Abstract
Ljungan virus (LV) was discovered 20 years ago in Swedish bank voles (Myodes glareolus, previously referred to as Clethrionomys glareolus) during the search for an infectious agent causing lethal myocarditis in young athletes. To date, the genomes of four LV isolates, including the prototype 87-012 strain, have been characterized. Three of these LV strains were isolated from bank voles trapped in Sweden. Sequence analysis of an American virus (M1146), isolated from a montane vole (Microtus montanus) in western USA, indicates that this strain represents a genotype that is different from the Swedish strains. Here, we present genomic analyses of a fifth LV strain (64-7855) isolated from a southern red-backed vole (Myodes gapperi) trapped during arbovirus studies in New York state in the north-eastern USA in the 1960s. Sequence analysis of the 64-7855 genome showed an LV-like genome organization and sequence similarity to other LV strains. Genetic and phylogenetic analyses of the evolutionary relationship between the 64-7855 strain and other viruses within the family Picornaviridae, including previously published LV strains, demonstrated that the 64-7855 strain constitutes a new genotype within the LV species. Analyses also showed that different regions of the 64-7855 genome have different phylogenetic relationships with other LV strains, indicating that previous recombination events have been involved in the evolution of this virus.
INTRODUCTION
Ljungan virus (LV) is one of two species within the genus Parechovirus, family Picornaviridae, a large family consisting of more than 300 virus serotypes divided into nine different genera (Stanway et al., 2005). Human parechovirus (HPeV), the other species in the genus Parechovirus, is a human pathogen often isolated from children with diarrhoea and gastroenteritis (Stanway & Hyypiä, 1999). Recently, several new HPeV genotypes have been characterized (Al-Sunaidi et al., 2007; Watanabe et al., 2007) and one of these genotypes, HPeV3, seems to be widely distributed (Abed & Boivin, 2005; Benschop et al., 2006; Boivin et al., 2005; Ito et al., 2004). LV was first isolated from bank voles (Myodes glareolus) trapped in Medelpad and Västerbotten counties in Sweden during the search for an infectious agent causing human disease (Niklasson et al., 1998, 1999). LV has been suggested as an aetiological agent of several human diseases, based on coinciding fluctuations of vole populations in northern Sweden and increasing incidences of type I diabetes mellitus, myocarditis and Guillain–Barré syndrome (Niklasson et al., 1998). Recently, LV antigens were detected by immunohistochemistry in fetal tissue samples in cases of human intrauterine fetal death (Niklasson et al., 2007). However, more information is needed before the role of LV as a possible zoonotic agent can be evaluated.
Three LV strains were initially isolated from Swedish bank voles: the prototype strain 87-012 and the strains 174F and 145SL. Genome sequence analyses showed that the 87-012 and 174F strain constitute genotype 1, since they are almost identical, while the 145SL strain was clearly related but more distant from the two other strains, thus representing a second genotype (Johansson et al., 2002). Phylogenetic analyses based on the 2C protease (2Cpro) and 3D polymerase (3Dpol) sequences showed that LV is closely related to HPeV, although monophyletic groups including these different viruses are clearly separated (Johansson et al., 2002; Lindberg & Johansson, 2002). Because of observed genetic differences, LV was assigned to a new species within the genus Parechovirus (Stanway et al., 2005). A fourth LV strain (M1146) isolated from a montane vole (Microtus montanus) in Oregon, USA, was recently characterized (Johansson et al., 2003). Analyses of the genomic P1 region showed that M1146 represents a different genotype than the Swedish LV strains. Molecular characterization of isolated LV strains has revealed that these viruses have unusual genomic features compared with other picornaviruses. These features include genes encoding only three different structural proteins (Ekström et al., 2007a; Johansson et al., 2004; Tolf et al., 2008), but also a rare combination with genomic sequences coding for two different 2A protein motifs (Johansson et al., 2002, 2003). As it is a recently discovered virus, studies of LV have, until now, mainly focused on basic characterization of its genomic features and replication capacity in cell culture. However, in order to achieve a more comprehensive understanding of LV biology, including its evolution, additional information is needed about its host range, geographical distribution and genetic variation.
In this study, we analysed the genomic sequence of a fifth LV strain, 64-7855, which was isolated from a southern red-backed vole (Myodes gapperi) trapped during arbovirus studies in the north-eastern USA (Whitney et al., 1970). Genetic characterization of the 64-7855 strain revealed an LV-like genome organization, and phylogenetic analyses of the VP1 and 3Dpol protein sequences demonstrated that the 64-7855 strain constitutes a fourth genotype within the LV species. Interestingly, sequence analyses also demonstrated that the 64-7855 genome contains different regions with shifting phylogenetic relationships towards other members of the LV species. In a central region of the genome, including parts of the 2C and 3A genes, 64-7855 is most closely related to the Swedish 145SL strain, while a closer genetic relationship to the other American strain M1146 was observed in the remainder of the genome. One possible explanation for these inconsistent genetic relationships in different regions of the 64-7855 genome is that previous recombination events have affected LV evolution.
METHODS
Viruses and sequences.
The 64-7855 strain was isolated in the early 1960s in New York state, USA (Whitney et al., 1970), and obtained from the World Reference Center for Emerging Viruses and Arboviruses at the University of Texas Medical Branch. Biophysical assays at that time suggested that the isolate was a member of the family Parvo- or Picornaviridae. Previously, the LV prototype and the American M1146 strain have been propagated in cell culture to enable molecular characterization (Johansson et al., 2003, 2004). In the current study, parallel attempts to propagate the 64-7855 strain in cell culture were unsuccessful. The reason for this inability of 64-7855 to replicate in cultured cells is not known. Thus, 64-7855 was generated by passages in infant mice by intracerebral inoculation, as described previously (Beaty et al., 1989). The GenBank accession numbers of the nucleotide (nt) and polyprotein sequences of the strains used in this study are given in Table 1. Two at present unclassified duck hepatitis viruses were also included in the phylogenetic analysis. The sequences of two picornavirus-related insect viruses were used as an outgroup in the phylogenetic analysis of the 3Dpol protein.
Table 1.
