Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) replication and transcription activator (RTA) is well established as a key transcriptional activator that regulates the KSHV life cycle from latency to lytic replication. It is expressed immediately after infection and activates a number of viral genes leading to virus replication. The RTA-responsive element (RRE) in the RTA target gene promoters is critical for RTA to mediate this transactivation. A number of non-conserved RREs have been identified in various RTA-responsive promoters, and AT-rich sequences have been proposed to serve as RTA targets, but no consensus RRE sequence has been identified so far. Two non-conserved RREs (RRE1 and RRE2) containing AT-rich sequences have been identified previously in the promoter of one of the KSHV lytic genes, ORF57, which can be strongly activated by RTA. Based on homology with the consensus sequence of the Epstein–Barr virus Rta RRE, this study identified a third RTA-responsive element (RRE3) in the ORF57 promoter. This RRE comprised a GC-rich sequence that could bind RTA both in vitro and in vivo, and plays a role in RTA-mediated transactivation of the ORF57 promoter. The presence of two of the three RREs in close proximity to each other was required for optimal RTA-mediated transactivation of the ORF57 promoter, even though the presence of only one RRE is needed for RTA binding. These results suggest that the ability of RTA to mediate transcriptional activation is distinct from its ability to bind to its target elements.
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV; human herpesvirus 8) was first identified from Kaposi's sarcoma tissue of an AIDS patient in 1994 and subsequently shown to be associated with Kaposi's sarcoma and other malignancies, including primary effusion lymphoma and multicentric Castleman's disease (Cesarman et al., 1995a, b; Chang et al., 1994). According to nucleotide sequence analysis, KSHV belongs to the genus Rhadinovirus in the subfamily Gammaherpesvirinae, with family members that include Epstein–Barr virus (EBV) and herpesvirus saimiri (Russo et al., 1996). As in all herpesviruses, KSHV goes through latent and lytic phases of infection, with latency predominantly found in infected B cells (Staskus et al., 1997). During latency, a small subset of viral genes is expressed in order to evade the immune response and maintain the viral genome in the host cells. These latent proteins have also been found to affect the survival and proliferation of infected cells in vitro (Friborg et al., 1999; Radkov et al., 2000; Rivas et al., 2001; Sharp et al., 2002; Sturzl et al., 1999). Virus lytic replication can be reactivated in vitro by treatment with chemical agents such as sodium butyrate or tetradecanoylphorbol acetate (Miller et al., 1997) and in vivo by the KSHV-encoded transcription factor RTA (replication and transcription activator) (Lukac et al., 1998). Upon reactivation from latency, lytic viral proteins are induced and expressed. These proteins are encoded by differentially expressed genes; the immediate-early genes are transcribed first, followed by expression of early genes, and the late genes are expressed after viral DNA replication (Sun et al., 1999).
RTA is an important viral regulatory protein encoded by the immediate-early gene ORF50. A striking feature of RTA is its ability to trigger KSHV lytic reactivation by itself in latently infected cells (Gradoville et al., 2000; Lukac et al., 1998, 1999; Staskus et al., 1997; Sun et al., 1998), and KSHV with an RTA gene deletion is incapable of lytic reactivation (Xu et al., 2005). Therefore, RTA has been implicated as an important switch to control the transition from KSHV latency to lytic replication. The RTA protein has 691 aa and is a potent transcriptional factor consisting of a DNA-binding domain at its N terminus, two putative nuclear localization signals and a transactivation domain at its C terminus. To date, a number of viral lytic genes, including ORF50 itself, that can be regulated by RTA have been identified (Bowser et al., 2006; Chang et al., 2002; Chen et al., 2000; Deng et al., 2000, 2002; Duan et al., 2001). RTA has been shown to transactivate its target promoters either by binding directly to an RTA-responsive element (RRE) or indirectly by interacting with cellular proteins, such as KSHV RTA-binding protein (K-RBP) (Wang et al., 2001b; Yang & Wood, 2007), CCAAT/enhancer-binding protein-α (CEBP-α) (Wang et al., 2003a, b), octamer-binding protein 1 (Oct-1) (Sakakibara et al., 2001) and recombination signal-binding protein Jκ (RBP-Jκ) (Liang & Ganem, 2004). RTA protein with its DNA-binding domain recognizes RREs in target gene promoters, and an AT-rich or A/T trinucleotide motif is considered to be a feature of KSHV RREs that can be found in the ORF57 and K8 promoters (Liao et al., 2003; Lukac et al., 2001). However, there is a lack of a consensus RRE sequence, as substantial diversity can be observed in the RREs identified in the promoters of various KSHV genes found to be responsive to RTA. These include the PAN (polyadenylated nuclear RNA) (Song et al., 2002), K12 (kaposin) (Chang et al., 2002), K2 (viral interleukin-6) (Deng et al., 2002) and K1 (Bowser et al., 2006) genes.
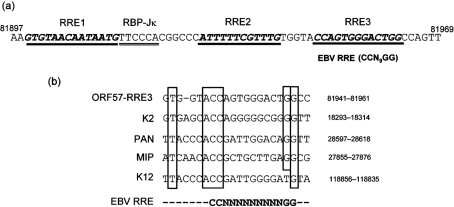
Epstein–Barr virus (EBV) Rta, a homologue of KSHV RTA, also functions as an activator of viral lytic promoters. A consensus EBV Rta-responsive element (EBV RRE), GNCCN9GGNG, where N can be any nucleotide, has been identified (Gruffat & Sergeant, 1994). Recently, the optimal EBV RRE sequence, GTCC(C/A)(T/C)(C/G)N(A/G)NC(A/G)(T/A)GGCG, was reported to be present in five EBV lytic cycle promoters (Chen et al., 2005). Interestingly, a CCN9GG-like sequence can also be found in the KSHV ORF57 promoter (Fig. 1a). ORF57 is one of the KSHV lytic genes and is highly responsive to RTA. It encodes a post-transcriptional activator involved in mRNA export and plays an important role in KSHV lytic replication. Previously, we and others (Duan et al., 2001; Lukac et al., 2001) have mapped a 40 bp segment of the ORF57 promoter encompassing nt 81904–81943 on the viral genome as the target site for RTA binding and upregulation of ORF57 transcription. Two RREs (ORF57-RRE1 and ORF57-RRE2) were identified in this 40 bp segment (Fig. 1a). ORF57-RRE1 was found to share some homology with the delayed early gene K8 promoter sequence (Lukac et al., 2001; Wang et al., 2001a). ORF57-RRE2 was found to bind not only RTA, but also interferon regulatory factor 7 (IRF-7), and competition between RTA and IRF-7 affects regulation of the ORF57 promoter by RTA (Wang et al., 2005). Even though these two RREs are AT-rich, as predicted for RREs, there is no similarity between them. However, upon inspection of the ORF57 promoter, a CCN9GG-like sequence distinct from ORF57-RRE1 and -RRE2 was also found. We termed this CCN9GG-like motif in the ORF57 promoter as ORF57-RRE3. In this study, we characterized this third RRE in the ORF57 promoter and found that it plays an important role in conferring responsiveness of the promoter to KSHV RTA.
