Abstract
The outer membrane of Gram-negative bacteria is an essential compartment containing a specific complement of lipids and proteins that constitute a protective, selective permeability barrier. Outer membrane β-barrel proteins are assembled into the membrane by the essential hetero-oligomeric BAM complex, which contains the lipoprotein BamE. We have identified a homologue of BamE, encoded by CC1365, which is located in the outer membrane of the stalked alpha-proteobacterium Caulobacter crescentus. BamE associates with proteins whose homologues in other bacteria are known to participate in outer membrane protein assembly: BamA (CC1915), BamB (CC1653) and BamD (CC1984). Caulobacter cells lacking BamE grow slowly in rich medium and are hypersensitive to anionic detergents, some antibiotics and heat exposure, which suggest that the membrane integrity of the mutant is compromised. Membranes of the ΔbamE mutant have normal amounts of the outer membrane protein RsaF, a TolC homologue, but are deficient in CpaC*, an aggregated form of the outer membrane secretin for type IV pili. ΔbamE membranes also contain greatly reduced amounts of three TonB-dependent receptors that are abundant in wild-type cells. Cells lacking BamE have short stalks and are delayed in stalk outgrowth during the cell cycle. Based on these findings, we propose that Caulobacter BamE participates in the assembly of outer membrane β-barrel proteins, including one or more substrates required for the initiation of stalk biogenesis.
INTRODUCTION
The outer membrane of Gram-negative bacteria is an important determinant of cell integrity, physiology and virulence, by virtue of its component lipids and proteins (Nikaido, 2003). The inner leaflet of the outer membrane contains phospholipids, while the outer leaflet contains glycolipids, primarily lipopolysaccharide (LPS). Lipoproteins, which are anchored in a membrane via amino-terminal lipid groups, can reside in either the cytoplasmic membrane or the outer membrane, and are sorted to the correct compartment by the LolAB system (Narita et al., 2004). Integral outer membrane proteins (OMPs) span the membrane as β-barrels composed of amphipathic β-strands. OMPs include porins (Delcour, 2002), TonB-dependent receptors (Braun & Endriss, 2007), TolC homologues (Koronakis et al., 2004), which transport proteins and small molecules, and secretins (Bayan et al., 2006), which are outer membrane channels for type II secretion systems and type IV pili.
The mechanism for targeting and assembling OMPs was largely unknown until 2003, when researchers studying Neisseria meningitidis found that the OMP Omp85 is essential for viability and for maintaining normal levels of folded, oligomerized β-barrel proteins in the outer membrane (Voulhoux et al., 2003). The Omp85 homologue in Escherichia coli was later discovered to be part of a hetero-oligomeric complex (the BAM complex) that assembles β-barrel proteins into the outer membrane (Knowles et al., 2009). The BAM complex contains the essential β-barrel protein BamA (formerly YaeT in E. coli and Omp85 in N. meningitidis) and the lipoproteins BamB (YfgL), BamC (NlpB), BamD (YfiO) and BamE (SmpA) (Sklar et al., 2007; Wu et al., 2005). In E. coli and N. meningitidis, BamA and BamD are essential for viability, and depletion of either one causes several OMPs to accumulate in their unfolded or monomeric forms (Doerrler & Raetz, 2005; Malinverni et al., 2006; Volokhina et al., 2009; Voulhoux et al., 2003; Werner & Misra, 2005). BamB, BamC and BamE are not essential for viability in E. coli (Bouvier et al., 1991; Rolhion et al., 2005; Ruiz et al., 2005; Sklar et al., 2007), and BamB is absent from neisserial genomes (Volokhina et al., 2009). In single bamB, bamC or bamE mutants, one or a few OMPs are incorrectly assembled, while double mutants have more severe OMP assembly defects or are inviable (Ruiz et al., 2005; Sklar et al., 2007; Volokhina et al., 2009; Wu et al., 2005). The N. meningitidis BAM complex contains an additional protein, RmpM (Volokhina et al., 2009), which also associates with the porins PorA and PorB and the TonB-dependent receptors TbpA and LbpA (Jansen et al., 2000; Prinz & Tommassen, 2000). Analysis of Neisseria rmpM mutants suggests that this protein stabilizes OMP complexes rather than participating in OMP assembly (Volokhina et al., 2009).
Caulobacter crescentus is a Gram-negative α-proteobacterium that lives in dilute aquatic environments (Poindexter, 1964). Although the Caulobacter outer membrane proteome has been characterized (Phadke et al., 2001), no proteins involved in the biogenesis of outer membrane components have been studied in this organism. Caulobacter has chiefly been the subject of research on cell cycle regulation and cell polarity (Collier & Shapiro, 2007). Each Caulobacter cell division is asymmetrical, yielding a flagellated swarmer cell that cannot initiate chromosome replication and a cell with a polar stalk that immediately begins a new round of chromosome replication and cell division. The swarmer progeny re-enters the cell division cycle when it differentiates into a stalked cell and initiates DNA replication (Iba et al., 1977; Stove & Stanier, 1962).
The investigation of OMP assembly in Caulobacter is likely to yield new insights, first because this organism encodes and expresses a distinctive set of OMPs (Ireland et al., 2002; Molloy et al., 2001; Nierman et al., 2001; Phadke et al., 2001). Caulobacter lacks genes for the trimeric porins OmpF/C and for the maltoporin LamB, which are highly expressed in E. coli. However, the genome encodes several proteins with homology to the monomeric porin OmpA as well as approximately 65 TonB-dependent receptors, which function in energy-dependent nutrient uptake (Postle & Larsen, 2007). TonB-dependent receptors transport siderophores (Pugsley & Reeves, 1976), cobalamin (Bassford & Kadner, 1977), maltodextrins (Neugebauer et al., 2005) and sucrose (Blanvillain et al., 2007), and members of this protein family may transport a wide variety of carbohydrates in plant pathogens and aquatic bacteria (Blanvillain et al., 2007). TonB-dependent receptors may also be particularly important for growth in dilute environments where passive diffusion through porins cannot satisfy the cell's nutritional requirements.
In addition, OMP assembly has not previously been studied in stalked bacteria. The Caulobacter stalk is an extension of the cell envelope that is devoid of cytoplasmic components and contains membrane proteins involved in nutrient uptake (Ireland et al., 2002; Poindexter & Cohen-Bazire, 1964). Stalk biogenesis is initiated once in the life of the cell, during differentiation of the swarmer progeny into a stalked cell, and stalk growth continues throughout the life of the cell (Stove & Stanier, 1962). Stalks become elongated under conditions of phosphate limitation (Gonin et al., 2000; Schmidt & Stanier, 1966), and isolated stalks take up the fluorogenic organophosphate compound fluorescein diphosphate (Wagner et al., 2006). Stalk growth could be mediated by the same proteins that build the cell envelope as a whole, but there may also be a dedicated pathway for the assembly of stalk-specific proteins.
Although little is known about the proteins directly involved in building the stalk, two regulatory pathways have been found to promote stalk growth. The alternative sigma factor σ54 (encoded by rpoN) and the response regulator TacA direct the transcription of genes needed for stalk biogenesis, and rpoN and tacA mutants are stalkless in rich medium (Biondi et al., 2006; Brun & Shapiro, 1992; Skerker et al., 2005). However, the need for σ54 and TacA can be overcome by growing cells in low-phosphate medium (Biondi et al., 2006; Brun & Shapiro, 1992). Under these conditions, a second regulatory pathway, possibly mediated by the response regulator PhoB, promotes stalk outgrowth (Gonin et al., 2000). In addition to these regulatory pathways, some proteins needed for elongation of the entire cell are involved in stalk biogenesis, including penicillin-binding protein 2, RodA and MreB (Seitz & Brun, 1998; Wagner et al., 2005).
Here, we identify a protein in C. crescentus that is homologous to E. coli BamE and Pseudomonas aeruginosa OmlA, and is encoded by CC1365. Caulobacter BamE is located in the outer membrane and is associated with homologues of other BAM complex proteins. The ΔbamE mutant grows slowly in rich and minimal media and is hypersensitive to heat, detergents and antibiotics. The membranes of cells lacking BamE contain normal levels of the TolC homologue RsaF (Toporowski et al., 2004), but contain greatly reduced levels of an aggregate of the secretin CpaC (Viollier et al., 2002), as well as three predicted TonB-dependent receptors. These results are consistent with the phenotypes of bamE or omlA mutants in other species (Lewis et al., 2008; Ochsner et al., 1999; Sklar et al., 2007). We further show that BamE is required for the timely initiation of stalk biogenesis during the Caulobacter cell division cycle. Together, our data strongly suggest that Caulobacter BamE functions in the assembly of OMPs which mediate membrane integrity, nutrient uptake and stalk biogenesis.
METHODS
Bacterial strains, plasmids and culture conditions.
Strains and plasmids used are listed in Supplementary Table S1, available with the online version of this paper. All experiments were performed using derivatives of C. crescentus strain CB15N (Evinger & Agabian, 1977) grown to mid-exponential phase. Plasmids were mobilized from E. coli to C. crescentus by conjugation using E. coli strain S17-1 (Ely, 1991). The sequences of primers used for amplification or introduction of restriction sites are available upon request. CB15N strains were grown in peptone-yeast extract (PYE; Ely, 1991), minimal medium (M2G; Ely, 1991) or M5G low-phosphate medium (10 mM PIPES, pH 7, 1 mM NaCl, 1 mM KCl, 0.05 % NH4Cl, 0.01 mM Fe/EDTA, 0.2 % glucose, 0.5 mM MgSO4, 0.5 mM CaCl2 and 0.03 mM phosphate) at the indicated temperatures. E. coli strains were grown in Luria Broth at 37 °C. Solid and liquid media were supplemented with antibiotics as described by Reisinger et al. (2007).
To construct the strain ΔbamE (ΔCC1365), we replaced the bamE open reading frame with a tetracycline resistance cassette flanked by FRT sites (McLeod et al., 1986) by two-step homologous recombination using pJT11. We transformed this strain (KR1694) with pBH474, which encodes Flp recombinase (House et al., 2004). Gentamicin-resistant colonies were selected, grown overnight in PYE medium without antibiotic selection and plated on PYE/3 % (w/v) sucrose to obtain colonies that had lost pBH474. Sucrose-resistant colonies were screened for those that had regained sensitivity to gentamicin and tetracycline.