Virus strains used in this study
| Genus | Virus/strain | Abbreviation | GenBank accession no. |
|---|---|---|---|
| Aphthovirus | Foot-and-mouth disease virus | FMDV | M10975 |
| Equine rhinitis A virus | ERAV | L43052 | |
| Cardiovirus | Encephalomyocarditis virus | EMCV | M22457 |
| Theiler's murine encephalomyelitis virus | TMEV | M20301 | |
| Enterovirus | Human poliovirus strain Sabin 1 | PV1S | V01150 |
| A-2 plaque virus | A2pV | NC_003988 | |
| Erbovirus | Equine rhinitis B virus | ERBV | X96871 |
| Hepatovirus | Hepatitis A virus | HAV | M59810 |
| Avian encephalomyelitis virus | AEV | AJ225173 | |
| Kobuvirus | Aichi virus | AiV | NC_001918 |
| Parechovirus | Human parechovirus 1 strain Harris | HPeV1 | L02971 |
| Human parechovirus 3 strain A308/99 | HPeV3 | AB084913 | |
| Human parechovirus 6 strain BNI-67/03 | HPeV6 | EU024629 | |
| Ljungan virus strain 87-012 | LV87-012 | AF327920 | |
| Ljungan virus strain 87-012G | LV87-012G | EF202833 | |
| Ljungan virus strain 174F | LV174F | AF327921 | |
| Ljungan virus strain 145SL | LV145SL | AF327922 | |
| Ljungan virus strain M1146 | LVM1146 | AF538689 | |
| Rhinovirus | Human rhinovirus 2 | HRV2 | X02316 |
| Teschovirus | Porcine teschovirus 1 | PTV | NC_003985 |
| Unclassified | Duck hepatitis virus 1 strain DRL-62 | DHV1DRL | DQ219396 |
| Duck hepatitis virus 1 strain AP-03337 | DHV1AP | DQ256132 | |
| Picornavirus-related insect viruses | Sacbrood virus | SBV | NC_002066 |
| Infectious flacherie virus | InFV | NC_003781 |
RNA isolation and RT-PCR.
Total RNA, extracted from virus-inoculated brain tissue using the Ultraspec II kit (Biotecx Laboratories) or a RiboPure kit (Ambion) according to the manufacturers' instructions, was reverse transcribed using SuperScript III RT enzyme (Invitrogen) and the primer NotdT27 (5′-ATAAGAATGCGGCCGCT27-3′) at 50 °C for 1 h before inactivating the enzyme at 70 °C. The cDNA was amplified by PCR using several different primer sets (all primer sequences are available in Supplementary Table S1, available in JGV Online). The primers were derived from aligned genomic sequences of previously published LV and HPeV strains and were later selected by a primer walking strategy. The PicoMaxx high fidelity PCR system (Stratagene) was used in all amplifications. Often, a nested PCR approach was required to generate a sufficient amount of amplicons. The need for nested PCR probably reflects the low amount of virus replicating in the brain tissues. Resulting specific and overlapping amplicons were isolated by agarose gel electrophoresis and directly sequenced or cloned into the pGEM-T Easy vector (Promega) and then sequenced. The nt sequence of 64-7855 was determined using the ABI Prism BigDye terminator cycle sequencing reaction kit (Applied Biosystems). Sequences derived from T/A cloned plasmids were generated by sequencing more than two clones for each region and by sequencing each clone in both directions. Sequence data were generated using a 3130 Genetic Analyzer (Applied Biosystems) and Sequencher 4.6 (Gene Codes Corporation) was used for assembly and editing of sequences.
Bioinformatic analyses.
The nt and amino acid (aa) sequences were aligned using the clustal w program (Thompson et al., 1994) or by pairwise alignment for sequence identity analyses (Needleman & Wunsch, 1970). The blosum substitution matrix was used to align protein sequences (Henikoff & Henikoff, 1992). Before phylogenetic analyses, aligned sequences were manually edited and phylogenetic information in each dataset was evaluated by likelihood mapping (Strimmer & von Haeseler, 1997); the appropriate model of sequence evolution for the RNA sequences was determined by the Modeltest program, version 3.7 (Posada & Crandall, 1998). Phylogenetic relationships were reconstructed using the maximum-likelihood method implemented in the HyPhy program and the JTT model for aa substitutions and the GTR model for nt substitutions (Jones et al., 1992; Pond et al., 2005; Tavaré, 1986). Phylogenetic reconstruction using distance-based methods resulted in similar relationships (data not shown). The significance of phylogenetic trees inferred by the maximum-likelihood method was evaluated by bootstrap analyses using the PhyML 3.0 program and 500 datasets (Guindon & Gascuel, 2003). The mega 4.0 program was used to visualize trees (Tamura et al., 2007). The RNAstructure version 4.6 software that includes the Dynalign method and the RNAalifold server (http://rna.tbi.univie.ac.at/cgi-bin/RNAalifold.cgi) were used for predictions of secondary RNA structures (Hofacker et al., 2002; Mathews, 2005; Mathews et al., 2004). Predictions by RNAalifold were based on alignments including all characterized LV strains, genotype 1, 3 and 6 of HPeV and members of the genera Aphtho- and Cardiovirus. The RnaViz 2 program was used to draw predicted secondary RNA structures (De Rijk et al., 2003).
To analyse the phylogenetic relationship within different parts of the 64-7855 genome, several different recombination detection methods were used; these were the recombination detection program (RDP) version 3.27 software (Martin & Rybicki, 2000), BootScanning (Martin et al., 2005; Salminen et al., 1995), the Maximum Chi method (MaxChi) (Smith, 1992), the Chimaera method (Posada & Crandall, 2001), the Sister Scanning method (SiScan) (Gibbs et al., 2000) and finally the 3Seq method (Boni et al., 2007). Aligned nt sequences of available LV strains were used together with the 64-7855 strain in recombination analyses. Default settings were used for all methods and general settings were applied in the RDP 3.27 software, including a highest acceptable P-value of 0.05, which were based on 1000 permutations.
RESULTS AND DISCUSSION
Strain 64-7855 is an LV
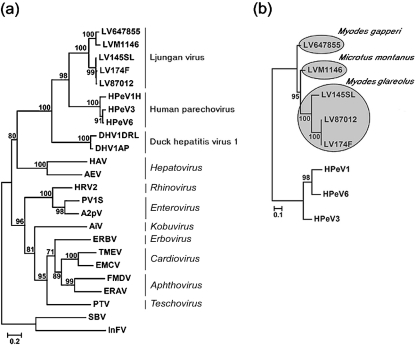
In order to position the 64-7855 strain within the family Picornaviridae, the 3Dpol nt sequence was determined and the aa sequence was compared with representative members of the various genera within the virus family. The phylogenetic relationship between strain 64-7855 and other viruses of the Picornaviridae demonstrated that this isolate is clearly related to previously characterized LV strains, and that 64-7855, based on the 3Dpol sequence, is most closely related to the other American LV strain, M1146, within the LV species (Fig. 1a). The complete coding sequence, the majority of the 5′ untranslated region (UTR) and the entire 3′ UTR of the 64-7855 genome were determined. Phylogenetic analyses based on VP1 protein sequences of previously characterized LV strains and members of the HPeVs verified that the 64-7855 is indeed an LV and suggested that this strain is a new genotype within the LV species (Fig. 1b).
Fig. 1.
