Abstract
LysB, a mycobacteriophage Ms6-encoded protein, was previously identified as a lipolytic enzyme able to hydrolyse the ester bond in lipase and esterase substrates. In the present work, we show that LysB can hydrolyse lipids containing mycolic acids from the outer membrane of the mycobacterial cell wall. LysB was shown to hydrolyse the mycolic acids from the mycolyl-arabinogalactan–peptidoglycan complex where the mycolates of the inner leaflet of the outer membrane are covalently attached to an arabinosyl head group. In addition, treatment of the extractable lipids from Mycobacterium smegmatis, Mycobacterium bovis BCG and Mycobacterium tuberculosis H37Ra with LysB showed that trehalose 6,6′-dimycolate (TDM), a trehalose diester of two mycolic acid molecules, was hydrolysed by the enzyme. We have also determined the structures of the mycolic acid molecules that form the M. smegmatis TDM. The identification of a phage-encoded enzyme that targets the outer membrane of the mycobacterial cell wall enhances our understanding of the mechanism of mycobacteriophage lysis.
INTRODUCTION
We have been undertaking bacteriophage studies with the aim of exploring their ability to destroy their host as a consequence of their lytic cycle. These studies are driven by either the use of bacteriophage itself as a therapeutic agent (phage therapy) or exploring the enzymes involved in cell lysis for enzyme therapy against pathogenic bacteria. At the end of their virulent cycle, all double-stranded DNA phages produce at least two enzymes: an endolysin, which hydrolyses the peptidoglycan, and a holin, a small membrane protein that defines the time of lysis and allows the phage endolysin to cleave its target. This leads to cell lysis and release of the progeny phage (Young et al., 2000; Young, 2005). Endolysins are also capable of degrading peptidoglycan when applied externally to the bacterial cell wall, resulting in a rapid lysis of the bacterial cell (Fischetti, 2005). These enzymes are normally very specific to the bacterial host of the phage from which they were derived (Fischetti, 2001) and this capacity renders them promising potential antibacterial agents. The use of purified bacteriophage lysins to destroy their bacterial hosts has been the subject of several recent reports (Loeffler et al., 2001; Schuch et al., 2002; Yoong et al., 2004; Zimmer et al., 2002). The data reported so far are all on Gram-positive bacteria, since direct contact of endolysins with the peptidoglycan is only possible in such bacteria; in Gram-negative bacteria, the presence of an outer membrane prevents this direct interaction. In addition to endolysins and holins, some bacteriophages encode additional genes within their lytic cassettes. Bacteriophage lambda encodes the Rz and Rz1 proteins. These proteins were recently described as being involved in the fusion of the inner and outer membranes, which thus eliminates all the conceivable barriers to the release of the progeny virions (Berry et al., 2008). Genes encoding similar gene products were identified in several other phages infecting Gram-negative hosts (Summer et al., 2007). Thus, our present knowledge of the mechanism of bacteriophage lysis suggests that the complexity of phage lytic cassettes depends on their hosts. Hosts with a simpler envelope require the phage to possess a simple lytic cassette, whereas hosts with a complex envelope require the phage to have a more complex lytic cassette.
In mycobacteriophage Ms6, a temperate phage that infects Mycobacterium smegmatis (Portugal et al., 1989), the lytic cassette comprises five genes (Garcia et al., 2002). In addition to the holin and the endolysin, Ms6 codes for a protein known as LysB. LysB was recently identified as a protein with lipolytic activity showing a higher affinity for substrates with long chain lengths (Gil et al., 2008).
Despite being classified as Gram-positive bacteria, mycobacteria have a complex cell wall. This cell wall consists of peptidoglycan covalently linked to arabinogalactan, which is in turn esterified to a variety of long chain (C60–C90), α-branched, β-hydroxy fatty acids (mycolic acids). This covalently linked complex, called mycolyl-arabinogalactan peptidoglycan (mAGP), is known as the cell wall core and forms the stable scaffolding to anchor the outer non-covalently associated lipids and glycolipids, including trehalose 6,6′-dimycolate (TDM or cord factor) (Daffé & Draper, 1998; Daffé, 2008). TDM, probably the most prominent and best-studied mycolic-acid-containing compound of mycobacteria, is composed of a trehalose molecule esterified with two mycolic acids and is implicated in the pathogenesis of certain mycobacterial species (Rao et al., 2005; Fujita et al., 2005). The mycolyl arabinogalactan together with TDM and trehalose 6′-monomycolate is responsible for the highly hydrophobic cell surface properties and acid-fastness of the mycobacterial cells, resulting in a resistance to dehydration and a natural impermeability to nutrients and antibacterial drugs (Barry et al., 1998; Crick & Brennan, 2008, Liu et al., 1996).
Given the complexity of the mycobacterial cell envelope, it is easy to understand why mycobacteriophages encode an additional lysis enzyme (LysB) that hydrolyses long chain esters (Gil et al., 2008). In the present work, we identify the substrates for Ms6 LysB activity in the cell wall of mycobacteria. Thus, we provide evidence that Ms6 LysB targets the M. smegmatis outer membrane by cleaving the ester bond between mycolic acids and arabinogalactan in the mAGP complex and between mycolic acids and trehalose in TDM. Moreover, its activity is not species-specific, as Ms6 LysB could also act on the TDM from Mycobacterium tuberculosis H37Ra and Mycobacterium bovis BCG.