For 3×FLAG fusions, we used PCR to generate a BamHI site either after the last codon or after codon 115 of bamE and cloned each fragment into SpeI/BamHI-digested pSK84, containing the 3×FLAG coding sequence. We digested these intermediate vectors with SpeI and EcoRV and ligated each insert into pMR10 to create pJH4, carrying bamE : : 3×FLAG or pLB40, carrying bamEΔC : : 3×FLAG. The 3×FLAG tag adds the amino acid residues DYKDHDGDYKDHDIDYKDDDDK to the C terminus of each protein.
Cell fractionation.
KR1735 was grown in PYE/kanamycin medium to an OD660 of 0.4. The cell pellet from 100 ml culture was harvested by centrifugation (10 min, 4 °C, 9000 r.p.m., Sorvall GSA rotor). Cells were resuspended in 3 ml TE (10 mM Tris/HCl, 1 mM EDTA, pH 8.0) and lysed by two passages through a French pressure cell at 6000 p.s.i. Unbroken cells were removed by centrifugation (10 min, 4 °C, 5000 r.p.m., Sorvall SS-34 rotor) and the supernatant was loaded onto a two-step gradient composed of 0.3 ml 65 % and 1.0 ml 25 % sucrose (w/v) in TE. After centrifugation (2.5 h, 4 °C, 50 000 r.p.m., Beckman SW60 Ti rotor), the top 3 ml was collected as a soluble fraction containing both cytoplasm and periplasm, the next 500 μl was discarded as cellular debris and the next 500 μl containing total cellular membranes was mixed with 1.5 ml TE. The membrane sample was loaded onto a discontinuous gradient composed of 0.5 ml 65 %, 0.5 ml 55 %, 1 ml 50 %, 2 ml 45 %, 2 ml 40 %, 2 ml 35 % and 1.5 ml 30 % sucrose (w/v) in TE and centrifuged (17 h, 4 °C, 36 000 r.p.m., Beckman SW41 rotor). After centrifugation, 400 μl samples were withdrawn from the top of the gradient, and 20 μl of each even-numbered fraction was assayed for total protein content using quick-start Bradford dye reagent (Bio-Rad) according to the manufacturer's instructions. Protein-containing fractions were analysed by SDS-PAGE and Western blotting using anti-McpA antiserum (Alley et al., 1992; 1 : 30 000), anti-FLAG M2 antibody (Sigma; 1 : 5000), anti-CpaC antiserum (Viollier et al., 2002; 1 : 2000) and anti-RsaF antiserum (Toporowski et al., 2004; 1 : 10 000).
Isolation of the BAM complex.
Cell pellets of strains KR1717, KR1735 and KR2473 grown to mid-exponential phase in PYE/kanamycin (30 ml culture at OD660=0.3) were resuspended using 1.0 ml buffer A (50 mM Tris/HCl, pH 7.5, 50 mM NaCl, 1 mM EDTA, 10 mM MgCl2, 0.5 % dodecyl maltoside). Cells were lysed by adding 10 μl of 10 mg lysozyme ml−1 and 2 μl benzonase nuclease (Novagen) and rotating end-over-end at room temperature for 15 min. Lysates were cleared by centrifugation for 10 min at 16 000 g. Anti-FLAG M2 agarose was prepared, added to cleared lysates and washed according to the manufacturer's instructions (FLAG immunoprecipitation kit, Sigma). To elute immunoprecipitates, 20 μl 2× SDS sample buffer was added, and samples were incubated for 5 min at 100 °C. To determine the efficiency of immunoprecipitating BamE–3×FLAG or BamEΔC–3×FLAG, we analysed 10 μl of each cleared lysate or flow-through fraction and 0.5 μl of each immunoprecipitate by SDS-PAGE and Western blotting with anti-FLAG M2 antibody (Sigma; 1 : 5000).
Identification of proteins by mass spectrometry.
To identify proteins that co-purified with BamE–3×FLAG or BamEΔC–3×FLAG, or proteins that were deficient in the membranes of ΔbamE cells, we analysed samples by separating using SDS-PAGE and staining with GelCode Blue (Thermo Scientific) according to the manufacturer's instructions. Proteins in each gel slice were excised and digested as described by Jimenez et al. (1998). Mass spectrometry was performed on peptides from band 1 of Fig. 5(d) and all BamE-associated bands (see Fig. 3) by the QB3 Proteomics/Mass Spectrometry Laboratory at UC Berkeley. Each sample was loaded onto a 10 cm nano LC column packed in a 100 μm inner diameter glass capillary with an emitter tip. The column consisted of Polaris c18 5 μm packing material (Varian). The column was directly coupled to an electrospray ionization source mounted on a Thermo-Finnigan Deca XP Plus ion trap mass spectrometer. The programmes sequest (Eng et al., 1994) and DTASelect (Tabb et al., 2002) were used to identify peptides. Statistical cutoffs for peptide identification were set at levels shown to give very low rates of false positive identifications (Elias et al., 2005). Peptides from bands 2 and 3 of Fig. 5(d) were analysed by the HHMI Mass Spectrometry Laboratory at UC Berkeley. The buffer was removed with C18 ZipTips (Millipore) and the resulting peptides were analysed by MALDI in reflector mode on an ABI 4800 TOF-TOF mass spectrometer (Applied Biosystems). A peptide mass fingerprint search of the NCBI database identified the principal component of band 3 as CC2010. The MS/MS spectra of three of the tryptic peptides obtained from that band were acquired and matched the theoretical tryptic peptide sequences, SSITQDFISR, [m/z 1153.6 (MH+, monoisotopic)], YNINPSNTGNLR (m/z 1163.5) and LTRPEPETTQAYDLGYR (m/z 2010.0) from CC2010. A similar search of the mass spectrum of band 2 identified it as primarily CC2819. Two of the tryptic peptides obtained from band 2 were consistent with the theoretical tryptic peptides FNNVGVNLVWSHLDDVVDDNPASR, (m/z 2682.3) and GFGVNPVPR (m/z 942.5) from that protein.
Fig. 5.
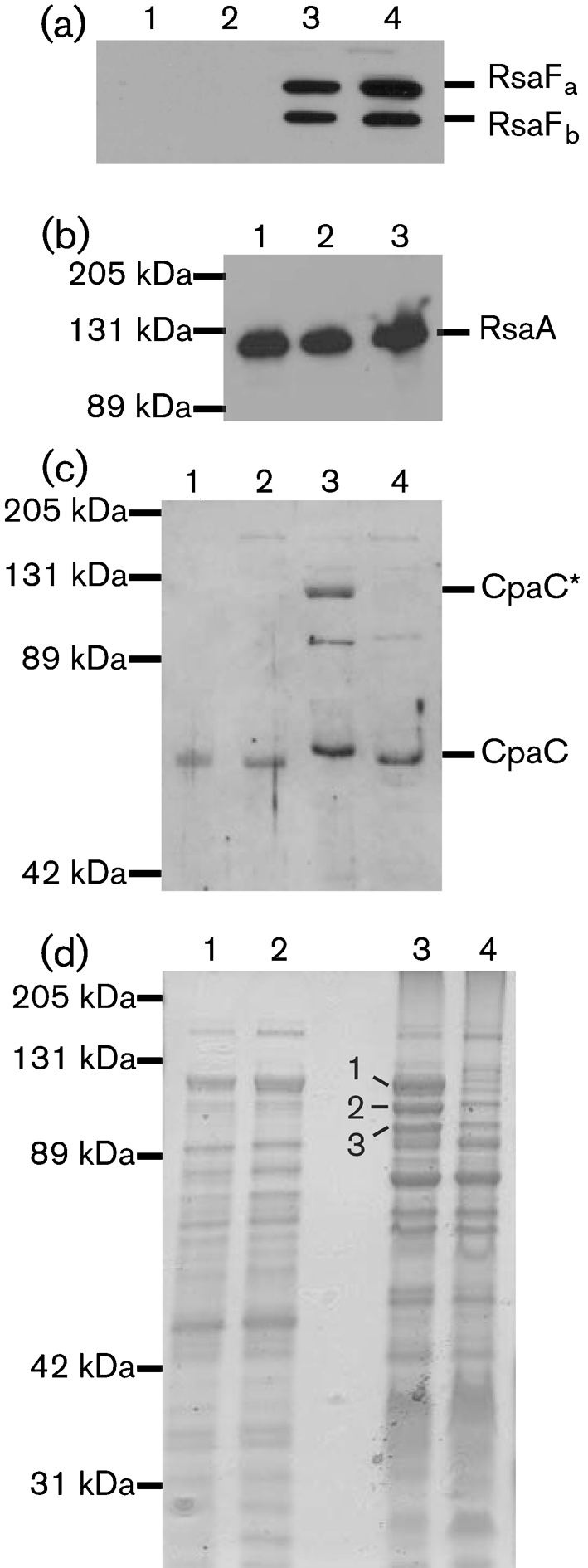
Levels of CpaC* and of three abundant membrane proteins are reduced in ΔbamE membranes. (a, c, d) Equal amounts of soluble and membrane proteins were analysed by SDS-PAGE. Lanes: 1, CB15N/pMR10 soluble; 2, ΔbamE/pMR10 soluble; 3, CB15N/pMR10 membrane; 4, ΔbamE/pMR10 membrane. RsaFa and RsaFb (a) and CpaC and CpaC* (c) were detected by Western blotting; proteins were visualized by GelCode Blue staining (d), and three indicated protein bands (1–3) were excised for trypsin digestion and analysis by mass spectrometry. (b) Surface-exposed RsaA was extracted from CB15N/pMR10 (lane 1), ΔbamE/pMR10 (2) and ΔbamE/pbamE (3) and detected by SDS-PAGE and Western blotting.
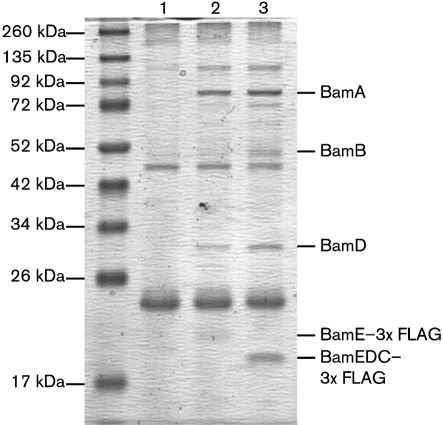
Fig. 3.