Phylogenetic relationships of the 64-7855 LV strain with representative members of the nine genera of the Picornaviridae and duck hepatitis virus type 1. (a) Phylogenetic tree based on 3Dpol protein sequences rooted with two picorna-like insect viruses: sacbrood virus and infectious flacherie virus. (b) Midpoint rooted tree based on VP1 protein sequence of members of the parechovirus. Grey spheres denote the vole species (indicated in italics) from which the LV strains were isolated. Numbers at nodes indicate the percentage of 500 bootstrap replicates supporting that node. Bootstrap values of 70 % or more are indicated. Bars, substitutions per site.
The genome of strain 64-7855
The genome sequence of 64-7855 (GenBank accession no. EU854568) was derived from virus propagated in mouse brain. Initial phylogenetic analyses of the 64-7855 genome confirmed that this virus belongs to the LV species of the Picornaviridae. In addition, molecular features associated with the LV genome including the presence of two different consecutive 2A protein motifs, a P1 region where the VP0 protein remains unprocessed and conserved secondary structures of the 5′ and 3′ UTR, were also identified in the 64-7855 sequence. Despite several attempts, we were not able to determine the sequence of the terminal 5′ end of the 64-7855 genome, probably due to advanced secondary structure (see below) and the minute amount of RNA genome present in the extracted mouse brain tissue. The recorded 5′ UTR contains a sequence of 584 nt, which is 178 nt shorter than the complete 5′ UTR sequence of a previously published infectious LV clone, pLV 87-012G (Ekström et al., 2007b). The 45 nt stretch of the most 5′ proximal part of the 5′ UTR in the pLV87-012G clone is predicted to fold into a stem–loop (SL) structure. This SL-A1 structure (Fig. 2b) is crucial for LV replication (Ekström et al., 2007b). In the 5′ UTR, 64-7855 shares the highest sequence identity with M1146 (Table 2). However, in the 5′ end of the 5′ UTR of M1146, 50 additional nucleotides have been determined compared with 64-7855 (Johansson et al., 2003). Taken together, comparison with other LV genomes suggests that the 5′ UTR of 64-7855 is not completely sequenced. The inability to determine the 5′ end of the American LV strains indicates that the sequence in the most 5′-proximal part of the 5′ UTR is not conserved between American and Swedish LV strains. The 5′ UTR sequence is followed by an open reading frame (ORF) coding for a polyprotein of 2254 aa and ends with a 3′ UTR of 87 nt (excluding the poly A tail). The 64-7855 genome has a GC content of 45 %, which is similar to the 42 % GC content of the LV prototype 87-012 and the closely related HPeV1 (39 %) (Johansson et al., 2002). Pairwise sequence comparisons demonstrated that the majority of the 64-7855 genome shares highest sequence identity with the M1146 strain, although part of the P2 and P3 regions displayed a different genetic relationship (see below and Table 2). Serotyping with specific antisera has traditionally been used to identify different types of enteroviruses and HPeVs. Today, typing by genetic analysis is an alternative method that is used to distinguish virus types. In genetic typing of enteroviruses, a clinical isolate is considered homologous to an existing genotype if the VP1 nt and aa sequence identities are 75 and 88 %, respectively, or more (Oberste et al., 1999). LV is a recently discovered virus and genetic and serological data for this virus are limited. However, if criteria for homology between enterovirus types are applied to the LV species, then 64-7855 should be considered as a new LV genotype (Table 2). Furthermore, based on pairwise genetic distances between VP1 sequences (Supplementary Table S2, available in JGV Online), we propose that the currently known LV strains represent four different genotypes, where 87-012 and 174F represent genotype 1, 145SL genotype 2, M1146 genotype 3 and 64-7855 represents a fourth genotype. This division into four genotypes was also supported by phylogenetic relationships based on LV VP1 aa and nt sequences (Figs 1b and 5b).
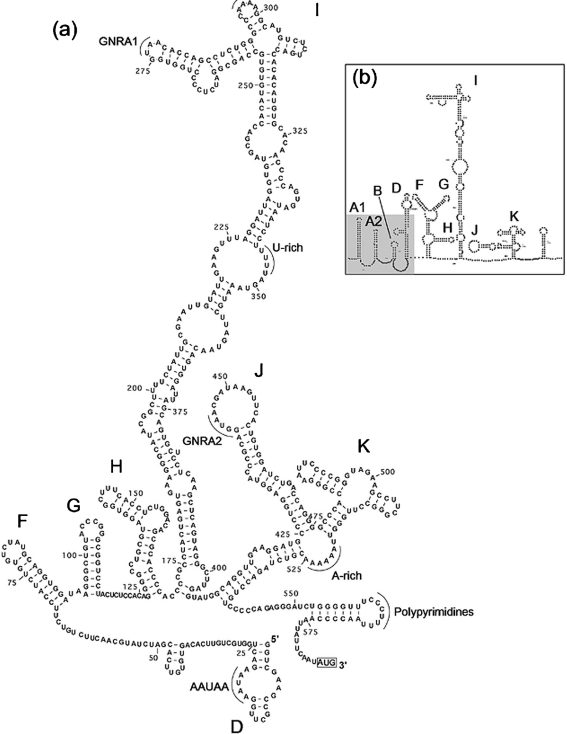
Fig. 2.
Predicted SL structures of the 64-7855 5′ UTR (a) compared with corresponding outlined structures of the 87-012G 5′ UTR (b). SL elements are labelled according to accepted notation of picornavirus type II IRES. The shaded area of the predicted 5′ UTR structure of 87-012G indicates the putative 5′ end sequence that remains to be determined for 64-7855. The first codon for initiation of translation is indicated at the 3′ end of the sequence.
Table 2.
