [Google Scholar]
- Markham, P. & Collinge, A. J. (1987). Woronin bodies in filamentous fungi. FEMS Microbiol Rev 46, 1–11. [Google Scholar]
- Mazzoni, C. & Falcone, C. (2008). Caspase-dependent apoptosis in yeast. Biochim Biophys Acta 1783, 1320–1327. [DOI] [PubMed] [Google Scholar]
- Metzenberg, R. L. (2004). Bird Medium: an alternative to Vogel Medium. Fungal Genet Newsl 51, 19–20. [Google Scholar]
- Michell, R. H. (2008). Inositol derivatives: evolution and functions. Nat Rev Mol Cell Biol 9, 151–161. [DOI] [PubMed] [Google Scholar]
- Modjtahedi, N., Giordanetto, F., Madeo, F. & Kroemer, G. (2006). Apoptosis-inducing factor: vital and lethal. Trends Cell Biol 16, 264–272. [DOI] [PubMed] [Google Scholar]
- Mousavi, S. A. A. & Robson, G. D. (2004). Oxidative and amphotericin B-mediated cell death in the opportunistic pathogen Aspergillus fumigatus is associated with an apoptotic-like phenotype. Microbiology 150, 1937–1945. [DOI] [PubMed] [Google Scholar]
- Mylyk, O. M. (1975). Heterokaryon incompatibility genes in Neurospora crassa detected using duplication-producing chromosome rearrangements. Genetics 80, 107–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newhouse, J. R. & MacDonald, W. L. (1991). The ultrastructure of hyphal anastomoses between vegetatively compatible and incompatible virulent and hypovirulent strains of Cryphonectria parasitica. Can J Bot 69, 602–614. [Google Scholar]
- Paoletti, M. & Clave, C. (2007). The fungus-specific HET domain mediates programmed cell death in Podospora anserina. Eukaryot Cell 6, 2001–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, D. D. (1984). Advantages of using the inactive-mating-type am1 strain as a helper component in heterokaryons. Fungal Genet Newsl 31, 41–42. [Google Scholar]
- Perkins, D. D. (1988a). The use of duplication-generating rearrangements for studying heterokaryon incompatibility genes in Neurospora. Fungal Genet Newsl 35, 44–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, D. D. (1988b). Main features of vegetative incompatibility in Neurospora crassa. Fungal Genet Newsl 35, 44–46. [Google Scholar]
- Phillips, A. J., Sudbery, I. & Ramsdale, M. (2003). Apoptosis induced by environmental stresses and amphotericin B in Candida albicans. Proc Natl Acad Sci U S A 100, 14327–14332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinan-Lucarre, B., Paoletti, M., Dementhon, K., Coulary-Salin, B. & Clave, C. (2003). Autophagy is induced during cell death by incompatibility and is essential for differentiation in the filamentous fungus Podospora anserina. Mol Microbiol 47, 321–333. [DOI] [PubMed] [Google Scholar]
- Pinan-Lucarre, B., Balguerie, A. & Clave, C. (2005). Accelerated cell death in Podospora autophagy mutants. Eukaryot Cell 4, 1765–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsdale, M. (2008). Programmed cell death in pathogenic fungi. Biochim Biophys Acta 1783, 1369–1380. [DOI] [PubMed] [Google Scholar]
- Reape, T. J., Molony, E. M. & McCabe, P. F. (2008). Programmed cell death in plants: distinguishing between different modes. J Exp Bot 59, 435–444. [DOI] [PubMed] [Google Scholar]
- Reed, J. C. (2000). Apoptosis. San Diego & London: Academic Press.
- Rong, Y. & Distelhorst, C. W. (2008). Bcl-2 protein family members: versatile regulators of calcium signaling in cell survival and apoptosis. Annu Rev Physiol 70, 73–91. [DOI] [PubMed] [Google Scholar]
- Ruepp, A., Zollner, A., Maier, D., Albermann, K., Hani, J., Mokrejs, M., Tetko, I., Güldener, U., Mannhaupt, G. & other authors (2004). The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res 32, 5539–5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler, A. J. & Williams, B. R. (2008). Interferon-inducible antiviral effectors. Nat Rev Immunol 8, 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar, S., Iyer, G., Wu, J. & Glass, N. L. (2002). Nonself recognition is mediated by HET-C heterocomplex formation during vegetative incompatibility. EMBO J 21, 4841–4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saupe, S. J. (2000). Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol Mol Biol Rev 64, 489–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saupe, S. J. & Glass, N. L. (1997). Allelic specificity at the het-c heterokaryon incompatibility locus of Neurospora crassa is determined by a highly variable domain. Genetics 146, 1299–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saupe, S. J., Kuldau, G. A., Smith, M. L. & Glass, N. L. (1996). The product of the het-C heterokaryon incompatibility gene of Neurospora crassa has characteristics of a glycine-rich cell wall protein. Genetics 143, 1589–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoldi, M., Malavazi, I., Soriani, F. M., Capellaro, J. L., Kitamoto, K., da Silva Ferreira, M. E., Goldman, M. H. & Goldman, G. H. (2008). Farnesol induces the transcriptional accumulation of the Aspergillus nidulans Apoptosis-Inducing Factor (AIF)-like mitochondrial oxidoreductase. Mol Microbiol 70, 44–59. [DOI] [PubMed] [Google Scholar]
- Sbrana, C., Nuti, M. & Giovannetti, M. (2007). Self-anastomosing ability and vegetative incompatibility of Tuber borchii isolates. Mycorrhiza 17, 667–675. [DOI] [PubMed] [Google Scholar]