Fig. 1.
Sequence comparison of RREs. (a) The sequences of the three potential KSHV RREs (RRE1, RRE2 and RRE3) in the ORF57 promoter are shown in bold italic and underlined. The RBP-Jκ-binding site is indicated with double underlining. (b) Alignment of potential KSHV RREs with homology to the EBV RRE CCN9GG sequence in the ORF57, K2, PAN, MIP and K12 promoters. Identical sequences are boxed. Numbers represent the nucleotide positions of the KSHV genome according to GenBank accession no. U75698. The sequence of EBV RRE is in bold, where N can be any nucleotide.
METHODS
Plasmids.
An RTA expression plasmid (pCMVtagORF50) encoding Flag-tagged full-length RTA has been described previously (Wang et al., 2001a). The β-galactosidase expression plasmid pCMVβ, which was used for normalization of transfection efficiency, was purchased from BD Clontech. All ORF57 reporter plasmids were cloned in the pGL3-Basic vector (Promega), and p57p1, p57B1, p57B1M1 and p57E have been described previously (Duan et al., 2001; Wang et al., 2001a). Plasmids p57-3RRE, p57-3RRE1, p57B2 and p57-3RRE4 contained ORF57 upstream regions spanning nt −184 to −20, −163 to −20, −68 to −184 and −151 to −20, respectively (the putative transcription start site at nt 82081 is designated +1). Plasmid p57p1m2 contained the same ORF57 promoter region from −525 to −73 as p57p1, but had mutations in ORF57-RRE2 (the T nucleotides in positions −141, −143 and −149 were replaced with C, and CTT at nt −145 to −147 were replaced with AGC). Plasmids p57-3RRE3, p57-3RRE2 and p57-3RRE03 contained the same ORF57 promoter region as p57-3RRE, but p57-3RRE2 had a substitution in the RBP-Jκ-binding site (nt −163 to −151) with the PstI restriction enzyme sequence, p57-3RRE3 had a deletion in ORF57-RRE2 (nt −151 to −138) and p57-3RRED3 had a deletion in ORF57-RRE3 (nt −122 to −137). Plasmid p57B2M contained the same ORF57 promoter region as p57B2, but had mutations in ORF57-RRE1 as in the p57B1M1 construct.
Electrophoretic mobility shift assay (EMSA).
Probes were obtained by labelling double-stranded oligonucleotides (see Figs 2a and 4a) with [α-32P]dATP (Amersham) using DNA polymerase I Klenow fragment (New England BioLabs). Labelled probes were incubated with purified His-tagged, full-length RTA protein, which was expressed in Sf9 cells and purified using Ni-NTA (Promega), for 30 min in binding buffer [10 mM Tris/HCl (pH 7.5), 150 mM KCl, 5.7 mM MgCl2, 1 mM EDTA, 0.1 μg poly(dI/dC), 5 μg BSA, 0.38 mM dithiothreitol, 0.38 mM PMSF, 50 mM β-mercaptoethanol, 5 % glycerol]. For competition assays, the purified Sf9/RTA protein was incubated with non-radioactive competitor DNA for 30 min; the labelled probe was then added and incubated for 30 min. The binding mixtures were loaded onto a 4.5 % polyacrylamide gel in 1× TGE buffer [5 mM Tris base, 190 mM glycine, 1 mM EDTA (pH 8.3)]. After electrophoretic separation at 200 V, gels were transferred to Whatman 3MM paper, dried and subjected to autoradiography with Kodak X-ray film.
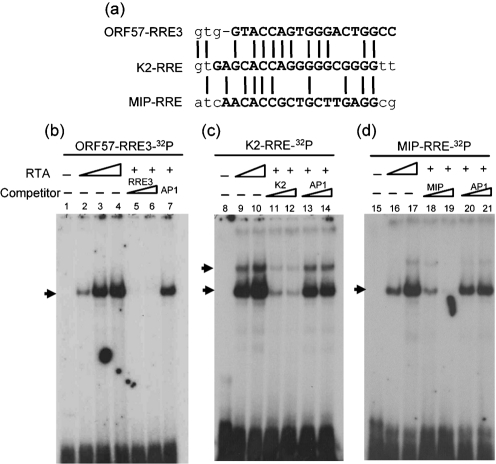
Fig. 2.
Binding of RTA to the CCN9GG core sequence in vitro. (a) Alignment of the ORF57-RRE3, K2-RRE and MIP-RRE sequences. Identical nucleotides are indicated by vertical lines. The synthetic oligonucleotide fragments of ORF57-RRE3, K2-RRE and MIP-RRE used for EMSAs are capitalized and in bold. (b–d) EMSA was carried out using full-length His-tagged RTA expressed in Sf9 cells and 32P-labelled dsDNA probes. RTA was shown to bind to ORF57-RRE3 and also to K2-RRE and MIP-RRE. Lanes 1, 8 and 15, 32P-labelled probe alone (control); lanes 2–4, 9, 10, 16, 17, increasing amounts of RTA incubated with probe; lanes 5–7, 11–14 and 18–21, 50- and 100-fold excess unlabelled specific or non-specific (AP1) competitor incubated in the presence of purified RTA. Arrows indicate the RTA–DNA complex.