Sequence identities between the 64-7855 strain and previously characterized LV strains 87-012, 174F, 145SL and M1146
| Region | Nt (aa) identity with 64-7855 (%) | |||
|---|---|---|---|---|
| 87-012 | 174F | 145SL | M1146 | |
| 5′ UTR | 59.8 | 59.6 | 60.0 | 70.3 |
| ORF | 72.1 (79.7) | 72.9 (79.7) | 74.0 (82.4) | 77.6 (89.0) |
| P1* | 67.0 (73.6) | 69.1 (73.6) | 68.8 (75.2) | 75.0 (85.9) |
| P2* | 73.3 (81.9) | 73.0 (82.0) | 77.4 (88.7) | 76.3 (88.9) |
| P3* | 76.4 (83.9) | 76.7 (83.9) | 76.6 (84.6) | 81.1 (92.2) |
| VP0 | 69.0 (78.4) | 71.9 (78.8) | 72.8 (76.8) | 78.5 (91.9) |
| VP3 | 65.7 (74.2) | 68.4 (74.2) | 68.7 (77.9) | 74.6 (85.2) |
| VP1† | 66.2 (69.1) | 66.4 (68.8) | 65.2 (71.9) | 71.8 (80.6) |
| 2A2 | 73.4 (84.4) | 74.0 (84.4) | 73.7 (85.9) | 70.5 (85.2) |
| 2B | 72.9 (84.3) | 72.6 (85.0) | 81.6 (93.6) | 79.9 (91.4) |
| 2C | 73.4 (80.5) | 72.7 (80.5) | 78.4 (88.6) | 78.2 (90.4) |
| 3A | 73.2 (78.5) | 73.6 (78.5) | 74.6 (81.5) | 78.6 (89.1) |
| 3B | 85.1 (86.2) | 83.9 (89.7) | 86.2 (89.7) | 83.9 (86.2) |
| 3C | 75.5 (86.4) | 76.8 (85.4) | 77.3 (86.4) | 81.3 (92.3) |
| 3D | 76.5 (84.0) | 76.9 (84.3) | 76.2 (84.3) | 81.5 (93.4) |
| 3′ UTR | 64.5 | 62.1 | 65.2 | 78.2 |
*Precursor protein region 1, 2 and 3.
†VP1-encoding gene including the 2A1 motif.
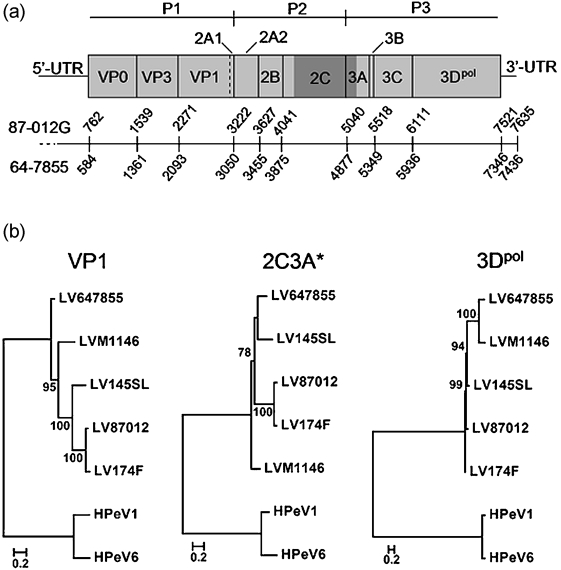
Fig. 5.
Phylogenetic analyses based on the VP1-, 2C3A*- and 3Dpol-encoding genes. (a) Schematic illustration of the LV genome organization, indicating P1–P3 regions of the ORF (grey boxes) and UTR sequences. Numbers indicate nt positions in the 87-012G and 64-7855 genomes; the dotted line indicates differences in the length of the 5′ UTR sequence between these virus strains. The position of the 2C3A* region is indicated by dark grey shading. (b) Phylogenetic analyses based on the indicated coding regions, using the corresponding sequences of HPeV1 and HPeV6 as an outgroup. Phylogenetic relationships were inferred by using the maximum-likelihood method. Numbers at nodes of midpoint-rooted trees indicate the percentage of 500 bootstrap replicates. Bootstrap values of 70 % or more are indicated. Bars, substitutions per site.
The viral polyprotein
Among picornaviruses, the majority of polyprotein cleavage sites are processed by the viral 3C protease (3Cpro). Cleavage by 3Cpro is confined to restricted recognition sites within the polyprotein (Racaniello, 2001). Primary, secondary and tertiary structures of these cleavage sites have been described previously (Dougherty & Semler, 1993; Palmenberg, 1990). Also, as previously predicted for 3Cpro of enteroviruses, rhinoviruses and aphthoviruses (Blom et al., 1996), the predicted cleavage sites of LV 3Cpro are characterized by a glutamine or glutamic acid residue at the P1 substrate position and a small residue at P1′ (Johansson et al., 2002). A bulky hydrophobic aa residue in the P4 position is also characteristic for LV 3Cpro cleavage sites. The 64-7855 polyprotein, derived from the nt sequence, shows 89 % identity to the American M1146 strain, while its identities to the Swedish LV strains are 79.7–82.4 % (Table 2). Corresponding identity values between Swedish strains are 88–99 % (data not shown). Comparative analysis of the 64-7855 polyprotein showed that predicted protein cleavage sites are generally confined to those in the polyprotein of previously characterized LV and HPeV1 (Supplementary Fig. S1, available in JGV Online). A more detailed comparison showed that the main sequence variations between the 64-7855 strain and previously characterized LV strains are found in the capsid proteins (Table 2), especially in predicted surface-exposed loop structures (Supplementary Fig. S2; see also Supplementary Methods for a description of secondary structure predictions; available in JGV Online). In addition, in these putative loop regions, there is less variation between the 64-7855 and M1146 sequences than between 64-7855 and the Swedish LV strains. For many picornaviruses, major neutralization antigenic sites are located in exposed BC- and EF-loops of the capsid proteins (Racaniello, 2001). The sequence variation within putative capsid loop structures may have implications for the serology of these viruses. However, at present, no conclusive serological data are available regarding members of the LV species.
5′ UTR of LV 64-7855
The 5′ UTR secondary structure of 64-7855 was predicted using the Dynalign method (Fig. 2a) (Mathews & Turner, 2002), a computer algorithm that combines free energy minimization and comparative sequence analysis to find a structure that applies to two related sequences. In addition, concordant secondary structures were obtained with a thermodynamic folding minimization algorithm (MFOLD) and the RNAalifold program (data not shown). For Dynalign predictions, the sequence of LV strain 87-012G was used together with the 64-7855 sequence (Fig. 2a, b). The 87-012G sequence was used, since its 5′ UTR sequence has been shown to be biologically active when included in an infectious viral cDNA clone (Ekström et al., 2007b). In the 64-7855 sequence, the initiation codon is located at position 585 in an optimal Kozak context (ANNAUGG) (Kozak, 1987). Considering that the most 5′-proximal part of the 5′ UTR of 64-7855 is not entirely sequenced, the predicted secondary structure clearly corresponds to a type II internal ribosomal entry site (IRES). Type II IRES is also present in aphthoviruses, cardioviruses and parechoviruses and has been predicted for Swedish LV strains (Ghazi et al., 1998; Johansson et al., 2002; Le et al., 1993; Pilipenko et al., 1989). Covariance of nt pairs in stems of predicted SL domains between different LV strains, including 64-7855, supports the predicted structure. Five pairs of nt substitutions in stems of the F and H SL domains and 21 substitutions in the stem of the I domain were detected (data not shown). This conservation between different LV genotypes makes the predicted structure of the type II IRES of LV more reliable. By analogy with aphthoviruses, cardioviruses and parechoviruses, parts of the predicted 5′ UTR structure of 64-7855 and other LV strains are likely to be involved in IRES functions (Racaniello, 2001). The 64-7855 sequence regions predicted to make up the top of the most extended SL domain I, as well as the J and K domains, show considerable primary and secondary sequence identity to both aphthoviruses, cardioviruses and parechoviruses (Fig. 2a) (Clarke et al., 1987; Ghazi et al., 1998; Palmenberg & Sgro, 1997).
