Abstract
Bacteria growing as surface-adherent biofilms are better able to withstand chemical and physical stresses than their unattached, planktonic counterparts. Using transcriptional profiling and quantitative PCR, we observed a previously uncharacterized gene, yjfO to be upregulated during Escherichia coli MG1655 biofilm growth in a chemostat on serine-limited defined medium. A yjfO mutant, developed through targeted-insertion mutagenesis, and a yjfO-complemented strain, were obtained for further characterization. While bacterial surface colonization levels (c.f.u. cm−2) were similar in all three strains, the mutant strain exhibited reduced microcolony formation when observed in flow cells, and greatly enhanced flagellar motility on soft (0.3 %) agar. Complementation of yjfO restored microcolony formation and flagellar motility to wild-type levels. Cell surface hydrophobicity and twitching motility were unaffected by the presence or absence of yjfO. In contrast to the parent strain, biofilms from the mutant strain were less able to resist acid and peroxide stresses. yjfO had no significant effect on E. coli biofilm susceptibility to alkali or heat stress. Planktonic cultures from all three strains showed similar responses to these stresses. Regardless of the presence of yjfO, planktonic E. coli withstood alkali stress better than biofilm populations. Complementation of yjfO restored viability following exposure to peroxide stress, but did not restore acid resistance. Based on its influence on biofilm maturation and stress response, and effects on motility, we propose renaming the uncharacterized gene, yjfO, as bsmA (biofilm stress and motility).
INTRODUCTION
When growing as surface-adherent, biofilm communities, bacteria are typically quite resistant to a variety of adverse environmental conditions, including antimicrobial agents, pH extremes and oxidative stresses (reviewed by Costerton et al., 1987). Biofilms have been implicated in a number of problems, including many infections, industrial fouling, and corrosion (Costerton et al., 1987; McLean et al., 1996). Several detailed studies have been conducted on biofilms to ascertain the differences between planktonic and biofilm bacteria. In Gram-negative bacteria, these studies include large-scale mutant screens using microtitre (O'Toole & Kolter, 1998b; Prigent-Combaret et al., 1999) and competition culture assays (Junker et al., 2006), promoter identification using in vivo expression technology (Finelli et al., 2003), proteomic analyses (Sauer et al., 2002), and transcription profiling (Domka et al., 2007; Hancock & Klemm, 2007; Junker et al., 2007; Ren et al., 2004; Schembri et al., 2003; Whiteley et al., 2001a). The use of these techniques has allowed the identification of genes important for biofilms, such as those associated with cell signalling involving acylated homoserine lactones (Herzberg et al., 2006; Lee et al., 2007a), osmotic stress and reduced oxygen (Prigent-Combaret et al., 1999), and the global regulators rpoS (Adams & McLean, 1999), relA and spoT (Balzer & McLean, 2002).
Many of the genes expressed at higher levels in biofilms than in planktonic cells encode proteins of unknown function. A number of these are now being associated with various biofilm phenotypes. In Pseudomonas aeruginosa, ndvB is responsible for the tobramycin resistance of biofilms (Mah et al., 2003). In Escherichia coli, ariR (ymgB) is needed for biofilm formation and for full expression of acid resistance genes in biofilms (Lee et al., 2007b). The E. coli bhsA (ycfR) gene is needed for stimulation of biofilm formation by stresses such as hydrogen peroxide, low pH and heat stress (Zhang et al., 2007). In the present study, we observed the uncharacterized gene yjfO (b4189) to be upregulated. This same gene has been found to be upregulated in at least two previous transcriptional profiling studies (Beloin et al., 2004; Junker et al., 2007). Here, we show that yjfO mutants are altered in biofilm structure and cell motility, and in their ability as biofilms to respond to pH and oxidative stresses.
METHODS
Bacterial strains and media.
The strains and plasmids used in this study are listed in Table 1. The insertion mutation in strain B4189 was confirmed upon receipt by PCR. Cultures were maintained on Luria–Bertani (LB) agar (MG1655), LB supplemented with 50 μg kanamycin ml−1 (mutant strain), or LB supplemented with 50 μg kanamycin ml−1 and 100 μg ampicillin ml−1 (complemented strain). For long-term preservation, overnight cultures were frozen at −80 °C using glycerol [final concentration 12.5 % (v/v)] as a cryoprotectant. Prior to experimentation, cultures were revived from frozen stock, cultured overnight on LB agar, and then transferred to MOPS serine medium (Neidhardt et al., 1974). This minimal medium, used in other biofilm studies (Junker et al., 2007; Schembri et al., 2003), contained serine (1 mg ml−1) as the carbon source. Additional amino acids (Ile, Arg, Gly, His, Leu, Met, Phe, Val and Thr, each at 40 μg ml−1) were also present. All cultures were grown at 37 °C, using laboratory facilities at Texas State University.
Table 1.
E. coli strains and plasmids used in this study
| Strain | Characteristics | Source and reference |
|---|---|---|
| MG1655 (DS291) | Wild-type (F−λ−rph-1) | M. Cashel, NIH (Hernandez & Cashel, 1995) |
| B4189 (FB22940) | MG1655 yjfO : : Tn5(KAN-I-SceI) at position 167 in minus orientation | F. R. Blattner, University of Wisconsin (Kang et al., 2004) |
| pMW201 | pGEM-T containing yjfO | This study |
Chemostat culture.