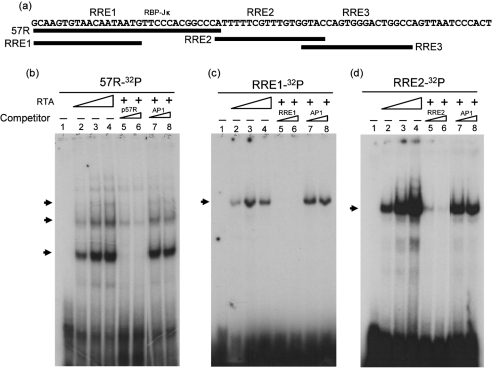
Fig. 4.
Binding of RTA to ORF57-RRE1 and -RRE2 is independent of the RBP-Jκ-binding site in vitro. (a) Solid lines show the four DNA fragments (57R, RRE1, RRE2 and RRE3) that were labelled with 32P and used as probes for EMSAs. (b–d) EMSA was performed with RTA and the three DNA fragments, p57R (b), RRE1 (c) and RRE2 (d). The conditions used for EMSA were as described in Fig. 2. Unlabelled probes were used as specific competitors and AP1 DNA was used as a negative control. Arrows indicate the RTA–DNA complex.
Transient chromatin immunoprecipitation (ChIP).
ORF57 reporter plasmid and RTA expression plasmid were co-transfected into 293T cells or TRExBJAB-RTA cells (provided by Dr Jae Jung, University of Southern California, CA, USA) using Lipofectamine 2000 reagent (Invitrogen) or Nucleofector (Amaxa). Transfection efficiency was approximately 90 % for 293T cells and approximately 70 % for TRExBJAB-RTA cells. At 24 h post-transfection, doxycycline was added to the TRExBJAB-RTA cells to induce RTA expression. At 40–48 h post-transfection, cells were fixed by the addition of 37 % formaldehyde to a final concentration of 1 % for 10 min. The cross-linking reaction was stopped by adding glycine to a final concentration of 0.125 M for 5 min. Cells were washed twice with 1× ice-cold PBS and resuspended in cell lysis buffer [5 mM PIPES (pH 8.0), 85 mM KCl, 0.5 % NP-40, 1 mM PMSF, protease inhibitor cocktail (Pierce)] for 10 min at 4 °C. After centrifugation, the cell pellets were resuspended in nuclear lysis buffer [50 mM Tris/HCl (pH 8.1), 10 mM EDTA, 1 % SDS] and incubated for 10 min at 4 °C. The resulting solution was diluted 10-fold with dilution buffer [16.7 mM Tris/HCl (pH 8.1), 167 mM NaCl, 1.2 mM EDTA, 0.01 % SDS, 1.1 % Triton X-100, 1 mM PMSF, protease inhibitor cocktail] and sonicated. After centrifugation, the supernatant was pre-cleared with mouse IgG (Santa Cruz) and immobilized protein A (Pierce). Immunoprecipitation was performed by incubation at 4 °C with anti-Flag M2 antibody (Stratagene) or anti-His antibody (Clontech). Immune complexes were collected by a further incubation with immobilized protein A for 1–2 h at 4 °C. The beads were washed four times with IP buffer [25 mM Tris/HCl (pH 7.2), 150 mM NaCl] and resuspended in ImmunoPure IgG elution buffer (Pierce). After centrifugation, the supernatant was neutralized with 1 M Tris/HCl (pH 7.2). RNase A and NaCl were added, followed by incubation at 65 °C overnight to reverse the cross-linked DNA–protein complex. Proteinase K (Stratagene) was added, followed by incubation at 55 °C for 1–2 h. DNA extraction was carried out using a miniprep kit (Qiagen) and analysed by PCR using specific primers located in the reporter vector. The DNA was quantified by TaqMan real-time PCR using TaqMan PCR core reagents (Applied Biosystems). For calculation of the relative DNA amount from quantitative real-time PCR, the Ct (threshold cycle) value of antibody-precipitated sample was normalized by the Ct value of the input, and the normalized Ct value of p57-3RRED3 was compared with the normalized Ct value of p57-3RRE. The mean±sd of the results for each promoter was calculated from the results of two reactions obtained from three different transfection experiments.
Cell culture, transfection and luciferase assay.
BJAB cells (a KSHV-negative cell line) were grown in RPMI 1640 (Gibco-BRL) supplemented with 10 % fetal bovine serum and 100 μg penicillin/streptomycin (Mediatech) ml−1 at 37 °C with 5 % CO2. Transfection of BJAB cells was carried out using Nucleofector (Amaxa) according to the manufacturer's recommendations. The transfection efficiency was normalized using the β-galactosidase expression plasmid pCMVβ as an internal control. Luciferase activities were determined using a Luciferase Assay System (Promega). Mean results were determined from three independent experiments.
RESULTS
A CCN9GG-like consensus sequence can be identified in a number of KSHV promoters
To examine whether KSHV RTA binds directly to the RRE3 sequence of the ORF57 promoter, we performed an EMSA using purified, His-tagged, full-length RTA protein expressed in recombinant baculovirus-infected Sf9 cells. Double-stranded ORF57-RRE3 oligonucleotides (18 bp; Fig. 2a) were labelled and incubated with binding buffer either alone or with increasing amounts of RTA. The ORF57-RRE3 oligonucleotide demonstrated dose-dependent binding to RTA (Fig. 2b, lanes 1–4). To demonstrate the specificity of RTA binding to ORF57-RRE3, excess amounts of unlabelled ORF57-RRE3 and AP1-binding sequence were used as specific and non-specific competitors, respectively, in competition experiments. The interaction of RTA and ORF57-RRE3 was shown to be specific as it could be competed out by homologous unlabelled oligonucleotide, but not by AP1-specific oligonucleotide (Fig. 2b, lanes 5–7).
To confirm further that RTA binds to a CCN9GG-like motif, 18 bp oligonucleotide fragments of K2-RRE and MIP (macrophage inflammatory protein)-RRE (Fig. 2a) were used for an EMSA. Similar to ORF57-RRE3, strong binding to RTA was observed, demonstrating that K2-RRE and MIP-RRE containing a CCN9GG-like sequence are also RTA targets, and this specificity was confirmed by EMSA using competitors (Fig. 2c, d). Our results indicated that CCN9GG is a potentially conserved motif of KSHV RRE in addition to AT-rich sequences identified previously, and that the CCN9GG-like motif could be a third RRE located in the ORF57 promoter.