Equal amounts of anti-FLAG M2 immunoprecipitates from strains KR1717 (ΔbamE/pbamE, lane 1), KR1735 (ΔbamE/pbamE : : 3×FLAG, lane 2) and KR2473 (ΔbamE/pbamEΔC : : 3×FLAG, lane 3) separated on SDS-PAGE and stained with GelCode Blue. Bands labelled BamA, BamB and BamD were excised from the gel and digested with trypsin, and the resulting peptides were identified by mass spectrometry.
Chemical sensitivity tests.
Exponential phase cells of each strain grown in PYE medium were diluted to OD660=0.25; 100 μl was added to 4 ml PYE medium containing 0.3 % agar at 42 °C. The agar was poured onto a PYE/kanamycin plate to solidify at room temperature. Ten microlitres of each tested compound was pipetted onto a sterile filter paper disc (3M) and each disc was transferred onto the overlaid agar using sterile forceps. Plates were incubated upright at 30 °C for 24 h and the zone of growth inhibition between each disc and the lawn of cells was measured.
Isolation of surface RsaA protein.
Surface-exposed RsaA protein was extracted with acid as described by Koronakis et al. (2000). RsaA was visualized using SDS-PAGE and Western blotting using anti-RsaA antiserum (Toporowski et al., 2004; 1 : 10 000).
Stalk length measurements.
To measure stalk lengths in PYE medium, cultures were grown at 30 °C to OD660 0.2–0.4. To measure stalk lengths in M5G-low phosphate medium, cells were initially grown in M2G medium to OD660 0.4–0.6, then diluted 1 : 500 into M5G low phosphate medium and grown at 30 °C to OD660 0.1–0.3. Cells were immobilized on agarose pads (1 % w/v agarose in distilled water), and differential interference contrast images were acquired as described by Reisinger et al. (2007). Pixel lengths of stalks were measured using the line region tool in Metavue (Universal Imaging) and all cells displaying a stalk were scored. Pixel lengths were converted to micrometres by photographing and measuring the distances between scored lines of a Petroff Hausser Counter (catalogue no. 3900, Hausser Scientific) in the same manner.
Time-lapse microscopy of synchronized cells.
Wild-type and mutant cells were grown in 50 ml M2G to OD660 0.3–0.5 and harvested by centrifugation (10 min at 12 000 r.p.m., Sorvall SS34 rotor). We resuspended each cell pellet in 5 ml M2 salts (6.0 mM Na2HPO4, 3.9 mM KH2PO4, 9.3 mM NH4Cl), added 5 ml Percoll (Sigma) and aliquoted the mixture into microcentrifuge tubes. After centrifuging for 20 min at 4 °C at 10 000 r.p.m. in a microcentrifuge, we harvested and pooled the swarmer cell bands. Isolated swarmer cells were washed twice in M2 salts and resuspended in 400–600 μl M2G medium. We spotted 0.6 μl onto an agarose pad [1 % (w/v) agarose in M2G medium], sealed the edges with Valap (equal weights lanolin, petroleum jelly and paraffin) and photographed the same field of cells at 30 min intervals. For each time point, we determined the percentage of cells that had grown a visible stalk and the percentage of cells that had divided. Cells on the agarose pad that failed to grow (i.e. were the same length at the beginning and end of the time-lapse) were not counted.
RESULTS
CC1365 has homology to the lipoprotein BamE
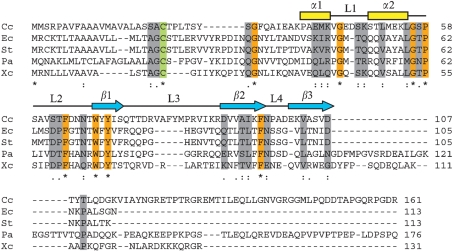
The C. crescentus gene CC1365 was originally predicted to encode a conserved hypothetical protein (Nierman et al., 2001) and was identified in a bioinformatics study as a signature protein of the α-proteobacteria (Kainth & Gupta, 2005). However, proteins closely related to CC1365 encoded in other α-proteobacterial genomes have been annotated as members of the SmpA/OmlA family of lipoproteins (Badger et al., 2006; DelVecchio et al., 2002), which is present in the alpha-, beta- and gammaproteobacteria. We aligned the amino acid sequence of CC1365 with BamE and OmlA proteins that have been studied experimentally from the organisms E. coli (Sklar et al., 2007), Salmonella enterica serovar Typhimurium (Lewis et al., 2008), P. aeruginosa PAO1 (Ochsner et al., 1999) and Xanthomonas axonopodis pv. citri (Vanini et al., 2008) using Kalign (Lassmann & Sonnhammer, 2005). These proteins share significant homology over the first approximately 107 aa of CC1365 (Fig. 1), corresponding to the Pfam domain SmpA_OmlA (PF04355). The regions of highest homology also correspond to distinct secondary structural elements in the solution structure of X. citri OmlA (Fig. 1; Vanini et al., 2008), including residues predicted to form the hydrophobic core of each protein (M41, L51, L55, W69, Y71 and F96 of CC1365). The N-terminal 22 aa of CC1365 are predicted to function as a lipoprotein secretion signal and lipid attachment site (Prosite PDOC00013). This sequence specifies lipid modification on the side chain of a conserved cysteine residue (Fig. 1, green shading), cleavage by signal peptidase II just before the cysteine and subsequent lipid modification of the α-amino group of the N-terminal cysteine (Sankaran et al., 1995). Based upon these conserved sequence features, we hereafter refer to CC1365 as Caulobacter BamE.
Fig. 1.
Sequence alignment of BamE/OmlA homologues generated by Kalign (Lassmann & Sonnhammer, 2005). C. crescentus CC1365 (Cc) was aligned with BamE/OmlA homologues that have been studied experimentally from E. coli (Ec), S. enterica serovar Typhimurium (St), P. aeruginosa (Pa) and X. axonopodis pv. citri (Xc). Identical (orange shading, asterisks) and similar (grey shading, dots) amino acid residues are highlighted. The conserved cysteine residue where signal cleavage and lipid modification are predicted to occur is indicated (green shading). Secondary structure elements indicated above the sequence correspond to the solution structure of X. citri OmlA (Vanini et al., 2008).
The OmlA proteins and Caulobacter BamE have non-homologous C-terminal extensions that are not found in E. coli or S. Typhimurium BamE (Fig. 1). BamE/OmlA homologues in alpha- and betaproteobacteria typically contain C-terminal extensions of approximately 60–100 aa beyond the region of conservation. Among the gamma-proteobacteria, BamE homologues in the enterobacteriales contain only a few amino acids beyond the SmpA_OmlA domain, while homologues in other groups possess tails up to 70 aa in length. When the C-terminal extension of Caulobacter BamE (beginning with T109) is used as a blast query (Altschul et al., 1990) against α-proteobacterial genomes, it identifies the same SmpA_OmlA proteins (and hypothetical proteins) as the full-length protein sequence, indicating that the C-terminal extension is conserved among alphaproteobacterial BamE proteins. In contrast, the C-terminal extension of Caulobacter BamE identifies no sequences of significant homology in beta- or gamma-proteobacterial genomes.
BamE is located in the Caulobacter outer membrane
Lipoproteins of Gram-negative bacteria are modified by three acyl chains on an N-terminal cysteine residue, and they can be anchored in either the cytoplasmic or the outer membrane (Narita et al., 2004). Rules for sorting lipoproteins have been established in some gammaproteobacteria (Lewenza et al., 2006; Narita & Tokuda, 2007; Seydel et al., 1999), but no similar studies have been undertaken in alpha-proteobacteria. To determine where BamE resides in the cell envelope, we deleted the chromosomal copy of bamE and expressed a version of BamE tagged at the C terminus with a 3×FLAG epitope for detection on Western blots. BamE–3×FLAG complements the chemical sensitivity (Table 2) and stalk length defects (Fig. 6a, h) of the deletion mutant, indicating that the fusion protein is functional. We harvested total cell membranes of this strain, separated them by sucrose density-gradient centrifugation, and probed the resulting fractions with anti-FLAG antibodies and antisera that recognize the inner membrane chemoreceptor McpA (Alley et al., 1992), the outer membrane secretin for the type IV pilus, CpaC (Skerker & Shapiro, 2000; Viollier et al., 2002), and the TolC homologue RsaF, which secretes the crystalline S-layer protein RsaA (Toporowski et al., 2004).
Table 2.
Susceptibilities of strains to detergents and antibiotics
nd, Not done.
| Compound* | Zone of growth inhibition (mm)† | ||||
|---|---|---|---|---|---|
| CB15N/pMR10 | ΔbamE/pMR10 | ΔbamE/pbamE | ΔbamE/pbamE : : 3×FLAG | ΔbamE/pbamEΔC : : 3×FLAG | |
| 10 % Tween 20 | 0 | 0 | 0 | nd | nd |
| 10 % Triton X-100 | 0 | 0 | 0 | 0 | nd |
| 10 % deoxycholate | 2 | 4 | 2 | 2 | 2 |
| 10 % SDS | 7 | 11 | 8 | 8 | 9 |
| 0.5 M EDTA | 5 | 11 | 5 | 5 | 7 |
| 2 mg oxytetracycline ml−1 | 13 | 14 | 13 | 13 | nd |
| 1 mg chloramphenicol ml−1 | 15 | 15 | 14 | 13 | nd |
| 10 mg nalidixic acid ml−1 | 1 | 10 | 1 | 1 | 2 |
| 50 mg carbenicillin ml−1 | 3 | 8 | 4 | 3 | 5 |
| 3 mg rifampicin ml−1 | 11 | 18 | 11 | 11 | 15 |
| 5 mg streptomycin ml−1 | 8 | 10 | 8 | 9 | nd |
*Ten microlitres of each compound was spotted onto a sterile disc placed on PYE top agar containing 5×107 cells.
†Zones were measured after 24 h growth at 30 °C. Each value is the mean of at least three experiments.
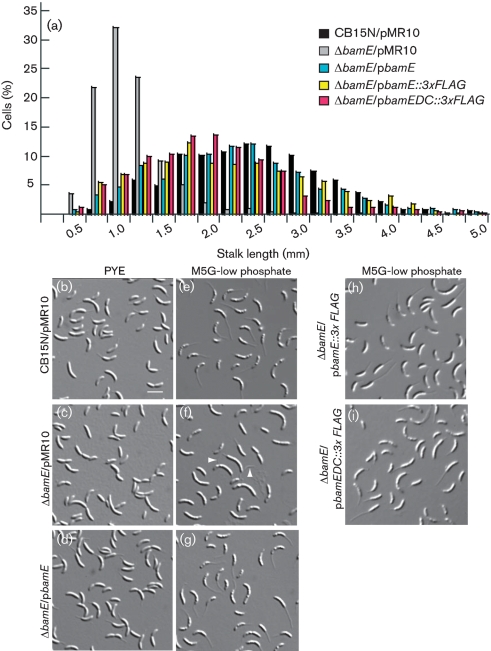
Fig. 6.