The biofilm culture chemostat apparatus has been previously described (Whiteley et al., 1997). Briefly, it consists of a chemostat from which the culture can be circulated through a biofilm culture device (in this case 4 m Tygon laboratory tubing or a flow cell). For culturing, the chemostat was filled with MOPS serine medium, as described above (Neidhardt et al., 1974), inoculated with 1 ml of an overnight culture of E. coli and grown at 37 °C as a batch culture for 24 h, after which continuous culture was initiated with a peristaltic pump at a dilution rate of 0.025 h−1. The culture was allowed to equilibrate for one full generation (40 h). At this point, biofilm growth was initiated as a second pump continuously circulated the chemostat culture through the 4 m length of attached tubing (100 ml h−1) for 96 h. Bacteria attached to the tubing were considered to be the biofilm culture, whereas unattached cells represented the planktonic culture.
Cell harvesting.
For planktonic cell harvesting, 200 ml culture was mixed with an equal volume of ice-cold (−20 °C) stop solution [5 % (v/v) water-saturated phenol in ethanol] to stop endogenous nuclease activity, and then placed into nuclease-free 50 ml centrifuge tubes (Falcon) (Arnold et al., 2001). Cells were harvested by centrifugation (3200 g, 4 °C for 20 min), frozen (−80 °C), and then transported in liquid nitrogen to Texas A&M University for RNA extraction and analysis.
For biofilm cell harvesting, the biofilm-colonized tubing was removed from the chemostat, drained to remove planktonic cells, and then filled with ice-cold stop solution. Following this, the tubing was cut into 2 cm sections, each section was cut in half and placed into a sterile Petri plate, and biofilm cultures were scraped from the tubing with a sterile scalpel into 200 ml ice-cold stop solution. The scraped tubing sections were then placed into 200 ml ice-cold stop solution and sonicated in a bath sonicator (Sonicor Instrument Corporation) at 60 Hz for 10 min to further dislodge biofilm cells. Biofilm cells were then harvested from the stop solution by centrifugation, frozen, and transported to Texas A&M University for RNA extraction and analysis as described above.
Biofilm growth measurements.
In order to measure biofilm and planktonic cell growth, duplicate chemostat cultures were established for each strain as described above. Following growth, the culture was analysed by dilution plating to enumerate the planktonic populations. For biofilm growth, the biofilm-colonized tubing was removed and rinsed with sterile medium, to remove loosely attached cells. The tubing was cut into five 2 cm sections, placed into sterile PBS, sonicated at 60 Hz for 5 min, and vortexed for 2 min to dislodge biofilm cells (McLean et al., 1999). Cell numbers were then measured by dilution plating onto LB agar.
RNA processing and gene array analysis.
RNA extraction and purification were conducted at Texas A&M University using a previously described hot phenol extraction protocol (Arnold et al., 2001) and enzymic purification. [33P]CTP (New England Biochemical) was used to label cDNA during reverse transcription (Arnold et al., 2001). Gene array analysis was conducted using the Sigma Genosys macroarray protocol previously described by Arnold et al. (2001). For differential expression patterns to be considered significant, a minimum twofold change in expression level (compared with the background) during both the original and replicate run was needed.
Real-time (quantitative) PCR.
Transcriptional profiling results for yjfO were validated using real-time PCR (Ju et al., 2007). RNA was purified as described above, and reverse-transcribed to generate cDNA using a high-capacity cDNA archive kit (Applied Biosystems) using the manufacturer's protocol. The primers used for quantitative PCR were: TM-B4189-204F (5′-ACC GCC AGT AAC GGA CCA T-3′) and TM-B4189-313R (5′-CTA ATG CGT CAT CCG GAG AAC-3′), and 5′-/5(6)-FAM/CCA TCG TGC TTA CGC TAC CTA TTC GCT GTA/36-TAMTph/-3′ (Integrated DNA Technologies) was used as a probe [5(6)-FAM=5′6-carboxyfluorescein; 36TAMpH=3′TAMRA]. No template and no reverse transcriptase controls were used. cDNA products were measured using an Applied Biosystems Prism 7500 real-time PCR system. Real-time analysis was conducted in duplicate.
Epifluorescence microscopy.
To compare biofilm formation of the various strains, each strain was grown in a chemostat coupled to a three-chamber flow cell (Stovall Life Science). The chemostat was inoculated and equilibrated as described above. For biofilm growth, the culture was pumped through the flow cell at 100 ml h−1. After 96 h biofilm growth, the flow cell was removed from the chemostat and each chamber was washed with 5 ml sterile water to remove planktonic cells. Cells were stained with 20 μM Syto 9 (Invitrogen) for 30 min, and then rinsed with 5 ml sterile water. Biofilms were viewed using a Nikon Eclipse 80 I microscope. Images were obtained using a Nikon DXM 1200F digital camera and images were analysed using Image-Pro Plus (version 5.1) (Mirza et al., 2007). Adobe Photoshop CS3 (version 10.0.1) was used to convert the images to greyscale, invert the image (to make bacteria appear dark against a light background), and optimize contrast. Each flow cell experiment was carried out in triplicate.
Transmission electron microscopy (TEM).
In order to observe whether the presence or absence of yjfO affected cell surface stability or flagella production, cells were examined by negative-stain preparations with TEM. Overnight cultures were grown on agar plates and then a loop-full of culture was suspended in a water droplet on Parafilm. The cell suspension was then mixed with 1 % (w/v) uranyl acetate, placed on a Formvar-coated grid, and examined by TEM (Merchant et al., 2007). TEM films were scanned (2700 dots per inch) using a photographic high-resolution scanner, and the images were inverted and optimized for contrast using Adobe Photoshop CS3 as described above.
Twitching and flagellar motility assays.