ORF57-RRE3 is required for RTA binding to the ORF57 promoter
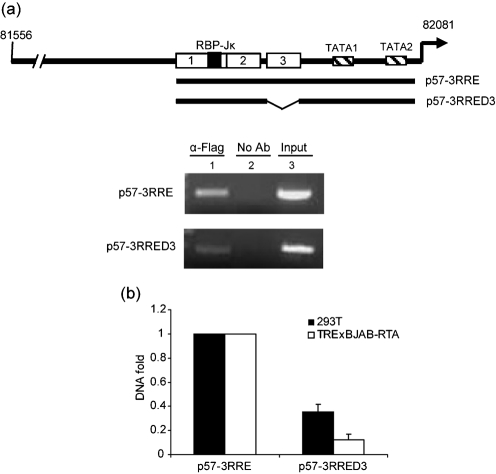
To confirm further that ORF57-RRE3 mediates RTA binding to the ORF57 promoter in vivo, transient ChIP was carried out in 293T and TRExBJAB-RTA cells. The reporter construct containing the ORF57 promoter, shown in Fig. 3(a), and the Flag-tagged, full-length RTA expression plasmid were co-transfected into 293T cells. The presence of ORF57 promoter DNA fragments within the immunoprecipitated complex was confirmed by PCR amplification. As shown in Fig. 3(a, upper panel, lane 1), a strong band was detected in the presence of RTA and the p57-3RRE plasmid containing all three RREs in the ORF57 promoter. The negative control with transfection in the absence of antibody failed to recover any PCR product (Fig. 3a, lane 2). In contrast to p57-3RRE, transfection of p57-3RRED3, which lacked RRE3 in the ORF57 promoter element, appeared to be less efficient in binding to RTA and resulted in a weaker amplified band (Fig. 3a, lower panel, lane 1). This was also confirmed by real-time PCR to quantify the amount of precipitated DNA from 293T and TRExBJAB-RTA cells, whereby the RRE3-lacking DNA fragment resulted in approximately 65 % reduction in binding to RTA in 293T cells and 85 % reduction in TRExBJAB-RTA cells (Fig. 3b). These results suggested that ORF57-RRE3 is required for efficient binding of RTA to the ORF57 promoter.
Fig. 3.
Binding of RTA to the ORF57 promoter RREs in vivo. (a) A transient ChIP assay was carried out in 293T cells transfected with the reporter plasmids p57-3RRE and p57-3RRED3 as indicated, together with Flag-tagged full-length RTA expression plasmid. RTA–DNA complexes were specifically precipitated with or without anti-Flag antibody. The precipitated DNA was recovered by PCR with specific primers located in the backbone of the reporter vector. (b) Taqman real-time PCR was performed to quantify the immunoprecipitated promoter DNA from transfected 293T and TRExBJAB-RTA cells.
RTA can bind to each of the three ORF57 RREs independently
In an earlier study, we demonstrated that RTA protein expressed in Escherichia coli can bind to the ORF57 promoter fragment 57R consisting of ORF57-RRE1 and the RBP-Jκ-binding site (Fig. 4a) (Wang et al., 2005). The results in Fig. 2 demonstrated that the RTA protein expressed in baculovirus-infected Sf9 cells could bind to RRE3 in vitro. To characterize further the binding of RTA to each of the three RRE elements, an EMSA was carried out with DNA fragments containing each of these elements (Fig. 4a). Multiple high-molecular-mass shifted bands were observed with the 57R labelled probe (Fig. 4b). Interestingly, we also observed interaction of RTA and RRE1, which lacked the RBP-Jκ consensus sequence, but only a single complex was detected (Fig. 4c). Similarly, only one band was detected with the ORF57-RRE2 probe (Fig. 4d). Figs 2 and 4 suggest that Sf9/RTA can bind directly to each of the three ORF57 RREs. This binding did not require the RBP-Jκ consensus sequence, but the presence of the RBP-Jκ-binding site may contribute to the formation of multiple complexes.
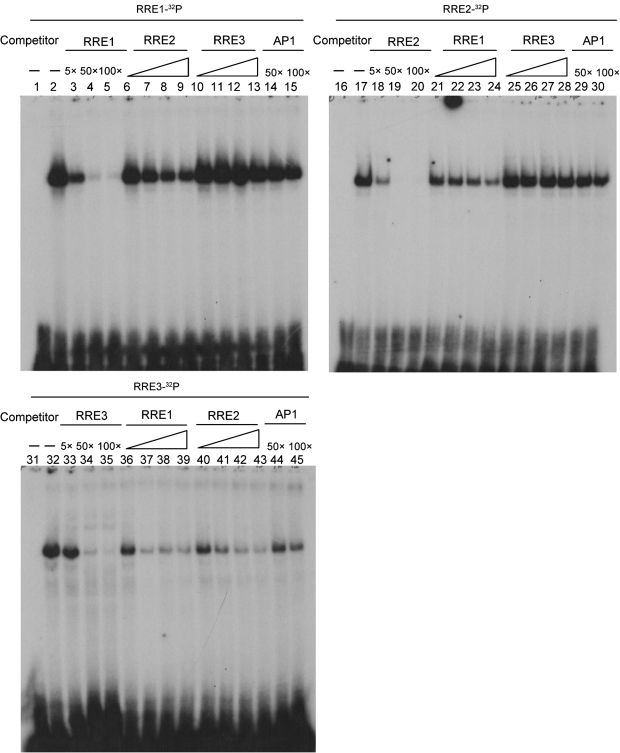
Binding of RTA to ORF57-RRE3 is weaker than to ORF57-RRE1 and -RRE2
To investigate further the differential ability of each ORF57 RRE to bind RTA, cross-competition assays were carried out. As expected, the binding of RTA to each ORF57 RRE probe was competed out efficiently by its homologous unlabelled oligonucleotide (Fig. 5, lanes 2–5, 17–20 and 32–35). ORF57-RRE1 binding to RTA was reduced by approximately 50 % with an excess of ORF57-RRE2 competitor (Fig. 5, lanes 6–9), and ORF57-RRE2 binding was reduced by approximately 80 % with an excess of ORF57-RRE1 competitor (Fig. 5, lanes 21–24) based on quantification of the bands. The binding of RTA to ORF57-RRE3 was efficiently competed out by unlabelled ORF57-RRE1 or ORF57-RRE2 (Fig. 5, lanes 36–43). However, ORF57-RRE3 was unable to compete for binding to RTA with either ORF57-RRE1 or ORF57-RRE2 (Fig. 5, lanes 10–13 and 25–28, respectively). Taken together, these results showed that the RTA-binding affinity of ORF57-RRE1 was comparable to that of ORF57-RRE2, but the RTA-binding affinity of ORF57-RRE1 and ORF57-RRE2 appeared to be much stronger compared with that of ORF57-RRE3.