Cells lacking BamE have short stalks. (a) Stalk lengths of the indicated cells grown in M5G-low phosphate medium were measured, and the distribution frequency is shown. (b–i) The indicated strains were grown in either rich medium (b–d) or minimal medium with limiting phosphate (e–i), immobilized on agarose pads and photographed. Bar [on (b)], 2.5 μm. White arrowheads denote debris resembling detached stalks.
McpA levels peaked in lower-density fractions and diminished steadily in the higher-density fractions (Fig. 2), consistent with the migration of inner-membrane proteins (Clancy & Newton, 1982). In contrast, BamE–3×FLAG abundance peaked in the high-density fractions, characteristic of OMPs. Both CpaC monomers (50 kDa) and the high-molecular-mass species CpaC* (∼110 kDa) were most abundant in high-density fractions. CpaC* is recognized by purified anti-CpaC antibodies and represents an aggregate of CpaC monomers (Viollier et al., 2002). Anti-RsaF antiserum recognizes two homologous proteins, RsaFa and RsaFb, each of which can secrete RsaA (Toporowski et al., 2004). RsaFa was distributed in both the inner and outer membrane fractions, while RsaFb was present exclusively in the high-density outer membrane fractions. Caulobacter BamE resides chiefly in the outer membrane, along with CpaC and RsaF. The slightly different distributions of the OMPs may be due to different assembly pathways or kinetics.
Fig. 2.
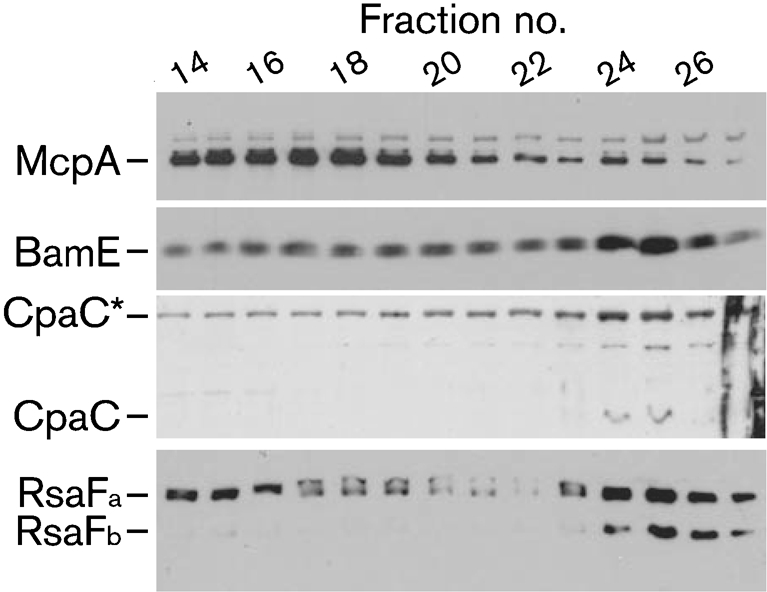
Total membranes of ΔbamE/pbamE : : 3×FLAG were separated by sucrose density centrifugation, and protein-containing fractions were analysed by SDS-PAGE and Western blotting with antibodies against McpA, CpaC, RsaF and the FLAG epitope.
Caulobacter BamE associates with other proteins implicated in OMP assembly
In E. coli, BamE is one subunit of a hetero-oligomeric complex (the BAM complex) responsible for assembling β-barrel proteins into the outer membrane (Sklar et al., 2007; Wu et al., 2005). Within the BAM complex, BamB interacts directly with BamA, while BamC, D and E depend on each other for interaction with BamA (Malinverni et al., 2006; Sklar et al., 2007; Vuong et al., 2008). Because Caulobacter lacks a BamC homologue (Gatsos et al., 2008), we wondered if BamE (and BamD) could still associate with BamA.
We solubilized whole cells of strain KR1717 (ΔbamE/pbamE) or strain KR1735 (ΔbamE/pbamE : : 3×FLAG), incubated the cleared lysates with anti-FLAG M2 agarose (Sigma), and eluted bound proteins. Western blots using anti-FLAG M2 antibodies showed that BamE–3×FLAG was completely removed from the initial cell lysate by this procedure (data not shown). Immunoprecipitates were analysed by SDS-PAGE and stained with GelCode Blue. FLAG-tagged BamE specifically coprecipitated with proteins of ∼85 and ∼30 kDa (Fig. 3, compare lanes 1 and 2). At ∼50 kDa, a faint doublet appeared, in which the lower band was non-specific, while the upper band was present only in the immunoprecipitate containing FLAG-tagged BamE (Fig. 3, compare lanes 1 and 2). The ∼85 kDa band was identified by mass spectrometry as Caulobacter BamA (CC1915), while the ∼30 kDa band corresponded to BamD (CC1984). Peptides from BamB (CC1653) were predominant in the 50 kDa region associated with BamE–3×FLAG, while this protein was absent from the 50 kDa region associated with untagged BamE. Thus, Caulobacter contains a protein complex including homologues of BamA, BamB, BamD and BamE, despite lacking a homologue of BamC.
Growth defect of the ΔbamE mutant
We measured the growth in rich (PYE) and minimal (M2G) media of the wild-type strain CB15N/pMR10, the mutant ΔbamE/pMR10 and ΔbamE containing the complementing low-copy plasmid pbamE. ΔbamE/pMR10 grew more slowly than either the wild-type strain or the complemented mutant in both media, but the growth rate discrepancy was reduced in M2G (Table 1). Further, ΔbamE/pMR10 cells grew at approximately the same rate in both rich and minimal media (Table 1). Thus, Caulobacter BamE is particularly important to sustain rapid growth on rich medium. Unlike the Caulobacter ΔbamE mutant, P. aeruginosa, E. coli and S. Typhimurium strains lacking OmlA or BamE have wild-type growth rates in both rich and minimal media (Lewis et al., 2008; Ochsner et al., 1999; Sklar et al., 2007). However, other combinations of mutations that affect components of the BAM complex in E. coli are only tolerated when the growth rate is reduced by propagation on minimal media or at low temperature (Ruiz et al., 2005; Wu et al., 2005).
Table 1.
Doubling times (h) of wild-type, ΔbamE and complemented strains
During exponential growth, the OD660 and c.f.u. ml−1 were measured for each indicated strain. The doubling time was calculated for each experiment, and the mean±sd for three independent experiments is reported.
| Strain | PYE | M2G | ||
|---|---|---|---|---|
| OD660 | c.f.u. ml−1 | OD660 | c.f.u. ml−1 | |
| CB15N/pMR10 | 1.67±.07 | 1.67±.12 | 2.46±.11 | 2.35±.07 |
| ΔbamE/pMR10 | 2.72±.34 | 2.91±.35 | 2.82±.10 | 2.86±.21 |
| ΔbamE/pbamE | 1.71±.12 | 1.62±.02 | 2.44±.10 | 2.66±.11 |
Heat and chemical sensitivity of the ΔbamE mutant
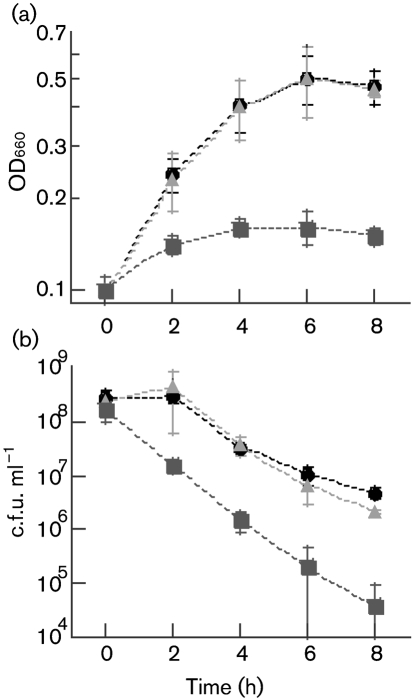
The P. aeruginosa mutant lacking OmlA is not thermosensitive (Ochsner et al., 1999), but the S. Typhimurium bamE mutant grows more slowly than a wild-type strain at 46 °C (Lewis et al., 2008). To determine if BamE is necessary for Caulobacter survival at elevated temperatures, we incubated the strains CB15N/pMR10, ΔbamE/pMR10 and ΔbamE/pbamE at 42 °C and measured the optical density (660 nm) and viable cell count of each culture. CB15N/pMR10 and ΔbamE/pbamE increased in optical density during the incubation (Fig. 4a), but the number of viable cells ml−1 decreased by approximately 100-fold (Fig. 4b). ΔbamE/pMR10 underwent only a minimal increase in optical density (Fig. 4a) and decreased approximately 10 000-fold in viability (Fig. 4b). Thus, in Caulobacter, BamE contributes to survival during exposure to high temperatures.
Fig. 4.
Cells lacking BamE have decreased growth and viability during heat exposure. Strains were grown to OD660=0.2–0.5 in PYE/kan medium at 30 °C, diluted to OD660=0.1, then incubated at 42 °C for the duration of the experiment. At the indicated times, samples were withdrawn from each culture, and OD660 (a) and viable cells (b) were measured. CB15N/pMR10, •; ΔbamE/pMR10, ▪; ΔbamE/pbamE, ▴. Error bars, ±sd.
Mutants in other species lacking components of the BAM complex are hypersensitive to some chemical agents due to membrane permeability defects (Fardini et al., 2009; Lewis et al., 2008; Ochsner et al., 1999; Ruiz et al., 2006; Volokhina et al., 2009). We tested the sensitivity of the wild-type and ΔbamE Caulobacter strains to a variety of detergents and antibiotics. ΔbamE cells displayed enhanced sensitivity to the anionic detergents deoxycholate and SDS, but not to the nonionic detergents Tween 20 and Triton X-100 (Table 2). Growth of ΔbamE cells was selectively inhibited by 0.5 M EDTA and by the antibiotics nalidixic acid, carbenicillin and rifampicin (Table 2), which target DNA gyrase, peptidoglycan transpeptidation and RNA polymerase, respectively (Raleigh et al., 2002). Hypersensitivity to each chemical was complemented by pbamE (Table 2). The ΔbamE mutation in Caulobacter causes hypersensitivity to antibiotics which affect disparate processes in different cellular compartments, consistent with a role for BamE in generating or maintaining the outer membrane permeability barrier.