In order to elicit differences in twitching motility between the strains, the methods outlined by Semmler et al. (1999) were employed. Each strain was stabbed into the centre of an LB agar plate containing 1 % (w/v) agar and incubated at 37 °C in an inverted position. To examine flagella-based motility, cultures were stab-inoculated into the centre of an LB soft agar (0.3 %, w/v, agar) plate (O'Toole & Kolter, 1998a) and the plates incubated in an upright position at 37 °C. Colony diameter due to twitching motility and flagella motility was measured after 16 h incubation.
Hydrophobicity assay.
Strains were tested for their ability to partition into hexane from an aqueous suspension as described by Zhang et al. (2007).
Biofilm stress assays.
We evaluated the bacterial responses to pH, hydrogen peroxide and elevated temperature stresses using the experimental strategy of Zhang et al. (2007). For this purpose, each strain was again cultured in a chemostat, coupled to laboratory tubing as described above. In order to evaluate biofilm stress responses, the biofilm-colonized tubing was removed and cut into 2 cm pieces. Sensitivity to acidic and alkaline conditions was assessed by placing five pieces of tubing into pH 2.5 or pH 12 MOPS serine medium, which was subsequently incubated at 37 °C for 20 min. To determine viability following exposure to hydrogen peroxide, five pieces of tubing were incubated for 5 min at 37 °C in 20 mM H2O2. To assess viability following exposure to heat, five pieces of tubing were placed into MOPS serine medium and incubated for 10 min at 65 °C. Following incubation, the tubing was placed into PBS, which was subsequently sonicated at 60 Hz for 5 min and vortexed for 2 min. Each sample was serially diluted and plated on LB. A total of three biological replicates were performed for each stress measurement.
Planktonic stress assays.
In order to determine strain viability following exposure to numerous environmental stressors, the methods outlined by Zhang et al. (2007) were employed. All strains were incubated in MOPS serine medium at 37 °C with shaking at 100 r.p.m. to OD600 0.3. To assess viability at altered pH, 2 ml culture was incubated for an additional hour at 37 °C without shaking in pH 2.5 or pH 12 MOPS serine medium. To determine culture viability following exposure to hydrogen peroxide, 1 ml culture was incubated with 20 mM H2O2 at 37 °C without shaking for 15 min. For heat sensitivity, 5 ml of each strain was removed and heated for 20 min at 65 °C. Following each treatment, cultures were serially diluted and plated on LB, and incubated at 37 °C for 24 h. A minimum of three biological replicates were performed.
Data analysis.
On the basis of dilution plating, the planktonic and biofilm cell concentrations were calculated as c.f.u. ml−1 and c.f.u. cm−2, respectively. For statistical analyses, the data were log-transformed (Whiteley et al., 2001b) and the data analysed by one-way ANOVA, with a minimum threshold of significance of P<0.05. Where applicable, all pair-wise comparisons were analysed by the Holm–Sidak method using SigmaStat v3.0 (Systat Software). Several measurements gave no detectable survival and in these cases, the c.f.u. data for that particular sample were assigned a value of 1 c.f.u. (the log10 of an undetectable sample was therefore zero). A minimum of three biological replicates was performed for each measurement. SigmaPlot v8.0 (Systat) was used to plot the results.
Complementation of E. coli mutant.
The yjfO gene was amplified from MG1655 using the primers yjfO-F1 (5′-GATGTGGGTTACGCTTTCGT-3′), yjfO-R1 (5′-CCACTGTCCTGTCACGATG-3′) using AmpliTaq Gold (Applied Biosystems). Purified yjfO gene products were cloned into the pGEM-T Easy Vector (Promega) to generate pMW201, where yjfO could be transcribed from the lac promoter through induction with 0.1 mM IPTG. The resulting plasmid was subsequently electroporated into the corresponding mutant strain. Verification of the inserted gene was accomplished through restriction digestion using EcoRI and plasmid sequencing. IPTG was present in the MOPS serine medium during chemostat experiments with the complemented strain.
RESULTS
Gene array
In order to investigate differences in biofilm and planktonic culture gene expression, we conducted transcriptional profiling on 96 h biofilms of E. coli. We found that 43 genes were differentially expressed between biofilm and planktonic culture. In the literature, the number of genes that are differentially expressed within biofilms in comparison with planktonic cultures ranges from a low of approximately 0.5 % of the genome (Whiteley et al., 2001a) to a high of almost 20 % (Hancock & Klemm, 2007; Ren et al., 2004). The values obtained in the present study (43 genes) (GenBank accession number GSE18362) represent approximately 1 % of the genome (Blattner et al., 1997) and are certainly consistent with other reports. As noted elsewhere (Beloin et al., 2004; Ren et al., 2004; Schembri et al., 2003), many of the differentially regulated genes were uncharacterized. Several initially uncharacterized genes, identified in earlier transcriptional profiling investigations, have since been shown to be important in biofilm functions (Domka et al., 2007; Zhang et al., 2007). Under our experimental conditions, expression of yjfO (b4189) was 3.3±0.3-fold higher in biofilms than in planktonic culture, as measured by transcriptional profiling. Upregulation of yjfO in biofilms was confirmed by quantitative PCR [Δ threshold cycle (Ct) 5.0±0.5]. Two previous studies have also shown yjfO to be upregulated in biofilms (Beloin et al., 2004; Junker et al., 2007).