- Schliebs, W., Wurtz, C., Kunau, W.-H., Veenhuis, M. & Rottensteiner, H. (2006). A eukaryote without catalase-containing microbodies: Neurospora crassa exhibits a unique cellular distribution of its four catalases. Eukaryot Cell 5, 1490–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader, M. & Fahimi, H. D. (2006). Peroxisomes and oxidative stress. Biochim Biophys Acta 1763, 1755–1766. [DOI] [PubMed] [Google Scholar]
- Semighini, C. P., Hornby, J. M., Dumitru, R., Nickerson, K. W. & Harris, S. D. (2006). Farnesol-induced apoptosis in Aspergillus nidulans reveals a possible mechanism for antagonistic interactions between fungi. Mol Microbiol 59, 753–764. [DOI] [PubMed] [Google Scholar]
- Severin, F. F. & Hyman, A. A. (2002). Pheromone induces programmed cell death in S. cerevisiae. Curr Biol 12, R233–R235. [DOI] [PubMed] [Google Scholar]
- Silverman-Gavrila, L. B. & Lew, R. R. (2002). An IP3-activated Ca2+ channel regulates fungal tip growth. J Cell Sci 115, 5013–5025. [DOI] [PubMed] [Google Scholar]
- Strahl, T. & Thorner, J. (2007). Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim Biophys Acta 1771, 353–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stridh, H., Kimland, M., Jones, D. P., Orrenius, S. & Hampton, M. B. (1998). Cytochrome c release and caspase activation in hydrogen peroxide- and tributyltin-induced apoptosis. FEBS Lett 429, 351–355. [DOI] [PubMed] [Google Scholar]
- Suzuki, C., Kawano, M., Kashiwagi, T., Arata, Y., Kawasumi, T. & Kashiwagi, Y. (2000). Lethal effect of the expression of a killer gene SMK1 in Saccharomyces cerevisiae. Protein Eng 13, 73–76. [DOI] [PubMed] [Google Scholar]
- Takemoto, D., Tanaka, A. & Scott, B. (2007). NADPH oxidases in fungi: diverse roles of reactive oxygen species in fungal cellular differentiation. Fungal Genet Biol 44, 1065–1076. [DOI] [PubMed] [Google Scholar]
- Tenney, K., Hunt, I., Sweigard, J., Pounder, J. I., McClain, C., Bowman, E. J. & Bowman, B. J. (2000). Hex-1, a gene unique to filamentous fungi, encodes the major protein of the Woronin body and functions as a plug for septal pores. Fungal Genet Biol 31, 205–217. [DOI] [PubMed] [Google Scholar]
- Tian, C., Kasuga, T., Sachs, M. S. & Glass, N. L. (2007). Transcriptional profiling of cross pathway control in Neurospora crassa and comparative analysis of the Gcn4 and CPC1 regulons. Eukaryot Cell 6, 1018–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend, J. P. (2003). Multifactorial experimental design and the transitivity of ratios with spotted DNA microarrays. BMC Genomics 4, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend, J. P. (2004). Resolution of large and small differences in gene expression using models for the Bayesian analysis of gene expression levels and spotted DNA microarrays. BMC Bioinformatics 5, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend, J. P. & Hartl, D. L. (2002). Bayesian analysis of gene expression levels: statistical quantification of relative mRNA level across multiple strains or treatments. Genome Biol 3, RESEARCH0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachova, L. & Palkova, Z. (2007). Caspases in yeast apoptosis-like death: facts and artefacts. FEMS Yeast Res 7, 12–21. [DOI] [PubMed] [Google Scholar]
- van Diepeningen, A. D., Debets, A. J. M. & Hoekstra, R. F. (1997). Heterokaryon incompatibility blocks virus transfer among natural isolates of black aspergilli. Curr Genet 32, 209–217. [DOI] [PubMed] [Google Scholar]
- Vercammen, D., Declercq, W., Vandenabeele, P. & Van Breusegem, F. (2007). Are metacaspases caspases? J Cell Biol 179, 375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videira, A., Kasuga, T., Tian, C., Lemos, C., Castro, A. & Glass, N. L. (2009). Transcriptional analysis of the Neurospora crassa response to phytosphingosine reveals links to mitochondrial function. Microbiology 155, 3134–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, H. J. (1956). A convenient growth medium for Neurospora. Microb Genet Bull 13, 42–46. [Google Scholar]
- Westergaard, M. & Mitchell, H. K. (1947). Neurospora V. A synthetic medium favoring sexual reproduction. Am J Bot 34, 573–577. [Google Scholar]
- Wu, J. & Glass, N. L. (2001). Identification of specificity determinants and generation of alleles with novel specificity at the het-c heterokaryon incompatibility locus of Neurospora crassa. Mol Cell Biol 21, 1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., Saupe, S. J. & Glass, N. L. (1998). Evidence for balancing selection operating at the het-c heterokaryon incompatibility locus in a group of filamentous fungi. Proc Natl Acad Sci U S A 95, 12398–12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, Q. & Glass, N. L. (2002). Identification of vib-1, a locus involved in vegetative incompatibility mediated by het-c in Neurospora crassa. Genetics 162, 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, N.-N., Dudgeon, D. D., Paliwal, S., Levchenko, A., Grote, E. & Cunningham, K. W. (2006). Multiple signaling pathways regulate yeast cell death during the response to mating pheromones. Mol Biol Cell 17, 3409–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]