Fig. 5.
Comparison of the binding affinity of RTA to ORF57-RRE1, -RRE2 and -RRE3. A cross-competition EMSA was used to determine the differential binding ability of RTA to the three RREs. Purified RTA protein (500 ng) was incubated with 40 fmol labelled probe in the absence or presence of homologous or non-homologous cold competitors. Increasing amounts (5×, 25×, 50× and 100×) of various non-homologous cold competitors were added. Non-specific competitor AP1 DNA was used a negative control and the fold excess relative to the labelled probes is indicated. Lanes 1, 16 and 31 represent probe alone (controls).
Two RREs are required for RTA-mediated transactivation of the ORF57 promoter
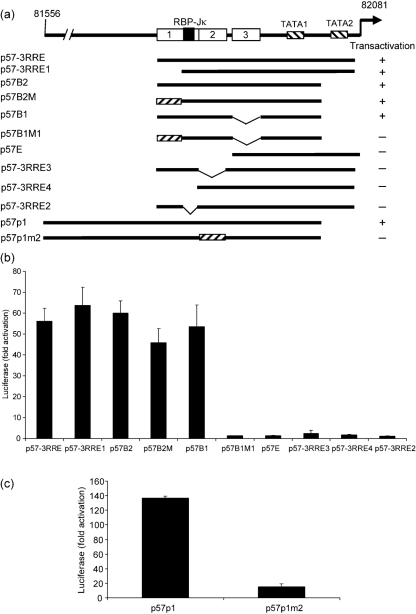
To verify that the binding of RTA to ORF57-RRE3 plays a role in transactivation of the ORF57 promoter, transient transfection experiments were carried out by co-transfection of the RTA expression plasmid with various reporter constructs of the ORF57 promoter as shown in Fig. 6(a). The p57-3RRE reporter, which contains the 40 bp segment, was induced effectively by RTA in BJAB cells (56-fold; Fig. 6b). Surprisingly, the p57-3RRE1 reporter, lacking ORF57-RRE1, was still transactivated effectively by RTA (63-fold), even though the AT-rich palindrome of ORF57-RRE1 has been considered a critical region for binding and transactivation by RTA (Lukac et al., 2001). Induction of the p57B2 reporter, with deletion of a 48 bp segment in the C-terminal ORF57 promoter, remained high (60-fold). The reporter p57B2M with mutations in ORF57-RRE1, and p57B1 reporter with a deleted ORF57-RRE3, remained highly responsive to RTA, comparable to the levels for p57B2. However, removal of ORF57-RRE3 and mutation of ORF57-RRE1 (p57B1M1) significantly reduced the promoter's responsiveness to RTA. These results suggest that ORF57-RRE1 or ORF57-RRE3 alone is not sufficient for efficient RTA transactivation, but that at least one of these two elements in combination with ORF57-RRE2 is required. This was confirmed by the p57E reporter containing ORF57-RRE3 alone, which showed a dramatic reduction in transactivation.
Fig. 6.
Two RTA-responsive elements and the RBP-Jκ-binding site are required for RTA-mediated transactivation. (a) Schematic representation of the ORF57 promoter reporter constructs and various deletion and site-directed mutagenesis clones used in the transient transfection analysis. Solid lines indicate different regions of the ORF57 promoter segment with various deletions (thin lines) or mutated regions (hatched boxes). The responsiveness of each reporter to RTA transactivation is indicated. (b, c) BJAB cells were used to analyse the promoter activity of different ORF57 reporter constructs. The total DNA amount used in each transfection was normalized by adding pCMVTag2A vector. Transfection of BJAB cells was carried out with 0.8 μg promoter reporter and 1 μg RTA expression plasmid. Luciferase activity was measured at 48 h post-transfection.
To confirm the role of ORF57-RRE2 in RTA-mediated transactivation, the ORF57-RRE2-deleted reporter p57-3RRE3 was used. Transactivation of p57-3RRE3 reporter by RTA was almost abolished (Fig. 6b). To confirm that the loss of transactivation response was not a result of the deletion, which shortened the length of the ORF57 promoter between ORF57-RRE1 and -RRE3, we tested the p57p1m2 reporter, which contained six nucleotide substitutions in ORF57-RRE2 but had overall promoter length that remained the same. As expected, the RTA responsiveness of p57p1m2 was still significantly reduced compared with p57p1, which acted as a positive control (Fig. 6c). These results suggested that ORF57-RRE2 is essential for RTA-mediated transactivation.
RTA-mediated transactivation of the ORF57 promoter requires the RBP-Jκ-binding site and two RREs
RBP-Jκ has been demonstrated to be a critical cellular factor for RTA-mediated transactivation of the ORF57 promoter by binding to the RBP-Jκ-binding site. The two promoter reporter constructs p57-3RRE1 and p57B1, both containing the RBP-Jκ-binding site, were strongly activated by RTA (Fig. 6b), suggesting that the RBP-Jκ-binding site is important in addition to the RREs. To define further the role of the RBP-Jκ-binding site and the two RREs in mediating RTA transactivation, the reporter p57-3RRE4, with a deletion of the upstream region of ORF57-RRE2 including the RBP-Jκ-binding site and ORF57-RRE1, was tested. As expected, this reporter lost its responsiveness to RTA, even though both ORF57-RRE2 and ORF57-RRE3 were intact. In addition, RTA inducibility was low with the p57-3RRE2 reporter, which has a deletion in the RBP-Jκ-binding site but an intact ORF57-RRE1, -RRE2 and -RRE3 (Fig. 6b). This was expected, as RBP-Jκ is well established as a co-activator of RTA that recognizes the consensus sequence located between ORF57-RRE1 and ORF57-RRE2 that regulates ORF57 transcription (Carroll et al., 2006; Liang et al., 2002). Interestingly, mutations in RRE2 (p57p1m2) and mutation/deletion in RRE1 and RRE3 (p57B1M1) affected the transcriptional induction of the ORF57 promoter by RTA (Fig. 6b, c). The responsiveness of these two promoter constructs to RTA was reduced substantially. Taken together, our results showed that RBP-Jκ plays an essential role in RTA-mediated ORF57 transactivation, but that the presence of two RREs (RRE1/RRE2 or RRE2/RRE3) is also necessary.