Biofilm formation
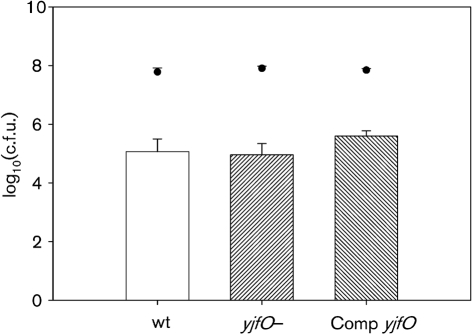
To investigate the role of yjfO in biofilm formation, we circulated a steady-state E. coli culture through a three-chambered flow cell for 96 h. The flow cell was removed after 96 h, stained with 20 μM Syto 9 and viewed using a Nikon Eclipse 80 I microscope at ×100 magnification. Clumping and microcolony formation were observed in the wild-type (MG1655) biofilms (Fig. 1a). This phenotype was absent in the yjfO mutant (Fig. 1b), but could be restored upon genetic complementation (Fig. 1c). In order to measure adherent cell populations, we grew each of the strains in a chemostat coupled to 4 m Tygon laboratory tubing for biofilm culture (Whiteley et al., 1997). Following 96 h of biofilm growth, the tubing was removed and assayed for colonization by dilution plating as described above. No statistically significant difference was noted in planktonic or adherent cell populations among any of the strains (Fig. 2). Based on these observations we concluded that yjfO expression is important in microcolony formation and biofilm maturation processes, but not necessary for planktonic growth or initial surface adhesion (Beloin et al., 2004; Sauer et al., 2002).
Fig. 1.
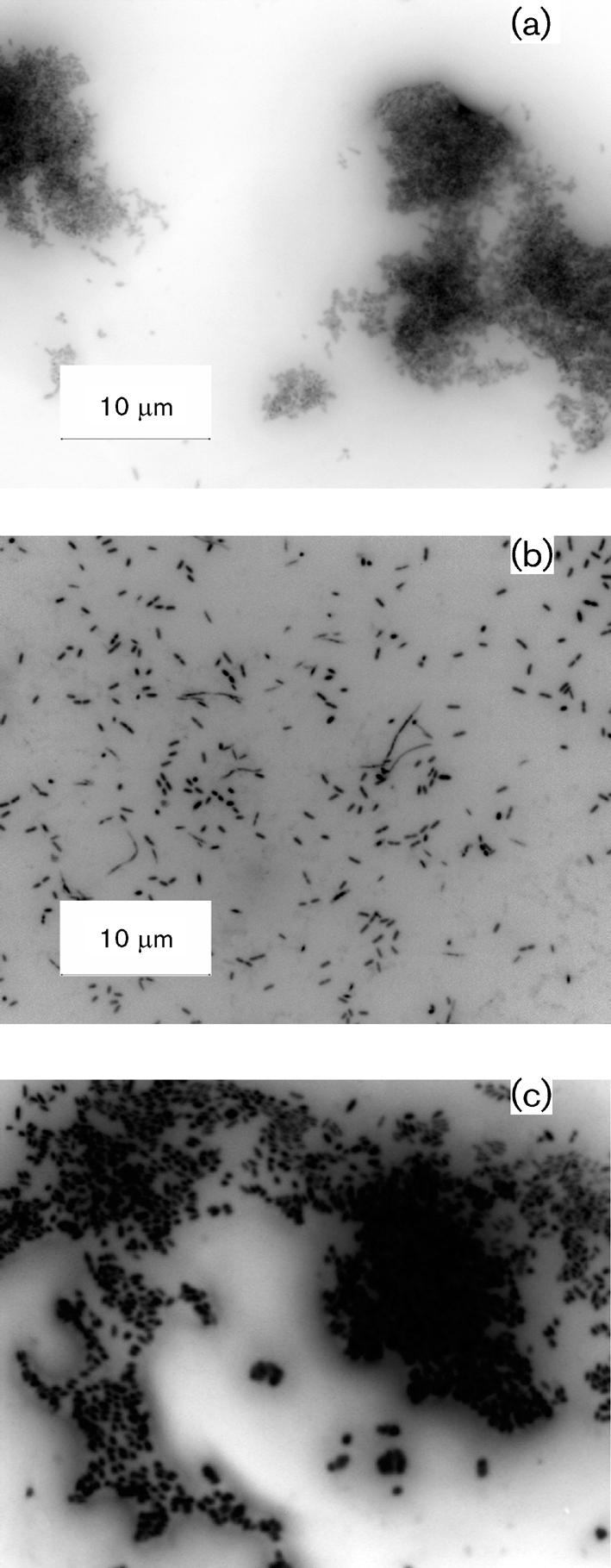
Representative epifluorescence microscopy images of 96 h biofilms from chemostat cultures grown in serine-limited MOPS medium: MG1655 (a), yjfO (b4189) (b) and complemented yjfO (c). The magnification is the same in all figures.
Fig. 2.
Planktonic (scatter plot) and biofilm growth (bar graph) of MG1655, yjfO (b4189) (yjfO−) and complemented yjfO (Comp yjfO) after 96 h chemostat growth. Values in all figures are expressed as log10(c.f.u. ml−1) for planktonic cultures and log10(c.f.u. cm−2) for biofilm cultures (±sem). No significant differences were seen among the planktonic populations or biofilm populations of the three strains.