Each species whose bamE/omlA mutant has been tested for chemical sensitivity defects displays a different basal level of sensitivity to different detergents and antibiotics (Fardini et al., 2009; Fuangthong et al., 2008; Lewis et al., 2008; Ochsner et al., 1999). For example, wild-type Xanthomonas campestris pv. phaseoli, P. aeruginosa and C. crescentus are growth-inhibited by 10 % SDS, but S. Typhimurium is unaffected by 20 % SDS. Furthermore, the bamE/omlA mutants of P. aeruginosa and C. crescentus are much more sensitive to SDS than their respective wild-type strains, but the omlA mutant of X. campestris is growth-inhibited to the same extent as the wild-type strain. It is therefore difficult to make broad statements about the effects of individual chemicals on the loss of BamE/OmlA. However, we noted two trends: (i) whether the species in question was growth-inhibited by non-ionic detergents or not, the bamE/omlA mutant of the same species was not affected to a greater extent than the wild-type (Fuangthong et al., 2008; Lewis et al., 2008; Ochsner et al., 1999); and (ii) all species tested were growth-inhibited by rifampicin, and all bamE/omlA mutants were more sensitive to rifampicin than the wild-type strains (Fardini et al., 2009; Lewis et al., 2008; Ochsner et al., 1999). Chemical sensitivity is influenced by the specific complement of porins and outer membrane transporters present in each wild-type strain, as well as which molecules are affected by the loss of BamE/OmlA, so it is not surprising to find distinct phenotypes in different species.
Membranes of ΔbamE cells are deficient in some OMPs
When proteins of the BAM complex are removed from N. meningitidis, E. coli or S. Enteritidis by mutation or depletion, OMPs of various types are reduced in abundance or accumulate in premature forms (Fardini et al., 2009; Ruiz et al., 2005; Sklar et al., 2007; Voulhoux et al., 2003; Wu et al., 2005). Depletion of BamA or BamD causes profound effects on OMP biogenesis, as only one OMP, PulD, has been reported to be normally assembled when BamA is depleted (Collin et al., 2007). In the case of BamC, a non-essential member of the complex in E. coli, the TolC protein is correctly assembled in the deletion mutant, but OmpA, LamB and OmpF/C are not (Charlson et al., 2006; Ruiz et al., 2005; Wu et al., 2005). An E. coli bamE mutant has reduced OmpA levels and accumulates an unfolded form of LamB (Sklar et al., 2007), but no other OMP defects have been reported. The non-uniform dependence of OMPs upon subunits of the BAM complex could indicate that some components are not strictly necessary for OMP assembly, but improve the efficiency of the process. Alternatively, individual OMPs may be channelled to the central component, BamA, using different subsets of the accessory lipoproteins, BamB–E.
We separated the membrane and soluble fractions of Caulobacter cells containing or lacking BamE to determine if known Caulobacter OMPs are present in reduced levels in the ΔbamE mutant. The RsaFa and RsaFb proteins are found exclusively in the Caulobacter membrane fraction, and they are present in similar amounts in wild-type and ΔbamE cells (Fig. 5a). RsaFa and RsaFb are expected to function as trimers, similar to TolC (Koronakis et al., 2000). To determine if the RsaF proteins in the ΔbamE mutant are competent to secrete RsaA, we isolated surface-exposed RsaA protein by low-pH extraction (Koronakis et al., 2000). Surface RsaA levels were similar in cells of CB15N/pMR10, ΔbamE/pMR10 and ΔbamE/pbamE (Fig. 5b), suggesting that RsaF is assembled into functional trimers in the absence of BamE.
We observed CpaC monomers in both the soluble and membrane fractions, in equal amounts in the wild-type and ΔbamE strains (Fig. 5c), indicating that ΔbamE cells can still express CpaC monomers and target them to the cell envelope. The high-molecular-mass form CpaC* was present only in the membrane fraction, and the level of CpaC* was greatly reduced in the mutant ΔbamE (Fig. 5c, lanes 3 and 4). These results suggest that BamE is necessary for the assembly or maintenance of CpaC multimers in the Caulobacter outer membrane.
To detect additional proteins affected by the loss of BamE, we analysed the soluble and membrane fractions of wild-type and ΔbamE cells by SDS-PAGE and staining with GelCode Blue. Both strains had roughly the same composition of soluble proteins (Fig. 5d, lanes 1 and 2). Unlike the P. aeruginosa omlA and S. Enteritidis bamE mutants that have little or no change in their OMP profiles (Fardini et al., 2009; Ochsner et al., 1999), ΔbamE membranes were severely deficient in three prominent, high molecular mass proteins (Fig. 5d, lanes 3 and 4). These three proteins were excised from the gel lane containing wild-type membranes, digested with trypsin and identified by mass spectrometry.
Each band was primarily composed of an OMP homologous to TonB-dependent receptors. Band 1 was identified as CC3013, band 2 as CC2819 and band 3 as CC0210. (For example, 83 % of the Caulobacter peptides identified in band 1 were derived from CC3013, covering 49.9 % of the CC3013 protein sequence. The next most abundant protein, CC0288 contributed only 10 % of the Caulobacter peptides in band 1, yielding 14.7 % sequence coverage.) All three proteins were identified in a proteomic study of Caulobacter OMPs (Phadke et al., 2001), and CC0210 and CC2819 were also identified in a study of proteins enriched in Caulobacter stalks (Ireland et al., 2002). Finally, CC3013 and CC0210 were identified as OMPs that are abundant during growth on rich medium but not on minimal medium (Neugebauer et al., 2005). Because the levels of CpaC* and three abundant TonB-dependent receptors are reduced in ΔbamE membranes, and because Caulobacter BamE associates with other BAM complex homologues, we propose that it participates in the assembly of these OMPs.
BamE is required for normal stalk biogenesis
In PYE medium, stalks of the mutant ΔbamE were shorter than those of wild-type cells, and this morphological defect was corrected by supplying bamE on a low-copy plasmid (Fig. 6b, d). The mean stalk lengths (μm) in PYE, calculated from measurements of >250 stalks for each strain, were 2.5±0.8 for CB15N/pMR10, 1.4±0.4 for ΔbamE/pMR10 and 2.6±0.7 for ΔbamE/pbamE.
Under conditions of phosphate limitation, wild-type Caulobacter cells grow elongated stalks (Gonin et al., 2000; Schmidt & Stanier, 1966) which enhance nutrient uptake (Wagner et al., 2006). Some strains, such as the mutants ctrA401 (Quon et al., 1996), rpoN (Brun & Shapiro, 1992) and tacA (Biondi et al., 2006; Skerker et al., 2005), are stalkless in rich medium but build polar stalks in low-phosphate medium. In M5G low-phosphate medium, the stalks of ΔbamE cells were longer than in PYE medium (Fig. 6c and f), indicating that this mutant can sense and respond to phosphate limitation. However, stalks of the mutant strain were still much shorter than those of wild-type cells or of the mutant complemented by pbamE (Fig. 6a and e–g). The stalk length distributions for the wild-type and complemented strains grown in low-phosphate medium are broad because measurements were performed using mixed cultures, which contain newly born stalked cells as well as older cells that have been engaged in stalk elongation for several generations.
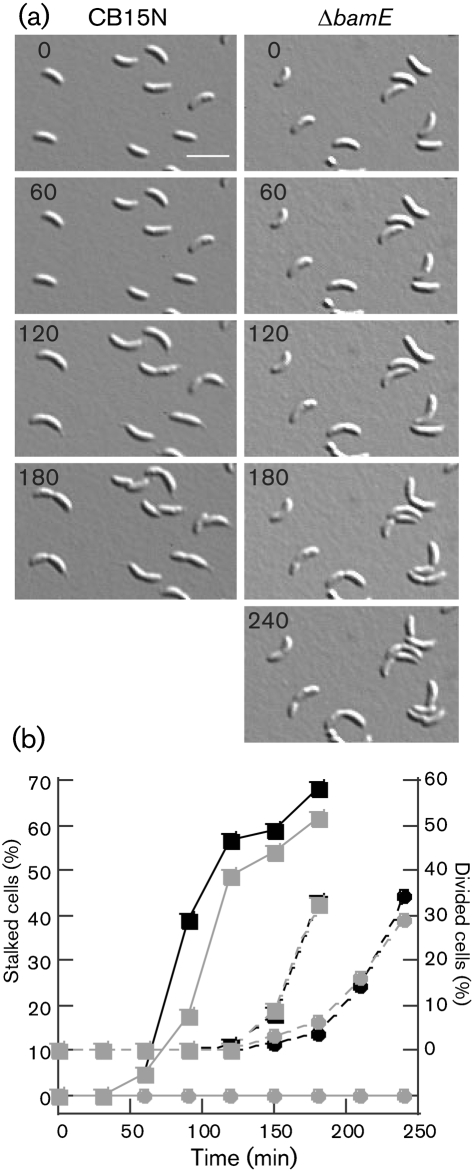
In micrographs of the mutant ΔbamE/pMR10 grown in low-phosphate medium, we observed debris that resembled broken stalks (Fig. 6f, white arrowheads). Short stalks could result from a defect in stalk growth, from a normal rate of growth followed by stalk breakage, or from a combination of breakage and impaired growth. The cells in these images were grown in shaking cultures, where shear forces could promote stalk breakage. To eliminate the effects of stalk breakage and to determine whether stalk growth is initiated at the proper time during differentiation, we performed time-lapse microscopy on cells growing on agarose pads. We isolated swarmer cells of the wild-type and ΔbamE strains and immobilized them on agarose pads containing minimal medium (M2G). We photographed the cells at 30 min intervals, noting the time of division and the first appearance of a stalk on each cell in the field. In these experiments, 33 % of wild-type cells divided completely by 180 min, and ∼60 % of cells had a visible stalk at this time (Fig. 7). ΔbamE cells took 210 min to achieve 30 % cell division, consistent with their slower growth in liquid cultures, but at this time, no cells had visible stalks (Fig. 7). We know that stalk growth is not completely inhibited in the mutant because liquid cultures contain cells with visible, albeit short, stalks (Fig. 6). Therefore, the short stalk phenotype of ΔbamE cells is caused at least in part by slow or delayed stalk growth.
Fig. 7.