DISCUSSION
Role of ORF57 RREs in RTA-mediated transactivation of the ORF57 promoter
In this study, we identified a third RRE, ORF57-RRE3, in the ORF57 promoter. It has a high G+C content, which distinguishes it from the other two previously characterized RREs in the ORF57 promoter. Similar RREs with high G+C content have been found in the promoter elements of several other KSHV genes that are responsive to KSHV RTA.
The three ORF57 RREs are located in close proximity to each other in the ORF57 promoter and each has a unique sequence. Among the three RREs analysed, RRE2 seems to be essential for RTA-mediated transactivation. However, the presence of RRE2 alone was insufficient for transactivation, and the presence of either RRE1 or RRE3 was also required for RRE2 to be fully responsive to RTA. It is possible that, for the ORF57 promoter, the RTA protein may require more than one RRE for it to regulate its expression; whether this is required for other RTA-responsive promoters needs to be determined. Our results also suggested that RRE1 and RRE3 may play similar roles, as each could be replaced by the other in combination with RRE2 to mediate RTA transactivation; the presence of at least two RREs was required for transactivation. We found that deletion of ORF57-RRE1 alone did not affect ORF57 transactivation mediated by RTA (Fig. 6a), which is in contrast to an earlier report, demonstrating that ORF57-RRE1 was critical for RTA binding and transactivation (Lukac et al., 2001). However, these authors tested an isolated RRE1/RBP-Jκ element, thus making any direct comparison with our ORF57 reporter studies difficult.
We also observed that the amount of RTA bound to the promoter element containing RRE1 and RRE2 alone was less than with the intact promoter containing all three RREs (Fig. 3), although their levels of transactivation by RTA were similar. It is possible that the amount of RTA bound to RRE1 and RRE2 without RRE3 is adequate to reach the threshold level of RTA needed to be recruited to the promoter element for it to mediate full transactivation. Alternatively, it is possible that two adjacent RREs are needed for the RTA complex to bind optimally, as RTA has recently been reported to multimerize in vitro (Bu et al., 2007; Liao et al., 2003). In fact, we observed RTA multimerization in EMSAs (Fig. 4b) with multiple high-molecular-mass shifted bands using a DNA probe containing ORF57-RRE1 and the RBP-Jκ-binding sites. These complexes could be due to spontaneous multimerization of RTA as described previously (Bu et al., 2007; Liao et al., 2003), but whether such complexes exist in vivo and participate in binding to the two RREs and in transactivation of the ORF57 promoter needs to be investigated further.
In spite of the similar roles played by ORF57-RRE1 and -RRE3 in mediating transactivation, the binding affinities of RTA to ORF57-RRE1 and -RRE3 were distinct, raising the possibility that the binding affinity of RTA to different RREs may not be directly proportional to the level of RTA-mediated transactivation. Transactivation function may require the presence of additional factors other than RTA; these may include both viral and cellular factors. One such factor is RBP-Jκ, which was shown to be a critical transcriptional factor interacting with RTA to bind to the RBP-Jκ-binding site between ORF57-RRE1 and -RRE2 to activate ORF57 transcription (Carroll et al., 2006; Liang et al., 2002). Indeed, our data revealed that RTA-mediated transcription of the ORF57 promoter could proceed only in the presence of all three elements – the RBP-Jκ-binding site, ORF57-RRE2 and either ORF57-RRE1 or -RRE3. Taken together, ORF57-RRE1 and -RRE3 may play a secondary but necessary role to enable the RTA–RBP-Jκ complex to bind to ORF57-RRE2 and the RBP-Jκ-binding site in the ORF57 promoter to mediate transactivation. We are investigating further the exact roles of ORF57-RRE1 and -RRE3 in RTA-mediated transactivation.
Conserved KSHV RTA-binding sequence
KSHV RREs have been studied extensively in recent years (Chang et al., 2005; Liao et al., 2003; Ziegelbauer et al., 2006). They have been found to be diverse and to lack a consensus sequence, but the exact motif mediating RTA binding and transactivation is still ambiguous. In contrast, the EBV Rta RRE sequence, GNCCN9GGNG, has been identified as being responsive to EBV Rta in several EBV lytic cycle promoters. Our finding that a CCN9GG-like sequence can also be used by KSHV RTA to mediate transactivation of the ORF57 promoter suggests that EBV and KSHV RREs may share sequence homology, and it is possible that the CCN9GG-like motif may also be found in other RTA-responsive promoters. Indeed, in the KSHV PAN promoter, the major sequence in the PAN RRE identified previously for RTA recognition and transactivation contains the ACCN9GG consensus sequence (Fig. 1b) (Song et al., 2002). In addition, it has been reported that another RTA-responsive promoter, MIP, lost its responsiveness following a deletion in the region encompassing nt 27 855–27 876 of the viral genome (Chang et al., 2005). We found that this region contains not only the RBP-Jκ-binding site, but also the ACCN9GG sequence, and have confirmed that RTA can bind to the MIP ACCN9GG motif alone using an EMSA. In contrast, Chang et al. (2005) failed to observe an interaction between the RTA protein and a DNA probe from the MIP promoter; this is not unexpected because of the absence of the ACCN9GG motif in the DNA probe that was used in the EMSA. Interestingly, the ACCN9GG motif can also be found in the K2-RRE previously identified by Deng et al. (2002), and we have now shown that RTA binds to the K2 DNA containing the ACCN9GG sequence (Fig. 2). Taken together, these results suggest that the KSHV ACCN9GG motif is an important RTA target.