Motility and ultrastructure
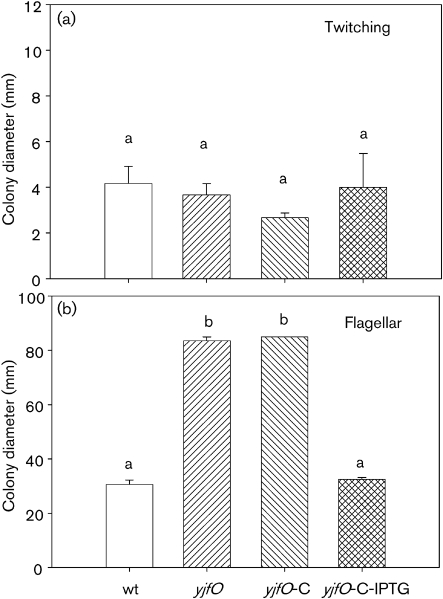
As twitching motility has been shown to be important for microcolony formation (O'Toole & Kolter, 1998a), we investigated whether the phenotype of the yjfO deletion could be explained by loss of this characteristic. Although there were slight differences in colony expansion (Fig. 3a), these differences were not significant when analysed by one-way ANOVA (P=0.63), indicating that yjfO had no effect on twitching motility. In contrast, flagella-based motility was enhanced in the yjfO mutant and the complemented but not induced strain (P<0.001 compared with the wild type), but could be restored to wild-type levels when the yjfO-complemented strains were induced by IPTG (Fig. 3b). Using TEM, we examined negatively stained preparations of the various strains for the presence of flagella and found an indication of enhanced flagella in some of the yjfO mutants (Fig. 4). In future studies, we will explore the mechanism(s) for this yjfO-enhanced flagella-based motility.
Fig. 3.
Twitching (a) and flagella-based (b) motility in MG1655 (wt), yjfO mutant (yjfO) and yjfO-complemented strains in the absence (yjfO-C) and presence (yjfO-C-IPTG) of IPTG induction. Values with the same letter, in all figures, are not significantly different (P<0.05).
Fig. 4.
TEM micrographs of negative-stain preparations of E. coli MG1655 (a), yjfO (b), and yjfO-complemented and IPTG-induced strains (c). Flagella (F), outer (OM) and inner (IM) membranes are indicated. Bars: 0.5 μm (a, b), 1.0 μm (c).
Based on its sequence, the YjfO protein is predicted to be a lipoprotein (Rudd et al., 1998), and as such has the potential to be associated with cell membranes. Cell preparation during negative-stain TEM involves suspending the bacteria in a heavy metal solution (to provide electron contrast), and then exposing the stained, unfixed cells to high vacuum during TEM examination. Other EM preparation approaches, such as conventional embedding or freeze substitution (Graham & Beveridge, 1990), employ chemical fixation and gradual dehydration, processes which stabilize and protect membranes and other structures from the high vacuum and electron beam in TEM. As fixation and gentle dehydration protocols are not used during routine negative-stain TEM preparation, one would anticipate that membranes lacking a key structural component would be much more prone to vacuum-induced damage. However, we did not observe any major differences in the cell envelope membranes, regardless of the presence (Fig. 4a, c) or absence of yjfO (Fig. 4b). However, we cannot rule out subtle, membrane-associated changes due to yjfO solely on the basis of negative-stain TEM examination.
Cell surface hydrophobicity
In order to assess the effect of yjfO on cell surface hydrophobicity, aqueous suspensions of the E. coli strains were mixed with an equal volume of hexane, and the fraction of cells remaining in the aqueous layer was measured as described elsewhere (Zhang et al., 2007). Following this procedure, the percentage of cells (±se) remaining in the aqueous phase was: wild-type 86.5 (0.5), yjfO 81.5 (0.5), complemented yjfO without IPTG 82.5 (0.5), and complemented and IPTG-induced yjfO 92 (3). Based on these results, yjfO appears to have had a limited effect on cell surface hydrophobicity in these experiments. However, recent work by Q. Ma and T. K. Wood (unpublished results, personal communication) has shown that yjfO overexpression in another E. coli strain results in greatly increased hydrophobicity. As a result, we cannot completely rule out a contribution of yjfO to cell surface hydrophobicity.
Planktonic and biofilm stress assays
In order to determine the viability of each strain following exposure to commonly encountered environmental stresses, chemostat-grown biofilm populations were exposed to acid, base, hydrogen peroxide (oxidative stress) or heat stress. As shown in Fig. 2, no statistically significant differences in overall cell numbers were noted between the strains in the absence of stress, thus allowing for standardization and efficient determination of biofilm cell viability following exposure to each stressor.
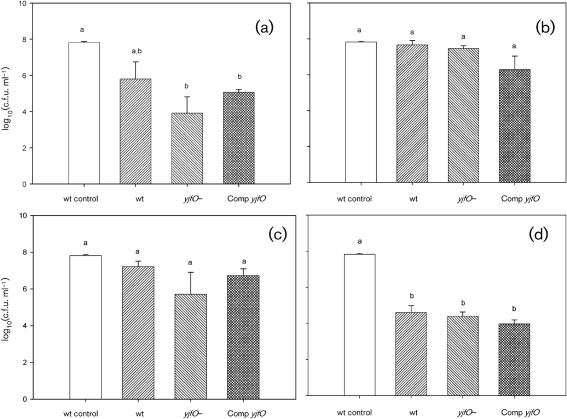
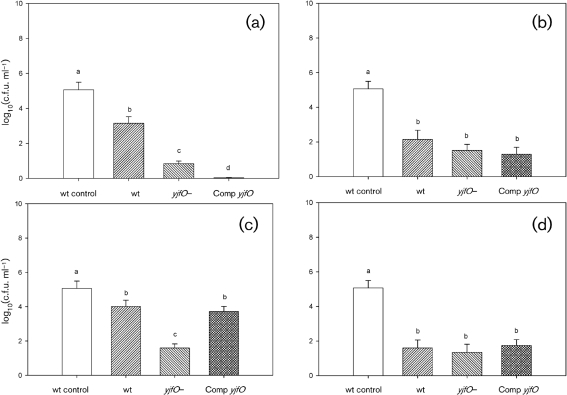
Exposure of 96 h biofilms to pH 2.5 MOPS serine medium (acid stress) resulted in a decrease in overall viability for all strains assayed. In the planktonic populations (Fig. 5a), the acid-treated wild-type cells were reduced in number in comparison with the untreated control; however, the difference was only marginally significant (P=0.099). The other two populations (yjfO mutant and complemented strain) were significantly reduced in comparison with the untreated control, with the yjfO strain showing the biggest reduction and the complemented strain showing a sensitivity intermediate between those of the wild-type and the yjfO mutant. However, the acid-treated planktonic strains did not differ significantly with respect to each other. In contrast to the planktonic results, the viability patterns in the biofilm cultures following acid stress were quite different (Fig. 6a), with all populations differing significantly from each other. In comparison with the acid-treated wild-type biofilms, yjfO mutant viability was reduced approximately 200-fold. The viability of the complemented strain was reduced even further (approximately sixfold in comparison with the mutant). The failure of pMW201 to complement the acid-sensitive phenotype of the mutant suggests that the yjfO insertion mutation has polar effects on expression of the adjacent downstream gene yjfN. Given that the complemented strain survives less well than the mutant, it is also possible that high levels of YjfO increase sensitivity to low pH.