BamE is required for normal stalk initiation during swarmer cell differentiation. (a) CB15N or ΔbamE swarmer cells were isolated, immobilized on M2G agarose pads and photographed at the indicated times (min) until divided cells were visible. Representative time points and fields for each strain are shown. Bar, 2.5 μm. (b) In time-lapse micrographs of CB15N (squares) and ΔbamE (circles), the percentage of cells that had grown a visible stalk (solid lines) or divided (dashed lines) was calculated at each time point. Black and grey symbols represent two independent experiments where >70 cells were observed.
The C-terminal extension of BamE is important for optimal BamE function, but not for stability of the BAM complex
To determine the importance of the C-terminal segment of BamE beyond the conserved BamE/OmlA domain, we truncated the protein after amino acid 115 and appended the 3×FLAG epitope. The resulting protein, BamEΔC–3×FLAG, was expressed in the ΔbamE mutant at levels comparable to the wild-type, tagged protein (data not shown). Cells expressing the truncated BamE protein were less sensitive to several chemicals than the ΔbamE mutant, but more sensitive than the mutant expressing full-length BamE (Table 2). When grown in M5G-low phosphate medium, cells expressing BamEΔC–3×FLAG grew extended stalks, similar to cells containing full-length versions of BamE (Fig. 6a, i). Thus, BamEΔC–3×FLAG complements different defects of the ΔbamE mutant to different extents.
A previous study of the E. coli BAM complex identified alleles of bamB (yfgL) that caused little or no phenotypic defect, yet weakened the interaction between BamB and BamA in co-immunoprecipitation experiments, suggesting that the affected amino acids are important for BAM complex stability (Vuong et al., 2008). To generate intermediate defects in chemical sensitivity (Table 2), either the function of Caulobacter BamEΔC or the stability of the resulting BAM complex could be compromised. We used anti-FLAG M2 agarose to isolate BamEΔC and its associated proteins from lysates of the strain ΔbamE/pbamEΔC : : 3×FLAG. BamA, BamB and BamD were present in coimmunoprecipitates of the truncated BamE protein, at levels comparable to the wild-type complex (Fig. 3, lane 3), and protein identities were confirmed by mass spectrometry. Thus, functional impairment of the truncated BamE protein is likely to cause the chemical sensitivity phenotype, rather than an inability of BamEΔC to associate with other BAM proteins.
DISCUSSION
Genes encoding predicted lipoproteins of the BamE/OmlA family are found in sequenced genomes of alpha-, beta- and gammaproteobacteria, and bamE or omlA mutants have been characterized in P. aeruginosa (Ochsner et al., 1999), E. coli (Sklar et al., 2007), S. Typhimurium (Lewis et al., 2008), S. Enteritidis (Fardini et al., 2009), X. campestris (Fuangthong et al., 2008) and N. meningitidis (Volokhina et al., 2009). The phenotypes of these mutants include slow growth, reduced virulence and enhanced sensitivity to detergents, antibiotics and heat. The E. coli bamE mutant also contained reduced levels of OmpA and accumulated an unfolded form of LamB. Here, we report the first studies of a BamE/OmlA homologue in an alphaproteobacterium, C. crescentus. The ΔbamE mutant is hypersensitive to anionic detergents and some antibiotics, has reduced survival during heat exposure and grows slowly in rich medium. BamE is located in the Caulobacter outer membrane and coprecipitates with three other proteins whose homologues in E. coli are involved in OMP assembly. Finally, the membranes of ΔbamE cells are deficient in three TonB-dependent receptors and CpaC*, an aggregated form of the secretin CpaC. CpaC monomers are present in the Caulobacter membrane fraction in equal amounts in wild-type and ΔbamE cells, but the aggregated form CpaC* is specifically deficient in cells lacking BamE. Taken together, these results strongly suggest that BamE performs a function in Caulobacter similar to its role in other bacteria, namely to assemble outer membrane β-barrel proteins as part of the BAM complex. The pilus assembly protein CpaE is also required to accumulate CpaC* (Viollier et al., 2002), so it is possible that BamE has an indirect effect upon CpaC via CpaE. However, CpaE itself is predicted to be located in the cytoplasm (Skerker & Shapiro, 2000; Viollier et al., 2002), so a more likely hypothesis is that BamE affects CpaC downstream of the step requiring CpaE. A second caveat is that we have not yet examined the assembly kinetics of the missing TonB-dependent receptors in wild-type and ΔbamE cells. The synthesis of one or more of these receptors could be downregulated in the ΔbamE mutant, either due to the slower growth rate of ΔbamE cells or to a specific mechanism that reduces OMP expression during envelope stress (reviewed by Vogel & Papenfort, 2006).
Although they have generally similar phenotypes, bamE and omlA mutations cause phenotypes of different severity in different species. For example, the E. coli bamE mutant is not impaired in either growth rate or heat survival (Sklar et al., 2007). The P. aeruginosa omlA mutant also grows at a normal rate and has no obvious changes in its OMP profile (Ochsner et al., 1999). In these respects, the Caulobacter ΔbamE mutant seems to have a more severe phenotype, because its growth rate, its heat and chemical sensitivity, and its complement of OMPs are all affected.
The composition of the BAM complex differs in the three species where it has been determined. Compared with E. coli, the Caulobacter complex lacks the lipoprotein BamC (Gatsos et al., 2008), while the N. meningitidis complex lacks the lipoprotein BamB and contains the additional protein RmpM (Volokhina et al., 2009). The C terminus of RmpM is predicted to bind peptidoglycan and is similar to the C-terminal region of OmpA, but RmpM lacks the N-terminal β-barrel-forming region of OmpA (Grizot & Buchanan, 2004). Instead, RmpM contains an approximately 40 aa N-terminal domain followed by a proline-rich region of approximately 20 residues. Three proteins encoded in Caulobacter (CC3229, CC0201 and CC2845) have features similar to RmpM (data not shown), but further studies will be necessary to determine if they associate with the Caulobacter BAM complex or are functionally similar to RmpM.
Differences in the composition of the BAM complex could render Caulobacter BamE more critical for membrane integrity than its counterparts in other bacteria. In E. coli, BamC, BamD and BamE interact with BamA as a group, while the BamA–BamB interaction is independent of the other lipoproteins (Malinverni et al., 2006; Sklar et al., 2007; Vuong et al., 2008). In mutants lacking either BamC or BamE, BamD levels are normal but less BamD is associated with BamA. A recent report demonstrated that an N. meningitidis bamE bamC double mutant is inviable, supporting the idea that these two lipoproteins function in the same process (Volokhina et al., 2009). Thus, because Caulobacter lacks a homologue of bamC, BamE may play a greater role in maintaining the interaction between BamA and BamD. We hypothesized that the C-terminal extension of Caulobacter BamE, beyond the conserved BamE/OmlA domain, might play an important role in stabilizing the BAM complex, particularly in the absence of a BamC homologue. When we truncated BamE, however, we found that the BAM complex was intact (Fig. 3). Thus, BamE itself may be more important for BAM complex stability in Caulobacter, but this is not the primary function of the BamE C-terminal segment.
Alternatively, the loss of BamE may cause a more severe phenotype in Caulobacter because this species contains a different complement of OMPs than E. coli, P. aeruginosa, N. meningitidis or Salmonella species. The Caulobacter outer membrane proteome is skewed toward OmpA homologues and TonB-dependent receptors (Phadke et al., 2001). While there is evidence that BamA interacts directly with β-barrel protein substrates (Habib et al., 2007; Knowles et al., 2008; Robert et al., 2006), different types of OMPs may be channelled to BamA via subsets of the accessory lipoproteins. If OmpA homologues or TonB-dependent receptors specifically require BamE for assembly, then bacteria whose outer membranes are rich in these proteins may be more susceptible to the loss of BamE.
Although ΔbamE cells have shorter stalks than wild-type cells in all media tested, the growth defect and chemical hypersensitivity of the mutant indicate that the function of BamE is not limited to stalk biogenesis. Consistent with these results, BamE and other members of the BAM complex are not enriched in the Caulobacter stalk proteome (Ireland et al., 2002). However, stalk biogenesis in the ΔbamE mutant may be impaired more than membrane biogenesis overall. During synchronous growth of the ΔbamE mutant in time-lapse experiments, swarmer cells elongated and divided, but unlike wild-type cells, they did not synthesize visible stalks before cell division (Fig. 7). Because ΔbamE cells grown in liquid media possess stalks, we interpret these results to mean that stalk outgrowth is delayed in the mutant, and the loss of BamE affects stalk growth relatively more than growth and division of the cell body. Stalk length is not always reduced in mutants with a slower growth rate; for example, null mutants in divJ grow more slowly than wild-type cells (Sciochetti et al., 2002), yet their stalks are as long as those of wild-type cells (Wheeler & Shapiro, 1999).
We propose that BamE, like its homologues in other species, participates in assembling a variety of β-barrel proteins into the outer membrane as part of the Caulobacter BAM complex. Without BamE, the complex may be inefficient at assembling many OMP substrates, causing a general loss of membrane integrity. To account for the delay in stalk outgrowth in ΔbamE cells, we also propose that the Caulobacter BAM complex assembles one or more substrates that specifically participate in stalk initiation or elongation. The assembly of these substrates may be particularly impaired in cells lacking BamE, causing cell division to occur before the first appearance of a stalk.
The lipoproteins of the E. coli BAM complex are transcriptionally regulated by σE (Dartigalongue et al., 2001; Kabir et al., 2005; Lewis et al., 2008; Rezuchova et al., 2003), an extracytoplasmic function (ECF) sigma factor that directs the transcription of chaperones, assembly factors and proteases to restore homeostasis after envelope stress (Ruiz & Silhavy, 2005). The Caulobacter genome encodes 13 ECF sigma factors (Nierman et al., 2001), and it is currently unclear which of them perform functions analogous to E. coli σE (Alvarez-Martinez et al., 2006, 2007). Caulobacter also contains an essential two-component system, composed of the histidine kinase CenK and the DNA-binding response regulator CenR (Skerker et al., 2005). Depletion of either CenK or CenR leads to severe membrane blebbing and death, suggesting that the transcriptional targets of this system are intimately involved in cell envelope integrity. Future studies will determine how the components of the BAM complex are transcriptionally regulated in Caulobacter.
Our studies have identified the Caulobacter BAM complex and show that the BamE homologue CC1365 plays a role in Caulobacter membrane integrity, stalk outgrowth and establishing or maintaining normal levels of some OMPs. It will be of interest to determine if TonB-dependent receptors, which are abundant in Caulobacter, have a particular requirement for BamE in their assembly. Additional experiments are also needed to identify substrates of the BAM complex that promote stalk outgrowth.