The GNRA tetranucleotide (GNRA1) loop, the following A/C-rich loop (CAAAA sequence at nt 295–299) of the SL I domain and the stem part of the J domain, and the UUAAAAAA sequence at the root of the SL K domain are well conserved among these picornaviruses. Previously, Johansson et al. (2002) reported a second GNRA sequence in the loop of the SL J domain of LV and HPeV. This GNRA2 is also present in the predicted structure of the 64-7855 5′ UTR; interestingly, it is present in the corresponding position of a structure predicted for the aphthovirus 5′ UTR (Fig. 2a and data not shown). The conservation of this GNRA tetranucleotide in the predicted 5′ UTR structures of different picornavirus genera suggests a functional significance. In the 5′-proximal part of the available 64-7855 5′ UTR sequence, an AAUAA sequence is found at nt 17–21 as a part of the truncated SL D domain (Fig. 2a). This sequence is conserved in the predicted 5′ UTR structures of LV and HPeV and also in the cardiovirus, EMCV (Ghazi et al., 1998; Palmenberg & Sgro, 1997). The main differences between the 5′ UTR structures of 64-7855 and previously published LV strains, except for the putative 5′ end sequence lacking in the determined 64-7855 sequence (i.e. the predicted secondary SL A1-B domains indicated by grey shading in Fig. 2b), were located in loops of the F and K domains. Apart from the GNRA2 tetranucleotide, substantial sequence variation was also found in the loop of the SL J domain (Fig. 2a). The sequence variation suggests that these regions are less important for viral replication.
3′ UTR structure
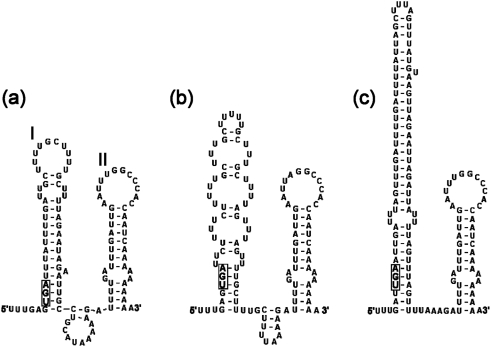
The 50–150 nt 3′ UTR of picornaviruses is important for genome replication and translation (Dobrikova et al., 2003; Rohll et al., 1995). The 3′ UTR of 64-7855 is only 87 nt compared with the 96–111 nt sequence determined for the other LV strains (Fig. 3) (Johansson et al., 2002, 2003). A comparison of 3′ UTRs of different LV genotypes showed that sequence variation, including deletions in 64-7855, M1146 and 145SL sequences compared with 87-012 and 174F, are found in the first of two predicted SL domains (Fig. 3). This part of the 3′ UTR is likely to be of less importance for viral replication, considering the variation of primary and predicted secondary structures of different LV strains. In contrast, the predicted SL II domain is highly conserved in all LV genomes, including the 64-7855 strain, suggesting that this putative hairpin plays a significant role during viral replication.
Fig. 3.
Predicted secondary structures for the 3′ UTRs of LV 64-7855 (a), M1146 (b) and 87-012 (c), representing three LV types. In order to simulate authentic viral RNA, two additional codons upstream from the 3′ UTR sequences and 10 adenosines of the poly-A tail were included in structure prediction analyses. SL domains I and II, depicted in the 3′ UTR structure of the 64-7855 strain, are designated according to previously established notation of structures predicted for LV 3′ UTR (Johansson et al., 2003). The stop codons of the polyprotein sequences are indicated by a boxed nucleotide triplet.
Analysis of a putative cis-acting replication element (cre) sequence in the VPg-encoding gene of 64-7855
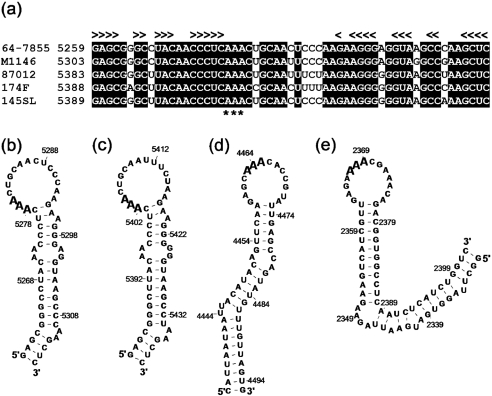
The presence of cre has been recognized in genomes of several picornaviruses (Goodfellow et al., 2000). The picornavirus cre is a short SL structure including an internal or a terminal loop with three unpaired adenine (A) nucleotides. This structure is critical for viral replication by its implication in the uridylylation of the viral VPg peptide (Paul et al., 2000; Rieder et al., 2000). These cre structures have been located in different regions of the picornavirus genome, including the 2C-encoding gene of poliovirus (Goodfellow et al., 2000) and the capsid-encoding regions of human rhinovirus type 14 (McKnight & Lemon, 1998). Recently, cre structures were proposed to reside in the VP0-encoding gene of HPeV and in the gene encoding VPg of the LV species (Al-Sunaidi et al., 2007). Sequence analyses of genomes representing four different LV genotypes revealed a conserved sequence stretch in the VPg gene that folds into a short hairpin structure (Fig. 4a–c). This structure is similar to those proposed for poliovirus and rhinovirus cre sequences (Goodfellow et al., 2003; McKnight & Lemon, 1998). The proposed LV cre, consisting of three base-paired stem regions divided by bulge loops and a terminal loop of 16 unpaired nt, corresponds to stem regions and the functional 14 nt unpaired loop in poliovirus and rhinovirus cre structures (Fig. 4b–e). However, the AAA sequence is located closer to the most terminal stem in the LV cre than it is in poliovirus and rhinovirus cre structures in which its position is conserved. These secondary structure predictions suggest the possibility of an LV VPg gene that does not only encode the VPg peptide but also contains a putative cre, which mediates post-translational modification of VPg in poliovirus (Paul et al., 2000).
Fig. 4.
Alignment (a) and secondary structures predicted for a putative LV cre in the VPg-encoding gene of the 64-7855 (b) and 87-012 prototype (c) strain, and secondary structures of cre sequences identified in the 2C-encoding gene of poliovirus type 3 (d) and the capsid-encoding region of human rhinovirus type 14 (e) (Goodfellow et al., 2003; McKnight & Lemon, 1998). In (a), shading indicates conserved nucleotides; < and > indicate conserved nucleotides participating in predicted stem structures. The AAA sequence, which is conserved in picornavirus cre, is indicated by asterisks (*) in the alignment and by bold type in secondary structures.