Fig. 5.
Survival of E. coli planktonic cells following exposure to acid (a), base (b), oxidative stress (c) and heat stress (d). Strains in this figure are designated untreated wild-type control (wt control), treated MG1655 (wt), yjfO (b4189) (yjfO−), and complemented and induced yjfO (Comp yjfO). The value for the untreated control corresponds to the average of the wild-type planktonic cells without exposure to any stressor and is included for viability comparisons.
Fig. 6.
Survival of E. coli biofilm cells following exposure to acid stress (a), base stress (b), oxidative stress (c) and heat stress (d). For strain designations, see legend to Fig. 5. The value for the untreated control corresponds to the average of the wild-type biofilm cells without exposure to any stressor and is included for viability comparisons.
Exposure of 96 h biofilms to pH 12 MOPS serine medium (base stress) resulted in decreased viability of E. coli biofilms (Fig. 6b). Here, all three strains (wild-type, mutant and complemented strain) showed a statistically similar reduction in viability. In contrast, there was no significant stress-induced change in viability in planktonic cultures (Fig. 5b). The increased susceptibility of biofilm populations to an environmental stress (in this case, alkaline pH), in comparison with planktonic populations, is highly unusual in that the converse is generally the case (Costerton et al., 1987; Davey & O'Toole, 2000). We will investigate this issue in future studies.
Exposure of planktonic and biofilm populations to oxidative stress (20 mM H2O2) resulted in a statistically significant decrease in viability only in the yjfO mutant biofilm, which was complemented by the reintroduction of the yjfO gene (Fig. 6c). In planktonic cultures, there was a modest, and statistically insignificant, reduction in the viability of the yjfO mutant (Fig. 5c). We interpret these results as demonstrating that yjfO is involved in the protection of biofilms against oxidative stress.
In contrast to the above-mentioned stresses, we observed no significant differences in planktonic (Fig. 5d) or biofilm (Fig. 6d) culture viability following exposure to heat (65 °C).
DISCUSSION
E. coli is routinely found in soil, water and intestinal mucus, all of which impose unique stresses on the organism in both biofilm and planktonic modes of growth (Fabich et al., 2008). In intestinal mucus, E. coli encounters acetate and other volatile fatty acids, produced as metabolic by-products of the resident flora (Arnold et al., 2001). These organic acids are capable of traversing the membrane into the bacterial cell, thus inducing acid stress (Arnold et al., 2001). Typically, these weak acids decrease bacterial viability, yet strains of E. coli, including MG1655 (Arnold et al., 2001), are capable of combating them. Acid resistance is one factor that allows E. coli to efficiently colonize and inhabit the intestinal tract. In this same largely anaerobic environment, E. coli would be exposed to transient concentrations of reactive oxygen species from mucosal innate defences (McLean et al., 1988). In contrast, E. coli does not experience alkaline conditions (above pH 8) in its normal environment, and so the low resistance to alkaline stress is anticipated. The surprising observation in the current study was the increased resistance of planktonic populations (Fig. 5b) to alkali stress when compared with biofilm populations (Fig. 6b). Also, heat susceptibility was unaffected by biofilm (Fig. 6d) or planktonic growth (Fig. 5d). Although biofilm growth is normally associated with stress resistance (Costerton et al., 1987; Davey & O'Toole, 2000), our current study certainly shows exceptions to this phenomenon.
In this study, we focused on yjfO, a member of the yhcN family. When first described (Rudd et al., 1998), the yhcN family consisted of nine parahomologous, uncharacterized genes (yjfO, yahO, ybiJ, ybiM, ycfR, ydgH, yhcN, yjfN and yjfY) of unknown function. Members of this family are predicted to have evolved from a common ancestor, based on the presence of a signal peptide and a shared motif in their N and C termini (Rudd et al., 1998). One of these genes, ycfR, has been shown to be upregulated in biofilm cultures and to be associated with biofilm stress responses (Zhang et al., 2007). On that basis, Zhang et al. (2007) have proposed that this gene be renamed to bhsA for influencing biofilms through hydrophobicity and stress response. In the present study, yjfO does contribute to the biofilm stress response but unlike ycfR does not appear to contribute to cell surface hydrophobicity.