Acknowledgments
We thank Arnold Falick of the HHMI Mass Spectrometry Laboratory and Lori Kohlstaedt and Daniela Mavrici of the QB3 Proteomics/Mass Spectrometry Laboratory at UC Berkeley for protein identification. We are grateful to Steven Hermiz, Jr, Joanne Hsu and Kourosh Kolahi for DNA constructs and to John Smit and Patrick Viollier for the gifts of antibodies. We thank the Komeili and Zusman laboratories for helpful discussions and members of the Ryan lab for critical reading of the manuscript. This work was supported by NSF grant MCB-0543801 to K. R. R. and NIH fellowship F32GM086951 to L. M. B.
Abbreviations
ECF, extracytoplasmic function
OMP, outer membrane protein
PYE, peptone-yeast extract
Footnotes
A supplementary table providing information about strains and plasmids is available with the online version of this paper.
References
- Alley, M. R., Maddock, J. R. & Shapiro, L. (1992). Polar localization of a bacterial chemoreceptor. Genes Dev 6, 825–836. [DOI] [PubMed] [Google Scholar]
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990). Basic local alignment search tool. J Mol Biol 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Alvarez-Martinez, C. E., Baldini, R. L. & Gomes, S. L. (2006). A Caulobacter crescentus extracytoplasmic function sigma factor mediating the response to oxidative stress in stationary phase. J Bacteriol 188, 1835–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Martinez, C. E., Lourenco, R. F., Baldini, R. L., Laub, M. T. & Gomes, S. L. (2007). The ECF sigma factor σT is involved in osmotic and oxidative stress responses in Caulobacter crescentus. Mol Microbiol 66, 1240–1255. [DOI] [PubMed] [Google Scholar]
- Badger, J. H., Hoover, T. R., Brun, Y. V., Weiner, R. M., Laub, M. T., Alexandre, G., Mrázek, J., Ren, Q., Paulsen, I. T. & other authors (2006). Comparative genomic evidence for a close relationship between the dimorphic prosthecate bacteria Hyphomonas neptunium and Caulobacter crescentus. J Bacteriol 188, 6841–6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassford, P. J., Jr & Kadner, R. J. (1977). Genetic analysis of components involved in vitamin B12 uptake in Escherichia coli. J Bacteriol 132, 796–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayan, N., Guilvout, I. & Pugsley, A. P. (2006). Secretins take shape. Mol Microbiol 60, 1–4. [DOI] [PubMed] [Google Scholar]
- Biondi, E. G., Skerker, J. M., Arif, M., Prasol, M. S., Perchuk, B. S. & Laub, M. T. (2006). A phosphorelay system controls stalk biogenesis during cell cycle progression in Caulobacter crescentus. Mol Microbiol 59, 386–401. [DOI] [PubMed] [Google Scholar]
- Blanvillain, S., Meyer, D., Boulanger, A., Lautier, M., Guynet, C., Denance, N., Vasse, J., Lauber, E. & Arlat, M. (2007). Plant carbohydrate scavenging through TonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One 2, e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier, J., Pugsley, A. P. & Stragier, P. (1991). A gene for a new lipoprotein in the dapA–purC interval of the Escherichia coli chromosome. J Bacteriol 173, 5523–5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, V. & Endriss, F. (2007). Energy-coupled outer membrane transport proteins and regulatory proteins. Biometals 20, 219–231. [DOI] [PubMed] [Google Scholar]
- Brun, Y. V. & Shapiro, L. (1992). A temporally controlled sigma-factor is required for polar morphogenesis and normal cell division in Caulobacter. Genes Dev 6, 2395–2408. [DOI] [PubMed] [Google Scholar]
- Charlson, E. S., Werner, J. N. & Misra, R. (2006). Differential effects of yfgL mutation on Escherichia coli outer membrane proteins and lipopolysaccharide. J Bacteriol 188, 7186–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy, M. J. & Newton, A. (1982). Localization of proteins in the inner and outer membranes of Caulobacter crescentus. Biochim Biophys Acta 686, 160–169. [DOI] [PubMed] [Google Scholar]
- Collier, J. & Shapiro, L. (2007). Spatial complexity and control of a bacterial cell cycle. Curr Opin Biotechnol 18, 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin, S., Guilvout, I., Chami, M. & Pugsley, A. P. (2007). YaeT-independent multimerization and outer membrane association of secretin PulD. Mol Microbiol 64, 1350–1357. [DOI] [PubMed] [Google Scholar]
- Dartigalongue, C., Missiakas, D. & Raina, S. (2001). Characterization of the Escherichia coli sigma E regulon. J Biol Chem 276, 20866–20875. [DOI] [PubMed] [Google Scholar]
- Delcour, A. H. (2002). Structure and function of pore-forming β-barrels from bacteria. J Mol Microbiol Biotechnol 4, 1–10. [PubMed] [Google Scholar]
- DelVecchio, V. G., Kapatral, V., Elzer, P., Patra, G. & Mujer, C. V. (2002). The genome of Brucella melitensis. Vet Microbiol 90, 587–592. [DOI] [PubMed] [Google Scholar]
- Doerrler, W. T. & Raetz, C. R. (2005). Loss of outer membrane proteins without inhibition of lipid export in an Escherichia coli YaeT mutant. J Biol Chem 280, 27679–27687. [DOI] [PubMed] [Google Scholar]
- Elias, J. E., Haas, W., Faherty, B. K. & Gygi, S. P. (2005). Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods 2, 667–675. [DOI] [PubMed] [Google Scholar]
- Ely, B. (1991). Genetics of Caulobacter crescentus. Methods Enzymol 204, 372–384. [DOI] [PubMed] [Google Scholar]
- Eng, J. K., McCormack, A. L. & Yates, J. R., III (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5, 976–989. [DOI] [PubMed] [Google Scholar]
- Evinger, M. & Agabian, N. (1977). Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol 132, 294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardini, Y., Trotereau, J., Bottreau, E., Souchard, C., Velge, P. & Virlogeux-Payant, I. (2009). Investigation of the role of the BAM complex and SurA chaperone in outer-membrane protein biogenesis and type III secretion system expression in Salmonella. Microbiology 155, 1613–1622. [DOI] [PubMed] [Google Scholar]
- Fuangthong, M., Sallabhan, R., Atichartpongkul, S., Rangkadilok, N., Sriprang, R., Satayavivad, J. & Mongkolsuk, S. (2008). The omlA gene is involved in multidrug resistance and its expression is inhibited by coumarins in Xanthomonas campestris pv. phaseoli. Arch Microbiol 189, 211–218. [DOI] [PubMed] [Google Scholar]
- Gatsos, X., Perry, A. J., Anwari, K., Dolezal, P., Wolynec, P. P., Likic, V. A., Purcell, A. W., Buchanan, S. K. & Lithgow, T. (2008). Protein secretion and outer membrane assembly in Alphaproteobacteria. FEMS Microbiol Rev 32, 995–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonin, M., Quardokus, E. M., O'Donnol, D., Maddock, J. & Brun, Y. V. (2000). Regulation of stalk elongation by phosphate in Caulobacter crescentus. J Bacteriol 182, 337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grizot, S. & Buchanan, S. K. (2004). Structure of the OmpA-like domain of RmpM from Neisseria meningitidis. Mol Microbiol 51, 1027–1037. [DOI] [PubMed] [Google Scholar]
- Habib, S. J., Waizenegger, T., Niewienda, A., Paschen, S. A., Neupert, W. & Rapaport, D. (2007). The N-terminal domain of Tob55 has a receptor-like function in the biogenesis of mitochondrial β-barrel proteins. J Cell Biol 176, 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House, B. L., Mortimer, M. W. & Kahn, M. L. (2004). New recombination methods for Sinorhizobium meliloti genetics. Appl Environ Microbiol 70, 2806–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iba, H., Fukuda, A. & Okada, Y. (1977). Chromosome replication in Caulobacter crescentus growing in a nutrient broth. J Bacteriol 129, 1192–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland, M. M. E., Karty, J. A., Quardokus, E. M., Reilly, J. P. & Brun, Y. V. (2002). Proteomic analysis of the Caulobacter crescentus stalk indicates competence for nutrient uptake. Mol Microbiol 45, 1029–1041. [DOI] [PubMed] [Google Scholar]
- Jansen, C., Wiese, A., Reubsaet, L., Dekker, N., de Cock, H., Seydel, U. & Tommassen, J. (2000). Biochemical and biophysical characterization of in vitro folded outer membrane porin PorA of Neisseria meningitidis. Biochim Biophys Acta 1464, 284–298. [DOI] [PubMed] [Google Scholar]
- Jimenez, C. R., Huang, L., Qiu, Y. & Burlingame, A. L. (1998). In-gel digestion of proteins for MALDI-MS fingerprint mapping. In Current Protocols in Protein Science, pp. 16.14.11–16.14.15. Edited by J. E. Coligan, B. M. Dunn, H. L. Ploegh, D. W. Speicher & P. T. Wingfield. New York: Wiley. [DOI] [PubMed]
- Kabir, M. S., Yamashita, D., Koyama, S., Oshima, T., Kurokawa, K., Maeda, M., Tsunedomi, R., Murata, M., Wada, C. & other authors (2005). Cell lysis directed by σE in early stationary phase and effect of induction of the rpoE gene on global gene expression in Escherichia coli. Microbiology 151, 2721–2735. [DOI] [PubMed] [Google Scholar]
- Kainth, P. & Gupta, R. S. (2005). Signature proteins that are distinctive of alpha proteobacteria. BMC Genomics 6, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles, T. J., Jeeves, M., Bobat, S., Dancea, F., McClelland, D., Palmer, T., Overduin, M. & Henderson, I. R. (2008). Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol Microbiol 68, 1216–1227. [DOI] [PubMed] [Google Scholar]
- Knowles, T. J., Scott-Tucker, A., Overduin, M. & Henderson, I. R. (2009). Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat Rev Microbiol 7, 206–214. [DOI] [PubMed] [Google Scholar]
- Koronakis, V., Sharff, A., Koronakis, E., Luisi, B. & Hughes, C. (2000). Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405, 914–919. [DOI] [PubMed] [Google Scholar]
- Koronakis, V., Eswaran, J. & Hughes, C. (2004). Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu Rev Biochem 73, 467–489. [DOI] [PubMed] [Google Scholar]