Phylogenetic analyses of the 64-7855 genome reveal evolutionary inconsistencies
Comparisons of phylogentic relationships between 64-7855 and other characterized LV strains in different regions of the viral genome by scanning methods showed that the clustering of the American LV strains recognized in P1 and P3 regions was not conserved in genes within the P2 region (Table 3 and Supplementary Fig. S3, available in JGV Online). This genetic inconsistency was supported by results from pairwise genetic distance analyses of genes in P2, showing that the sequence identity between 64-7855 and the Swedish strain 145SL was higher compared with the identity between 64-7855 and the other Swedish strains (Table 2). Extensive genetic variation is a hallmark of RNA viruses and is caused by point mutations (Holland et al., 1982) but can also be due to genomic rearrangements such as deletions, insertions and recombination (Agol, 2002). It has been shown previously that recombination drives genetic diversification in multiple genera of the picornavirus family (Andersson et al., 2002; Carrillo et al., 2005; Lindberg et al., 2003; Simmonds, 2006), including the genus Parechovirus (Al-Sunaidi et al., 2007; Benschop et al., 2008). The changes in genetic relationships in the LV genome were investigated further by using bioinformatic methods developed for detection of recombination within homologous sequences. Using these methods, a fragment in the LV genome sequences spanning a part of P2 and the beginning of the P3 region was identified (Table 3 and Supplementary Fig. S3). This region included parts of the 2C- and 3A-encoding genes (herein referred to as 2C3A*) in the LV genome (Fig. 5a). Phylogenetic relationships among LVs based on the nt sequence of the 2C3A* region (nt 4044–4956; see dark grey shading on Fig. 5a), as well as the VP1- and 3Dpol-encoding genes in surrounding genome regions, confirm previous indications of phylogenetic incongruence in different parts of the LV genome (Table 3 and Supplementary Fig. S3). The Swedish LV strains 87-012, 174F and 145SL and the American 64-7855 and M1146 strains clustered in trees according to isolation sites, showing phylogenetic relationships based on the VP1 and 3Dpol genes. The observed distinction between Swedish and American LV strains was not observed in the phylogenetic tree based on the 2C3A* region (Fig. 5b). In this tree, the 64-7855 strain and the Swedish 145SL strain form a cluster which is separated from the other LV strains. The observed incongruence suggests that the 64-7855 genome contains a region, 2C3A*, that has a different evolutionary history compared with the remaining parts of the genome. Taken together, these results imply that recombination has taken place between different strains of the LV species.
Table 3.
Putative recombination events detected in the LV genome by different recombination detection methods. The genome region from nt 4044 to 4956 (numbered according to the strain 64-7855) was tested
| Method | P-value |
|---|---|
| RDP | 1.4×10−9 |
| BootScan | 7.1×10−9 |
| MaxChi | 2.8×10−8 |
| Chimaera | 6.9×10−9 |
| SiScan | 1.2×10−11 |
| 3Seq | 2.0×10−18 |
The significance of virus–host relations was recently highlighted when the evolution of viruses belonging to the Caliciviridae was connected to the evolution of their host species (Etherington et al., 2006). It is not known at present whether Swedish strains representing LV genotype 1 and 2 circulate in the Americas or if members of the American strains constituting genotype 3 and 4 are present in European rodents. However, it is worth noting that 64-7855 and 145SL, which are more related in a central part of the genome, were both isolated from arvicoline rodents of the genus Myodes. In the remaining regions of the genome, 64-7855 is more closely related to M1146, a virus isolated from a Microtus rodent trapped in North America. The limited number of LV genomes available makes inference of the evolutionary history difficult. However, it is possible that a central part of the 64-7855 genome originated with an ancestral Myodes rodent, while the remainder of the genome was formed due to more recent ecological interactions of North American rodents (Fig. 5b).
Concluding remarks
Genetic and phylogenetic analyses of the 64-7855 genome clearly demonstrate that this virus belongs to the LV species and is thus a member of the genus Parechovirus within the family Picornaviridae. Sequence analyses demonstrated that the 64-7855 strain represents the first member of a novel fourth genotype within the LV species. Furthermore, the additional sequence information provided by this fourth LV genotype enabled refined analyses of a putative LV cre structure identified within the VPg-encoding gene in the LV genome. The 64-7855 sequence also facilitated genetic and phylogenetic analyses that showed, for the first time, that phylogenetic relationships vary in different parts of the LV genome. Previous recombination events between diverse members of the LV species are a plausible explanation for observed genetic variation in the 64-7855 genome. However, more sequence information from additional LV strains is needed in order to investigate this finding further. Sequences of additional LV isolates will also aid in the understanding of LV evolution and how this virus is transmitted in nature.
Supplementary Material
Acknowledgments
We thank Kjell Edman, Jens-Ola Ekström and Bo Niklasson for support and collaboration. This work was supported by grants from the Swedish Knowledge Foundation, Carl Trygger Foundation, Apodemus AB and the PhD program in Medical Bioinformatics at Karolinska Institutet, Stockholm, Sweden. R. B. T. was supported by contract NO1-AI30027 from the US National Institutes of Health.
Footnotes
The GenBank/EMBL/DDBJ accession number of the sequence reported in this paper is EU854568.
A supplementary method, two tables and three figures are available with the online version of this paper.
References
- Abed, Y. & Boivin, G. (2005). Molecular characterization of a Canadian human parechovirus (HPeV)-3 isolate and its relationship to other HPeVs. J Med Virol 77, 566–570. [DOI] [PubMed] [Google Scholar]
- Agol, V. I. (2002 ). Genomic instability in picornaviruses. Mol Biol (Mosk) 36, 286–295 (in Russian). [PubMed] [Google Scholar]
- Al-Sunaidi, M., Williams, C. H., Hughes, P. J., Schnurr, D. P. & Stanway, G. (2007). Analysis of a new human parechovirus allows the definition of parechovirus types and the identification of RNA structural domains. J Virol 81, 1013–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, P., Edman, K. & Lindberg, A. M. (2002). Molecular analysis of the echovirus 18 prototype: evidence of interserotypic recombination with echovirus 9. Virus Res 85, 71–83. [DOI] [PubMed] [Google Scholar]
- Beaty, B. J., Calisher, C. H. & Shope, R. E. (1989). Arboviruses. In Diagnostic procedures for viral, rickettsial and chlamydial infections, pp. 797–855. Edited by N. J. Schmidt, D. A. Lennette, E. T. Lennette, E. H. Lennette & R. W. Emmons. Washington, DC: American Public Health Associations.