Studies of the EBV Rta-responsive promoters that contain the recognition sequence GNCCN9GGNG have shown that the differences in their binding affinities and responsiveness to Rta transcription activation could be attributed to variation in the N9 sequence within each promoter element (Chen et al., 2005). Similarly, for KSHV, the five promoters containing the consensus sequence CACC(A/G)NTNGGN(C/G)NGG(T/C) that we identified here displayed variations in their N9 sequences (Fig. 1b), and this variation may play a role in their differential responsiveness to RTA transactivation. Indeed, it was found that the PAN-RRE had a stronger binding affinity and transactivation by RTA compared with the K12- and K2-RREs, although all of these RREs contained the CACC(A/G)NTNGGN(C/G)NGG(T/C) motif (Song et al., 2003). It is possible that the PAN N9 sequence is optimal for RTA binding; it is also possible that cellular factors may play a role in the differential binding and responsiveness of these promoters to RTA. A number of cellular proteins, such as K-RBP, RBP-Jκ, Oct-1, AP1, SP1 and CEBP-α, are known to regulate RTA-mediated transactivation (Chen et al., 2000; Liang & Ganem, 2004; Sakakibara et al., 2001; Wang et al., 2003a, b; Yang & Wood, 2007), and some of their binding motifs are found to be present in the vicinity of the CACC(A/G)NTNGGN(C/G)NGG(T/C) motif. Thus, the combination of RTA binding to these motifs with the presence of cellular factors may confer optimal activation of the promoter. Taken together, it is likely that the ability of RTA to mediate transactivation is distinct from its ability to bind to RREs and its binding affinity to these elements. Its ability to uncouple these two functions may play a role in its ability to regulate lytic gene expression at different stages of the KSHV life cycle.
In summary, we have identified a new RRE sequence containing a GC-rich sequence in five KSHV lytic gene promoters based on sequence homology to the consensus EBV RRE. Whether this consensus RRE sequence, CACC(A/G)NTNGGN(C/G)NGG(T/C), is present in other RTA target genes and whether there are optimal N sequences required remain to be determined. This is also the first report to demonstrate that KSHV RTA-mediated transcription requires two RREs in close proximity to each other. A better understanding of the role played by RTA and the various RREs will be needed to elucidate the mechanism of RTA-mediated transactivation and induction of KSHV lytic replication.
Acknowledgments
This study was supported by PHS grant CA76958 and NCRR COBRE grant RR15635 to C. W. We thank Dr Jae Jung at University of Southern California, CA, USA, for providing the TRExBJAB-RTA cell line and Dr Janos Zempleni at University of Nebraska-Lincoln, NE, USA, for his assistance in the ChIP experiments.
References
- Bowser, B. S., Morris, S., Song, M. J., Sun, R. & Damania, B. (2006). Characterization of Kaposi's sarcoma-associated herpesvirus (KSHV) K1 promoter activation by Rta. Virology 348, 309–327. [DOI] [PubMed] [Google Scholar]
- Bu, W., Carroll, K. D., Palmeri, D. & Lukac, D. M. (2007). Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 ORF50/Rta lytic switch protein functions as a tetramer. J Virol 81, 5788–5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, K. D., Bu, W., Palmeri, D., Spadavecchia, S., Lynch, S. J., Marras, S. A., Tyagi, S. & Lukac, D. M. (2006). Kaposi's sarcoma-associated herpesvirus lytic switch protein stimulates DNA binding of RBP-Jk/CSL to activate the Notch pathway. J Virol 80, 9697–9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesarman, E., Chang, Y., Moore, P. S., Said, J. W. & Knowles, D. M. (1995a). Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med 332, 1186–1191. [DOI] [PubMed] [Google Scholar]
- Cesarman, E., Moore, P. S., Rao, P. H., Inghirami, G., Knowles, D. M. & Chang, Y. (1995b). In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86, 2708–2714. [PubMed] [Google Scholar]
- Chang, Y., Cesarman, E., Pessin, M. S., Lee, F., Culpepper, J., Knowles, D. M. & Moore, P. S. (1994). Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266, 1865–1869. [DOI] [PubMed] [Google Scholar]
- Chang, P. J., Shedd, D., Gradoville, L., Cho, M. S., Chen, L. W., Chang, J. & Miller, G. (2002). Open reading frame 50 protein of Kaposi's sarcoma-associated herpesvirus directly activates the viral PAN and K12 genes by binding to related response elements. J Virol 76, 3168–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, P. J., Shedd, D. & Miller, G. (2005). Two subclasses of Kaposi's sarcoma-associated herpesvirus lytic cycle promoters distinguished by open reading frame 50 mutant proteins that are deficient in binding to DNA. J Virol 79, 8750–8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Ueda, K., Sakakibara, S., Okuno, T. & Yamanishi, K. (2000). Transcriptional regulation of the Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor gene. J Virol 74, 8623–8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. W., Chang, P. J., Delecluse, H. J. & Miller, G. (2005). Marked variation in response of consensus binding elements for the Rta protein of Epstein–Barr virus. J Virol 79, 9635–9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, H., Young, A. & Sun, R. (2000). Auto-activation of the rta gene of human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus. J Gen Virol 81, 3043–3048. [DOI] [PubMed] [Google Scholar]
- Deng, H., Song, M. J., Chu, J. T. & Sun, R. (2002). Transcriptional regulation of the interleukin-6 gene of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J Virol 76, 8252–8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, W., Wang, S., Liu, S. & Wood, C. (2001). Characterization of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF57 promoter. Arch Virol 146, 403–413. [DOI] [PubMed] [Google Scholar]
- Friborg, J., Jr, Kong, W., Hottiger, M. O. & Nabel, G. J. (1999). p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402, 889–894. [DOI] [PubMed] [Google Scholar]