- Lassmann, T. & Sonnhammer, E. L. L. (2005). Kalign – an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics 6, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewenza, S., Vidal-Ingigliardi, D. & Pugsley, A. P. (2006). Direct visualization of red fluorescent lipoproteins indicates conservation of the membrane sorting rules in the family Enterobacteriaceae. J Bacteriol 188, 3516–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, C., Skovierova, H., Rowley, G., Rezuchova, B., Homerova, D., Stevenson, A., Sherry, A., Kormanec, J. & Roberts, M. (2008). Small outer-membrane protein lipoprotein, SmpA, is regulated by σE and has a role in cell envelope integrity and virulence of Salmonella enterica serovar Typhimurium. Microbiology 154, 979–988. [DOI] [PubMed] [Google Scholar]
- Malinverni, J. C., Werner, J., Kim, S., Sklar, J. G., Kahne, D., Misra, R. & Silhavy, T. J. (2006). YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol 61, 151–164. [DOI] [PubMed] [Google Scholar]
- McLeod, M., Craft, S. & Broach, J. R. (1986). Identification of the crossover site during FLP-mediated recombination in the Saccharomyces cerevisiae plasmid μ circle. Mol Cell Biol 6, 3357–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy, M. P., Phadke, N. D., Maddock, J. R. & Andrews, P. C. (2001). Two-dimensional electrophoresis and peptide mass fingerprinting of bacterial outer membrane proteins. Electrophoresis 22, 1686–1696. [DOI] [PubMed] [Google Scholar]
- Narita, S. & Tokuda, H. (2007). Amino acids at positions 3 and 4 determine the membrane specificity of Pseudomonas aeruginosa lipoproteins. J Biol Chem 282, 13372–13378. [DOI] [PubMed] [Google Scholar]
- Narita, S., Matsuyama, S. & Tokuda, H. (2004). Lipoprotein trafficking in Escherichia coli. Arch Microbiol 182, 1–6. [DOI] [PubMed] [Google Scholar]
- Neugebauer, H., Herrmann, C., Kammer, W., Schwartz, G., Nordheim, A. & Braun, V. (2005). ExbBD-dependent transport of maltodextrins through the novel MalA protein across the outer membrane of Caulobacter crescentus. J Bacteriol 187, 8300–8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman, W. C., Feldblyum, T. V., Laub, M. T., Paulsen, I. T., Nelson, K. E., Eisen, J. A., Heidelberg, J. F., Alley, M. R., Ohta, N. & other authors (2001). Complete genome sequence of Caulobacter crescentus. Proc Natl Acad Sci U S A 98, 4136–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido, H. (2003). Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67, 593–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner, U. A., Vasil, A. I., Johnson, Z. & Vasil, M. I. (1999). Pseudomonas aeruginosa fur overlaps with a gene encoding a novel outer membrane lipoprotein, OmlA. J Bacteriol 181, 1099–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadke, N. D., Molloy, M. P., Steinhoff, S. A., Ulintz, P. J., Andrews, P. C. & Maddock, J. R. (2001). Analysis of the outer membrane proteome of Caulobacter crescentus by two-dimensional electrophoresis and mass spectrometry. Proteomics 1, 705–720. [DOI] [PubMed] [Google Scholar]
- Poindexter, J. S. (1964). Biological properties and classification of the Caulobacter group. Bacteriol Rev 28, 231–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poindexter, J. L. S. & Cohen-Bazire, G. (1964). The fine structure of stalked bacteria belonging to the family Caulobacteraceae. J Cell Biol 23, 587–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postle, K. & Larsen, R. A. (2007). TonB-dependent energy transduction between outer and cytoplasmic membranes. Biometals 20, 453–465. [DOI] [PubMed] [Google Scholar]
- Prinz, T. & Tommassen, J. (2000). Association of iron-regulated outer membrane proteins of Neisseria meningitidis with RmpM (class 4) protein. FEMS Microbiol Lett 183, 49–53. [DOI] [PubMed] [Google Scholar]
- Pugsley, A. P. & Reeves, R. (1976). Iron uptake in colicin B-resistance mutants of Escherichia coli K-12. J Bacteriol 126, 1052–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quon, K. C., Marczynski, G. T. & Shapiro, L. (1996). Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84, 83–93. [DOI] [PubMed] [Google Scholar]
- Raleigh, E. A., Elbing, K. & Brent, R. (2002). Selected topics from classical bacterial genetics. In Current Protocols in Molecular Biology, pp. 1.4.1–1.4.14. Edited by F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith & K. Struhl. New York: Wiley. [DOI] [PubMed]
- Reisinger, S. J., Huntwork, S., Viollier, P. H. & Ryan, K. R. (2007). DivL performs critical cell cycle functions in Caulobacter crescentus independent of kinase activity. J Bacteriol 189, 8308–8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezuchova, B., Miticka, H., Homerova, D., Roberts, M. & Kormanec, J. (2003). New members of the Escherichia coli σE regulon identified by a two-plasmid system. FEMS Microbiol Lett 225, 1–7. [DOI] [PubMed] [Google Scholar]
- Robert, V., Volokhina, E. B., Senf, F., Bos, M. P., Van Gelder, P. & Tommassen, J. (2006). Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol 4, e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolhion, N., Barnich, N., Claret, L. & Darfeuille-Michaud, A. (2005). Strong decrease in invasive ability and outer membrane vesicle release in Crohn's disease-associated invasive Escherichia coli strain Lf82 with the yfgL gene deleted. J Bacteriol 187, 2286–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz, N. & Silhavy, T. J. (2005). Sensing external stress: watchdogs of the Escherichia coli cell envelope. Curr Opin Microbiol 8, 122–126. [DOI] [PubMed] [Google Scholar]
- Ruiz, N., Falcone, B., Kahn, D. & Silhavy, T. J. (2005). Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 121, 307–317. [DOI] [PubMed] [Google Scholar]
- Ruiz, N., Wu, T., Kahne, D. & Silhavy, T. J. (2006). Probing the barrier function of the outer membrane with chemical conditionality. ACS Chem Biol 1, 385–395. [DOI] [PubMed] [Google Scholar]
- Sankaran, K., Gupta, S. D. & Wu, H. C. (1995). Modification of bacterial lipoproteins. Methods Enzymol 250, 683–697. [DOI] [PubMed] [Google Scholar]
- Schmidt, J. M. & Stanier, R. Y. (1966). The development of cellular stalks in bacteria. J Cell Biol 28, 423–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciochetti, S. A., Lane, T., Ohta, N. & Newton, A. (2002). Protein sequences and cellular factors required for polar localization of a histidine kinase in Caulobacter crescentus. J Bacteriol 184, 6037–6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz, L. C. & Brun, Y. V. (1998). Genetic analysis of mecillinam-resistant mutants of Caulobacter crescentus deficient in stalk biosynthesis. J Bacteriol 180, 5235–5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydel, A., Gounon, P. & Pugsley, A. P. (1999). Testing the ‘+2 rule’ for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol Microbiol 34, 810–821. [DOI] [PubMed] [Google Scholar]
- Skerker, J. M. & Shapiro, L. (2000). Identification and cell-cycle control of a novel pilus system in Caulobacter crescentus. EMBO J 19, 3223–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker, J. M., Prasol, M. S., Perchuk, B. S., Biondi, E. G. & Laub, M. T. (2005). Two-component signal transduction pathways regulating growth and cell cycle progresion in a bacterium: A systems-level analysis. PLoS Biol 3, e334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar, J. G., Wu, T., Gronenberg, L. S., Malinverni, J. C., Kahne, D. & Silhavy, T. J. (2007). Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci U S A 104, 6400–6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stove, J. L. & Stanier, R. Y. (1962). Cellular differentiation in stalked bacteria. Nature 196, 1189–1192. [Google Scholar]
- Tabb, D. L., McDonald, W. H. & Yates, J. R. (2002). DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res 1, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toporowski, M. C., Nomellini, J. F., Awram, P. & Smit, J. (2004). Two outer membrane proteins are required for maximal type I secretion of the Caulobacter crescentus S-layer protein. J Bacteriol 186, 8000–8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanini, M. M. T., Spisini, A., Sforca, M. L., Pertinhez, T. A. & Benedetti, C. E. (2008). The solution structure of the outer membrane lipoprotein OmlA from Xanthomonas axonopodis pv. cirtri reveals a protein fold implicated in protein–protein interaction. Proteins 71, 2051–2064. [DOI] [PubMed] [Google Scholar]
- Viollier, P. H., Sternheim, N. & Shapiro, L. (2002). A dynamically localized histidine kinase controls the asymmetric distribution of polar pili proteins. EMBO J 21, 4420–4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J. & Papenfort, K. (2006). Small non-coding RNAs and the bacterial outer membrane. Curr Opin Microbiol 9, 605–611. [DOI] [PubMed] [Google Scholar]
- Volokhina, E. B., Beckers, F., Tomassen, J. & Bos, M. P. (2009). The β-barrel outer membrane protein assembly complex of Neisseria meningitidis. J Bacteriol 191, 7074–7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voulhoux, R., Bos, M. P., Geurtsen, J., Mols, M. & Tommassen, J. (2003). Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299, 262–265. [DOI] [PubMed] [Google Scholar]
- Vuong, P., Bennion, D., Mantei, J., Frost, D. & Misra, R. (2008). Analysis of YfgL and YaeT interactions through bioinformatics, mutagenesis, and biochemistry. J Bacteriol 190, 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, J. K., Galvani, C. D. & Brun, Y. V. (2005). Caulobacter crescentus requires RodA and MreB for stalk synthesis and prevention of ectopic pole formation. J Bacteriol 187, 544–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, J. K., Setayeshgar, S., Sharon, L. A., Reilly, J. P. & Brun, Y. V. (2006). A nutrient uptake role for bacterial envelope extensions. Proc Natl Acad Sci U S A 103, 11772–11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, J. & Misra, R. (2005). YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent proteins of Escherichia coli. Mol Microbiol 57, 1450–1459. [DOI] [PubMed] [Google Scholar]
- Wheeler, R. T. & Shapiro, L. (1999). Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol Cell 4, 683–694. [DOI] [PubMed] [Google Scholar]
- Wu, T., Malinverni, J., Ruiz, N., Kim, S., Silhavy, T. J. & Kahne, D. (2005). Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121, 235–245. [DOI] [PubMed] [Google Scholar]