- Benschop, K. S. M., Schinkel, J., Luken, M. E., van den Broek, P. J. M., Beersma, M. F. C., Menelik, N., van Eijk, H. W. M., Zaaijer, H. L., VandenBroucke-Grauls, C. M. J. E. & other authors (2006). Fourth human parechovirus serotype. Emerg Infect Dis 12, 1572–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benschop, K. S. M., Williams, C. H., Wolthers, K. C., Stanway, G. & Simmonds, P. (2008). Widespread recombination within human parechoviruses: analysis of temporal dynamics and constraints. J Gen Virol 89, 1030–1035. [DOI] [PubMed] [Google Scholar]
- Blom, N., Hansen, J., Blaas, D. & Brunak, S. (1996). Cleavage site analysis in picornaviral polyproteins: discovering cellular targets by neural networks. Protein Sci 5, 2203–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin, G., Abed, Y. & Boucher, F. D. (2005). Human parechovirus 3 and neonatal infections. Emerg Infect Dis 11, 103–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni, M. F., Posada, D. & Feldman, M. W. (2007). An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics 176, 1035–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo, C., Tulman, E. R., Delhon, G., Lu, Z., Carreno, A., Vagnozzi, A., Kutish, G. F. & Rock, D. L. (2005). Comparative genomics of foot-and-mouth disease virus. J Virol 79, 6487–6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, B. E., Brown, A. L., Currey, K. M., Newton, S. E., Rowlands, D. J. & Carroll, A. R. (1987). Potential secondary and tertiary structure in the genomic RNA of foot and mouth disease virus. Nucleic Acids Res 15, 7067–7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rijk, P., Wuyts, J. & De Wachter, R. (2003). RnaViz 2: an improved representation of RNA secondary structure. Bioinformatics 19, 299–300. [DOI] [PubMed] [Google Scholar]
- Dobrikova, E. Y., Florez, P. & Gromeier, M. (2003). Structural determinants of insert retention of poliovirus expression vectors with recombinant IRES elements. Virology 311, 241–253. [DOI] [PubMed] [Google Scholar]
- Dougherty, W. G. & Semler, B. L. (1993). Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol Rev 57, 781–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekström, J. O., Tolf, C., Edman, K. A. & Lindberg, A. M. (2007a). Physicochemical properties of the Ljungan virus prototype virion in different environments: inactivated by heat but resistant to acidic pH, detergents and non-physiological environments such as Virkon®-containing solutions. Microbiol Immunol 51, 841–850. [DOI] [PubMed] [Google Scholar]
- Ekström, J. O., Tolf, C., Fahlgren, C., Johansson, E. S., Arbrandt, G., Niklasson, B., Edman, K. A. & Lindberg, A. M. (2007b). Replication of Ljungan virus in cell culture: the genomic 5′-end, infectious cDNA clones and host cell response to viral infections. Virus Res 130, 129–139. [DOI] [PubMed] [Google Scholar]
- Etherington, G. J., Ring, S. M., Charleston, M. A., Dicks, J., Rayward-Smith, V. J. & Roberts, I. N. (2006). Tracing the origin and co-phylogeny of the caliciviruses. J Gen Virol 87, 1229–1235. [DOI] [PubMed] [Google Scholar]
- Ghazi, F., Hughes, P. J., Hyypiä, T. & Stanway, G. (1998). Molecular analysis of human parechovirus type 2 (formerly echovirus 23). J Gen Virol 79, 2641–2650. [DOI] [PubMed] [Google Scholar]
- Gibbs, M. J., Armstrong, J. S. & Gibbs, A. J. (2000). Sister-Scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16, 573–582. [DOI] [PubMed] [Google Scholar]
- Goodfellow, I., Chaudhry, Y., Richardson, A., Meredith, J., Almond, J. W., Barclay, W. & Evans, D. J. (2000). Identification of a cis-acting replication element within the poliovirus coding region. J Virol 74, 4590–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow, I. G., Kerrigan, D. & Evans, D. J. (2003). Structure and function analysis of the poliovirus cis-acting replication element (CRE). RNA 9, 124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon, S. & Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52, 696–704. [DOI] [PubMed] [Google Scholar]
- Henikoff, S. & Henikoff, J. G. (1992). Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A 89, 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker, I. L., Fekete, M. & Stadler, P. F. (2002). Secondary structure prediction for aligned RNA sequences. J Mol Biol 319, 1059–1066. [DOI] [PubMed] [Google Scholar]
- Holland, J., Spindler, K., Horodyski, F., Grabau, E., Nichol, S. & VandePol, S. (1982). Rapid evolution of RNA genomes. Science 215, 1577–1585. [DOI] [PubMed] [Google Scholar]
- Ito, M., Yamashita, T., Tsuzuki, H., Takeda, N. & Sakae, K. (2004). Isolation and identification of a novel human parechovirus. J Gen Virol 85, 391–398. [DOI] [PubMed] [Google Scholar]
- Johansson, S., Niklasson, B., Maizel, J., Gorbalenya, A. E. & Lindberg, A. M. (2002). Molecular analysis of three Ljungan virus isolates reveals a new, close-to-root lineage of the Picornaviridae with a cluster of two unrelated 2A proteins. J Virol 76, 8920–8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson, E. S., Niklasson, B., Tesh, R. B., Shafren, D. R., da Rosa, A. P. A. T. & Lindberg, A. M. (2003). Molecular characterization of M1146, an American isolate of Ljungan virus (LV) reveals the presence of a new LV genotype. J Gen Virol 84, 837–844. [DOI] [PubMed] [Google Scholar]
- Johansson, E. S., Ekström, J. O., Shafren, D. R., Frisk, G., Hyypiä, T., Edman, K. & Lindberg, A. M. (2004). Cell culture propagation and biochemical analysis of the Ljungan virus prototype strain. Biochem Biophys Res Commun 317, 1023–1029. [DOI] [PubMed] [Google Scholar]
- Jones, D. T., Taylor, W. R. & Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8, 275–282. [DOI] [PubMed] [Google Scholar]
- Kozak, M. (1987). At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian-cells. J Mol Biol 196, 947–950. [DOI] [PubMed] [Google Scholar]
- Le, S. Y., Chen, J. H., Sonenberg, N. & Maizel, J. V., Jr (1993). Conserved tertiary structural elements in the 5′ nontranslated region of cardiovirus, aphthovirus and hepatitis A virus RNAs. Nucleic Acids Res 21, 2445–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg, A. M. & Johansson, S. (2002). Phylogenetic analysis of Ljungan virus and A-2 plaque virus, new members of the Picornaviridae. Virus Res 85, 61–70. [DOI] [PubMed] [Google Scholar]
- Lindberg, A. M., Andersson, P., Savolainen, C., Mulders, M. N. & Hovi, T. (2003). Evolution of the genome of human enterovirus B: incongruence between phylogenies of the VP1 and 3CD regions indicates frequent recombination within the species. J Gen Virol 84, 1223–1235. [DOI] [PubMed] [Google Scholar]