- Gradoville, L., Gerlach, J., Grogan, E., Shedd, D., Nikiforow, S., Metroka, C. & Miller, G. (2000). Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J Virol 74, 6207–6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruffat, H. & Sergeant, A. (1994). Characterization of the DNA-binding site repertoire for the Epstein–Barr virus transcription factor R. Nucleic Acids Res 22, 1172–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Y. & Ganem, D. (2004). RBP-J (CSL) is essential for activation of the K14/vGPCR promoter of Kaposi's sarcoma-associated herpesvirus by the lytic switch protein RTA. J Virol 78, 6818–6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Y., Chang, J., Lynch, S. J., Lukac, D. M. & Ganem, D. (2002). The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jκ (CSL), the target of the Notch signaling pathway. Genes Dev 16, 1977–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, W., Tang, Y., Kuo, Y. L., Liu, B. Y., Xu, C. J. & Giam, C. Z. (2003). Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 transcriptional activator Rta is an oligomeric DNA-binding protein that interacts with tandem arrays of phased A/T-trinucleotide motifs. J Virol 77, 9399–9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukac, D. M., Renne, R., Kirshner, J. R. & Ganem, D. (1998). Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252, 304–312. [DOI] [PubMed] [Google Scholar]
- Lukac, D. M., Kirshner, J. R. & Ganem, D. (1999). Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J Virol 73, 9348–9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukac, D. M., Garibyan, L., Kirshner, J. R., Palmeri, D. & Ganem, D. (2001). DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J Virol 75, 6786–6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, G., Heston, L., Grogan, E., Gradoville, L., Rigsby, M., Sun, R., Shedd, D., Kushnaryov, V. M., Grossberg, S. & Chang, Y. (1997). Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein–Barr virus in dually infected body cavity lymphoma cells. J Virol 71, 314–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radkov, S. A., Kellam, P. & Boshoff, C. (2000). The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma–E2F pathway and with the oncogene Hras transforms primary rat cells. Nat Med 6, 1121–1127. [DOI] [PubMed] [Google Scholar]
- Rivas, C., Thlick, A. E., Parravicini, C., Moore, P. S. & Chang, Y. (2001). Kaposi's sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. J Virol 75, 429–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo, J. J., Bohenzky, R. A., Chien, M. C., Chen, J., Yan, M., Maddalena, D., Parry, J. P., Peruzzi, D., Edelman, I. S. & other authors (1996). Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc Natl Acad Sci U S A 93, 14862–14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara, S., Ueda, K., Chen, J., Okuno, T. & Yamanishi, K. (2001). Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J Virol 75, 6894–6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, T. V., Wang, H. W., Koumi, A., Hollyman, D., Endo, Y., Ye, H., Du, M. Q. & Boshoff, C. (2002). K15 protein of Kaposi's sarcoma-associated herpesvirus is latently expressed and binds to HAX-1, a protein with antiapoptotic function. J Virol 76, 802–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, M. J., Li, X., Brown, H. J. & Sun, R. (2002). Characterization of interactions between RTA and the promoter of polyadenylated nuclear RNA in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol 76, 5000–5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, M. J., Deng, H. & Sun, R. (2003). Comparative study of regulation of RTA-responsive genes in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol 77, 9451–9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskus, K. A., Zhong, W., Gebhard, K., Herndier, B., Wang, H., Renne, R., Beneke, J., Pudney, J., Anderson, D. J. & other authors (1997). Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol 71, 715–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturzl, M., Hohenadl, C., Zietz, C., Castanos-Velez, E., Wunderlich, A., Ascherl, G., Biberfeld, P., Monini, P., Browning, P. J. & Ensoli, B. (1999). Expression of K13/v-FLIP gene of human herpesvirus 8 and apoptosis in Kaposi's sarcoma spindle cells. J Natl Cancer Inst 91, 1725–1733. [DOI] [PubMed] [Google Scholar]
- Sun, R., Lin, S. F., Gradoville, L., Yuan, Y., Zhu, F. & Miller, G. (1998). A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc Natl Acad Sci U S A 95, 10866–10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, R., Lin, S. F., Staskus, K., Gradoville, L., Grogan, E., Haase, A. & Miller, G. (1999). Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J Virol 73, 2232–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., Liu, S., Wu, M., Geng, Y. & Wood, C. (2001a). Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF50 gene product contains a potent C-terminal activation domain which activates gene expression via a specific target sequence. Arch Virol 146, 1415–1426. [DOI] [PubMed] [Google Scholar]
- Wang, S., Liu, S., Wu, M. H., Geng, Y. & Wood, C. (2001b). Identification of a cellular protein that interacts and synergizes with the RTA (ORF50) protein of Kaposi's sarcoma-associated herpesvirus in transcriptional activation. J Virol 75, 11961–11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. E., Wu, F. Y., Fujimuro, M., Zong, J., Hayward, S. D. & Hayward, G. S. (2003a). Role of CCAAT/enhancer-binding protein alpha (C/EBPα) in activation of the Kaposi's sarcoma-associated herpesvirus (KSHV) lytic-cycle replication-associated protein (RAP) promoter in cooperation with the KSHV replication and transcription activator (RTA) and RAP. J Virol 77, 600–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. E., Wu, F. Y., Yu, Y. & Hayward, G. S. (2003b). CCAAT/enhancer-binding protein-α is induced during the early stages of Kaposi's sarcoma-associated herpesvirus (KSHV) lytic cycle reactivation and together with the KSHV replication and transcription activator (RTA) cooperatively stimulates the viral RTA, MTA, and PAN promoters. J Virol 77, 9590–9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Zhang, J., Zhang, L., Harrington, W., Jr, West, J. T. & Wood, C. (2005). Modulation of human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus replication and transcription activator transactivation by interferon regulatory factor 7. J Virol 79, 2420–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., AuCoin, D. P., Huete, A. R., Cei, S. A., Hanson, L. J. & Pari, G. S. (2005). A Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 ORF50 deletion mutant is defective for reactivation of latent virus and DNA replication. J Virol 79, 3479–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. & Wood, C. (2007). The transcriptional repressor K-RBP modulates RTA-mediated transactivation and lytic replication of Kaposi's sarcoma-associated herpesvirus. J Virol 81, 6294–6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelbauer, J., Grundhoff, A. & Ganem, D. (2006). Exploring the DNA binding interactions of the Kaposi's sarcoma-associated herpesvirus lytic switch protein by selective amplification of bound sequences in vitro. J Virol 80, 2958–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]