Cell aggregation and microcolony formation of surface-adherent bacteria via twitching motility and other processes are hallmarks of early biofilm maturation (O'Toole & Kolter, 1998a; Sauer et al., 2002). In the current study (Fig. 1), we noted that wild-type E. coli formed microcolonies within flow cells. This feature was absent in the yjfO mutant biofilm, although it was restored upon genetic complementation. However, adherent cell concentrations were similar (Fig. 2). Aside from twitching and flagella motility (O'Toole & Kolter, 1998a), other characteristics involved in cell aggregation include cell–cell adhesion, cell signalling (Domka et al., 2006) and hydrophobic interactions (McEldowney & Fletcher, 1986). The most striking feature of yjfO deletion was the loss of microcolony formation (Fig. 1) and greatly enhanced flagella motility (Fig. 3). Other investigators have shown the importance of microcolonies, water channels and other biofilm structures in the resistance of the component organisms to various stresses (Matz et al., 2004; Pamp & Tolker-Nielsen, 2007). Although flagella motility has been shown to be important in biofilm structure development (Wood et al., 2006), our present study suggests that unchecked flagella motility disrupts microcolonies, a feature that certainly provides a plausible explanation for the greatly reduced biofilm stress response. Due to its contribution to biofilm stress response as well as its contribution to flagella motility, we propose renaming yjfO as bsmA (biofilm stress and motility).
Acknowledgments
This work was funded by grants from the National Institutes of Health (1R15 AI050638) to R. J. C. M. and (RO1 GM55154) to D. A. S., and from the Texas Higher Education Coordinating Board Advanced Research Program (003615-0037-2007) to R. J. C. M. We thank Kerry Fuson, Qun Ma and Patricia Zenker for assistance, and Jean Marc Ghigo, Anthony Hay, Karl Klose, Ron Walter, Marvin Whiteley and Tom Wood for helpful discussions and advice. R. J. C. M. would like to dedicate this paper to the memory of Blanche A. V. McLean.
Abbreviations
TEM, transmission electron microscopy
Footnotes
The gene array data discussed in this paper have been deposited in Gene Expression Omnibus (GEO) under accession number GSE18362.
References
- Adams, J. L. & McLean, R. J. C. (1999). The impact of rpoS deletion on Escherichia coli biofilms. Appl Environ Microbiol 65, 4285–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, C. N., McElhanon, J., Lee, A., Leonhart, R. & Siegele, D. A. (2001). Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. J Bacteriol 183, 2178–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzer, G. J. & McLean, R. J. C. (2002). The stringent response genes relA and spoT are important for Escherichia coli biofilms under slow-growth conditions. Can J Microbiol 48, 675–680. [DOI] [PubMed] [Google Scholar]
- Beloin, C., Valle, J., Latour-Lambert, P., Faure, P., Kzreminski, M., Balestrino, D., Haagensen, J. A., Molin, S., Prensier, G. & other authors (2004). Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol Microbiol 51, 659–674. [DOI] [PubMed] [Google Scholar]
- Blattner, F. R., Plunkett, G., III, Bloch, C. A., Perna, N. T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J. D., Rode, C. K. & other authors (1997). The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- Costerton, J. W., Cheng, K.-J., Geesey, G. G., Ladd, T. I., Nickel, J. C., Dasgupta, M. & Marrie, T. J. (1987). Bacterial biofilms in nature and disease. Annu Rev Microbiol 41, 435–464. [DOI] [PubMed] [Google Scholar]
- Davey, M. E. & O'Toole, G. A. (2000). Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64, 847–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domka, J., Lee, J. & Wood, T. K. (2006). YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl Environ Microbiol 72, 2449–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domka, J., Lee, J., Bansal, T. & Wood, T. K. (2007). Temporal gene-expression in Escherichia coli K-12 biofilms. Environ Microbiol 9, 332–346. [DOI] [PubMed] [Google Scholar]
- Fabich, A. J., Jones, S. A., Chowdhury, F. Z., Cernosek, A., Anderson, A., Smalley, D., McHargue, J. W., Hightower, G. A., Smith, J. T. & other authors (2008). Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun 76, 1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finelli, A., Gallant, C. V., Jarvi, K. & Burrows, L. L. (2003). Use of in-biofilm expression technology to identify genes involved in Pseudomonas aeruginosa biofilm development. J Bacteriol 185, 2700–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, L. L. & Beveridge, T. J. (1990). Evaluation of freeze-substitution and conventional embedding protocols for routine electron microscopic processing of eubacteria. J Bacteriol 172, 2141–2149. [DOI] [PMC free article] [PubMed]
- Hancock, V. & Klemm, P. (2007). Global gene expression profiling of asymptomatic bacteriuria Escherichia coli during biofilm growth in human urine. Infect Immun 75, 966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, V. J. & Cashel, M. (1995). Changes in conserved region 3 of Escherichia coli σ70 mediate ppGpp-dependent functions in vivo. J Mol Biol 252, 536–549. [DOI] [PubMed] [Google Scholar]
- Herzberg, M., Kaye, I. K., Peti, W. & Wood, T. K. (2006). YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J Bacteriol 188, 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju, Z., Wells, M. C., Heater, S. J. & Walter, R. B. (2007). Multiple tissue gene expression analyses in Japanese medaka (Oryzias latipes) exposed to hypoxia. Comp Biochem Physiol C Toxicol Pharmacol 145, 134–144. [DOI] [PubMed] [Google Scholar]