- Martin, D. & Rybicki, E. (2000). RDP: detection of recombination amongst aligned sequences. Bioinformatics 16, 562–563. [DOI] [PubMed] [Google Scholar]
- Martin, D. P., Posada, D., Crandall, K. A. & Williamson, C. (2005). A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res Hum Retroviruses 21, 98–102. [DOI] [PubMed] [Google Scholar]
- Mathews, D. H. (2005). Predicting a set of minimal free energy RNA secondary structures common to two sequences. Bioinformatics 21, 2246–2253. [DOI] [PubMed] [Google Scholar]
- Mathews, D. H. & Turner, D. H. (2002). Dynalign: an algorithm for finding the secondary structure common to two RNA sequences. J Mol Biol 317, 191–203. [DOI] [PubMed] [Google Scholar]
- Mathews, D. H., Disney, M. D., Childs, J. L., Schroeder, S. J., Zuker, M. & Turner, D. H. (2004). Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci U S A 101, 7287–7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight, K. L. & Lemon, S. M. (1998). The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA 4, 1569–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman, S. B. & Wunsch, C. D. (1970). A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 48, 443–453. [DOI] [PubMed] [Google Scholar]
- Niklasson, B., Hörnfeldt, B. & Lundman, B. (1998). Could myocarditis, insulin-dependent diabetes mellitus, and Guillain–Barré syndrome be caused by one or more infectious agents carried by rodents? Emerg Infect Dis 4, 187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklasson, B., Kinnunen, L., Hörnfeldt, B., Hörling, J., Benemar, C., Hedlund, K. O., Matskova, L., Hyypiä, T. & Winberg, G. (1999). A new picornavirus isolated from bank voles (Clethrionomys glareolus). Virology 255, 86–93. [DOI] [PubMed] [Google Scholar]
- Niklasson, B., Samsioe, A., Papadogiannakis, N., Kawecki, A., Hörnfeldt, B., Saade, G. R. & Klitz, W. (2007). Association of zoonotic Ljungan virus with intrauterine fetal deaths. Birth Defects Res Part A Clin Mol Teratol 79, 488–493. [DOI] [PubMed] [Google Scholar]
- Oberste, M. S., Maher, K., Kilpatrick, D. R., Flemister, M. R., Brown, B. A. & Pallansch, M. A. (1999). Typing of human enteroviruses by partial sequencing of VP1. J Clin Microbiol 37, 1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmenberg, A. C. (1990). Proteolytic processing of picornaviral polyprotein. Annu Rev Microbiol 44, 603–623. [DOI] [PubMed] [Google Scholar]
- Palmenberg, A. C. & Sgro, J.-Y. (1997). Topological organization of picornaviral genomes: statistical prediction of RNA structural signals. Semin Virol 8, 231–241. [Google Scholar]
- Paul, A. V., Rieder, E., Kim, D. W., van Boom, J. H. & Wimmer, E. (2000). Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J Virol 74, 10359–10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko, E. V., Blinov, V. M., Chernov, B. K., Dmitrieva, T. M. & Agol, V. I. (1989). Conservation of the secondary structure elements of the 5′-untranslated region of cardio- and aphthovirus RNAs. Nucleic Acids Res 17, 5701–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond, S. L., Frost, S. D. & Muse, S. V. (2005). HyPhy: hypothesis testing using phylogenies. Bioinformatics 21, 676–679. [DOI] [PubMed] [Google Scholar]
- Posada, D. & Crandall, K. A. (1998). modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818. [DOI] [PubMed] [Google Scholar]
- Posada, D. & Crandall, K. A. (2001). Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci U S A 98, 13757–13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racaniello, V. R. (2001). Picornaviridae: the viruses and their replication. In Fields Virology, 4th edn, pp. 685–722. Edited by D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Riozman & S. E. Straus. Philadelphia: Lippincott Williams & Wilkins.
- Rieder, E., Paul, A. V., Kim, D. W., van Boom, J. H. & Wimmer, E. (2000). Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J Virol 74, 10371–10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohll, J. B., Moon, D. H., Evans, D. J. & Almond, J. W. (1995). The 3′-untranslated region of picornavirus RNA: features required for efficient genome replication. J Virol 69, 7835–7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen, M. O., Carr, J. K., Burke, D. S. & McCutchan, F. E. (1995). Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res Hum Retroviruses 11, 1423–1425. [DOI] [PubMed] [Google Scholar]
- Simmonds, P. (2006). Recombination and selection in the evolution of picornaviruses and other mammalian positive-stranded RNA viruses. J Virol 80, 11124–11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J. M. (1992). Analyzing the mosaic structure of genes. J Mol Evol 34, 126–129. [DOI] [PubMed] [Google Scholar]
- Stanway, G. & Hyypiä, T. (1999). Parechoviruses. J Virol 73, 5249–5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanway, G., Brown, F., Christian, P., Hovi, T., Hyypiä, T., King, A. M. Q., Knowles, N. J., Lemon, S. M., Minor, P. D. & other authors (2005). Family Picornaviridae. In Virus Taxonomy, Eighth Report of the International Committee on Taxonomy of Viruses, pp. 757–778. Edited by C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger & L. A. Ball. London: Elsevier.
- Strimmer, K. & von Haeseler, A. (1997). Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc Natl Acad Sci U S A 94, 6815–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., Dudley, J., Nei, M. & Kumar, S. (2007). mega4: molecular evolutionary genetics analysis (mega) software version 4.0. Mol Biol Evol 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tavaré, S. (1986). Some probabilistic and statistical problems in the analysis of DNA sequences. In Some Mathematical Questions in Biology – DNA Sequence Analysis, pp. 57–86. Edited by R. M. Miura. Providence, RI: American Mathematical Society.
- Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994). clustal w: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolf, C., Ekström, J. O., Gullberg, M., Arbrandt, G., Niklasson, B., Frisk, G., Liljeqvist, J. A., Edman, K. & Lindberg, A. M. (2008). Characterization of polyclonal antibodies against the capsid proteins of Ljungan virus. J Virol Methods 150, 34–40. [DOI] [PubMed] [Google Scholar]
- Watanabe, K., Oie, M., Higuchi, M., Nishikawa, M. & Fujii, M. (2007). Isolation and characterization of novel human parechovirus from clinical samples. Emerg Infect Dis 13, 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney, E., Roz, A. P. & Rayner, G. A. (1970). Two viruses isolated from rodents (Clethrionomys gapperi and Microtus pennsvlvanicus) trapped in St. Lawrence County, New York. J Wildl Dis 6, 48–55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.