- Junker, L. M., Peters, J. E. & Hay, A. G. (2006). Global analysis of candidate genes important for fitness in a competitive biofilm using DNA-array-based transposon mapping. Microbiology 152, 2233–2245. [DOI] [PubMed] [Google Scholar]
- Junker, L. M., Toba, F. A. & Hay, A. G. (2007). Transcription in Escherichia coli PHL628 biofilms. FEMS Microbiol Lett 268, 237–243. [DOI] [PubMed] [Google Scholar]
- Kang, Y., Durfee, T., Glasner, J. D., Qiu, Y., Frisch, D., Winterberg, K. M. & Blattner, F. R. (2004). Systematic mutagenesis of the Escherichia coli genome. J Bacteriol 186, 4921–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J., Jayaraman, A. & Wood, T. K. (2007a). Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol 7, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J., Page, R., García-Contreras, R., Palermino, J. M., Zhang, X. S., Doshi, O., Wood, T. K. & Peti, W. (2007b). Structure and function of the Escherichia coli protein YmgB: a protein critical for biofilm formation and acid-resistance. J Mol Biol 373, 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah, T. F., Pitts, B., Pellock, B., Walker, G. C., Stewart, P. S. & O'Toole, G. A. (2003). A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426, 306–310. [DOI] [PubMed] [Google Scholar]
- Matz, C., Bergfeld, T., Rice, S. A. & Kjelleberg, S. (2004). Microcolonies, quorum sensing and cytotoxicity determine the survival of Pseudomonas aeruginosa biofilms exposed to protozoan grazing. Environ Microbiol 6, 218–226. [DOI] [PubMed] [Google Scholar]
- McEldowney, S. & Fletcher, M. (1986). Variability of the influence of physicochemical factors affecting bacterial adhesion to polystyrene substrata. Appl Environ Microbiol 52, 460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean, R. J. C., Nickel, J. C., Cheng, K.-J. & Costerton, J. W. (1988). The ecology and pathogenicity of urease-producing bacteria in the urinary tract. Crit Rev Microbiol 16, 37–79. [DOI] [PubMed] [Google Scholar]
- McLean, R. J. C., Fortin, D. & Brown, D. A. (1996). Microbial metal binding mechanisms and their relation to nuclear waste disposal. Can J Microbiol 42, 392–400. [Google Scholar]
- McLean, R. J. C., Whiteley, M., Hoskins, B. C., Majors, P. D. & Sharma, M. M. (1999). Laboratory techniques for studying biofilm growth, physiology, and gene expression in flowing systems and porous media. Methods Enzymol 310, 248–264. [DOI] [PubMed] [Google Scholar]
- Merchant, M. M., Welsh, A. K. & McLean, R. J. C. (2007). Rheinheimera texasensis sp. nov., a halointolerant freshwater oligotroph. Int J Syst Evol Microbiol 57, 2376–2380. [DOI] [PubMed] [Google Scholar]
- Mirza, B. S., Welsh, A. & Hahn, D. (2007). Saprophytic growth of inoculated Frankia sp. in soil microcosms. FEMS Microbiol Ecol 62, 280–289. [DOI] [PubMed] [Google Scholar]
- Neidhardt, F. C., Bloch, P. L. & Smith, D. F. (1974). Culture medium for Enterobacteria. J Bacteriol 119, 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole, G. A. & Kolter, R. (1998a). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30, 295–304. [DOI] [PubMed] [Google Scholar]
- O'Toole, G. A. & Kolter, R. (1998b). Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol Microbiol 28, 449–461. [DOI] [PubMed] [Google Scholar]
- Pamp, S. J. & Tolker-Nielsen, T. (2007). Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. J Bacteriol 189, 2531–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent-Combaret, C., Vidal, O., Dorel, C. & Lejeune, P. (1999). Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J Bacteriol 181, 5993–6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, D., Bedzyk, L. A., Thomas, S. M., Ye, R. W. & Wood, T. K. (2004). Gene expression in Escherichia coli biofilms. Appl Microbiol Biotechnol 64, 515–524. [DOI] [PubMed] [Google Scholar]
- Rudd, K. E., Humphrey-Smith, I., Wasinger, V. C. & Bairoch, A. (1998). Low molecular weight proteins: a challenge for post-genomic research. Electrophoresis 19, 536–544. [DOI] [PubMed] [Google Scholar]
- Sauer, K., Camper, A. K., Ehrlich, G. D., Costerton, J. W. & Davies, D. G. (2002). Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol 184, 1140–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schembri, M. A., Kjærgaard, K. & Klemm, P. (2003). Global gene expression in Escherichia coli biofilms. Mol Microbiol 48, 253–267. [DOI] [PubMed] [Google Scholar]
- Semmler, A. B. T., Whitchurch, C. B. & Mattick, J. S. (1999). A reexamination of twitching motility in Pseudomonas aeruginosa. Microbiology 145, 2863–2873. [DOI] [PubMed] [Google Scholar]
- Whiteley, M., Brown, E. & McLean, R. J. C. (1997). An inexpensive chemostat apparatus for the study of microbial biofilms. J Microbiol Methods 30, 125–132. [Google Scholar]
- Whiteley, M., Bangera, M. G., Bumgarner, R. E., Parsek, M. R., Teitzel, G. M., Lory, S. & Greenberg, E. P. (2001a). Gene expression in Pseudomonas aeruginosa biofilms. Nature 413, 860–864. [DOI] [PubMed] [Google Scholar]
- Whiteley, M., Ott, J. R., Weaver, E. A. & McLean, R. J. C. (2001b). Effects of community composition and growth rate on aquifer biofilm bacteria and their susceptibility to betadine disinfection. Environ Microbiol 3, 43–52. [DOI] [PubMed] [Google Scholar]
- Wood, T. K., Gonzalez Barrios, A. F., Herzberg, M. & Lee, J. (2006). Motility influences biofilm architecture in Escherichia coli. Appl Microbiol Biotechnol 72, 361–367. [DOI] [PubMed] [Google Scholar]
- Zhang, X. S., Garcia-Contreras, R. & Wood, T. K. (2007). YcfR (BhsA) influences Escherichia coli biofilm formation through stress response and surface hydrophobicity. J Bacteriol 189, 3051–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]