METHODS
Plasmids, bacterial strains and culture media.
Plasmid pMP302, which encodes LysB, has been described previously (Gil et al., 2008). Escherichia coli JM109 (pMP302) was grown at 37 °C in Luria–Bertani broth or agar containing 100 μg ampicillin ml−1. M. smegmatis mc2155 (Snapper et al., 1990), M. bovis BCG (Pasteur strain ATCC 35734) and M. tuberculosis H37Ra (ATCC 25177) were grown in Middlebrook 7H9 (supplemented with 0.05 % Tween 80, 0.2 % glycerol) at 37 °C. For the growth of M. bovis BCG and M. tuberculosis H37Ra, the medium was also supplemented with 10 % oleic acid/albumin/dextrose/catalase enrichment (BD Biosciences).
Chemical and lipid standards.
The TDM from M. tuberculosis (MTb TDM) and the trehalose standards were purchased from Sigma. All solvents were HPLC-grade and were purchased from Fisher Scientific.
Expression and purification of LysB protein.
6×His-tagged LysB was expressed and purified from E. coli JM109 (pMP302) as described previously (Gil et al., 2008).
Preparation of M. smegmatis cell wall components.
mAGP from M. smegmatis was obtained as described previously (Bhamidi et al., 2008). In addition, the purified mAGP was extracted with diethyl ether at room temperature, overnight, in order to remove all the free mycolic acids. The mixture was centrifuged at 2500 g</bold> for 5 min and the pellet was washed once more with diethyl ether. The pellet was dried under nitrogen and left in a chemical hood for a couple of hours.
For total lipid extraction, the bacterial culture was centrifuged after growing for 24 h and the wet cell mass was extracted with chloroform/methanol [2 : 1, v/v, 15 ml (g wet mass)−1] at 55 °C overnight. The extracts were evaporated by rotary evaporation. The dried crude extract was stored at room temperature for further use.
Treatment of mAGP with LysB.
Purified mAGP (1 mg) was treated with LysB (360 μg) in PBS pH 7.4, containing 0.2 % Triton X-100 in a total volume of 400 μl. The mAGP was suspended using an ultrasonic water bath for 1 min and the incubation was continued for 24 h at room temperature with shaking. The mycolic acids were extracted with 1 ml diethyl ether and washed with water. The organic and water layers were separated by centrifugation and the organic phase (upper layer) was collected and washed once with 1 ml water. After drying, the samples were analysed by liquid chromatography-mass spectrometry (LC-MS) in negative ion mode.
To test the effect of LysB on mycobacterium lipids, 200 μg total lipids (extracted from M. smegmatis, M. bovis BCG or M. tuberculosis H37Ra) were dried in a glass tube and resuspended in PBS buffer using an ultrasonic water bath. LysB (360 μg) was added and the samples were incubated at 37 °C for 24 h. After treatment, chloroform and methanol were added to the samples such that the ratio was 8 : 4 : 3 (chloroform/methanol/water); the organic layer was then transferred to a new tube and dried. For analysis, total lipids were redissolved in chloroform to 10 mg ml−1 and then separated by 1D thin-layer chromatography (TLC) on aluminium-backed silica 60 F254 gel plates (EMD Chemicals) developed with chloroform/methanol/water (20 : 4 : 0.5; by vol.) and visualized with 10 % CuSO4 in 8 % H3PO4 followed by charring at 200 °C. In parallel, control assays were performed in the same conditions using PBS instead of the Ms6 LysB protein.
Separation and purification of the lipid hydrolysed from M. smegmatis by LysB (Msmeg_1).
The crude lipid extract (20 mg) was applied to a Silica Gel 60 (Merck, 70–230 mesh) column (2×50 cm) and eluted successively with chloroform (4×100 ml), acetone (4×100 ml) and methanol (4×100 ml). Fractions of 100 ml were collected, dried and monitored for Msmeg_1 by TLC.
Determination of Msmeg_1 structure.
Purified Msmeg_1 was analysed for the presence of mycolic acids as described by Phetsuksiri et al. (1999). Briefly, the lipid was resuspended in 2 ml of 15 % tetrabutylammonium hydroxide (TBAH; Sigma) and saponified at 100 °C overnight. After cooling, 2 ml water, 3 ml dichloromethane and 300 μl iodomethane (Aldrich) were added to the entire reaction mixture, which was then left on a shaking platform for 4 h. After centrifugation, the upper layer was discarded and the lower organic phase was washed three times with 3 ml water. The washed lower phase was dried by nitrogen flow, extracted with 4 ml diethyl ether, sonicated for 5 min and centrifuged at 2500 g. The ether extract was transferred into new glass tubes and dried. The mycolic acid methyl esters (MAMEs) from intact M. smegmatis were obtained in a similar fashion, starting with whole cells. MAMEs derived from Msmeg_1 and from M. smegmatis were fully separated into subclasses by threefold TLC developed with n-hexane/ethyl acetate (95 : 5; v/v) and then silica scrapings were extracted in diethyl ether overnight. The ether extracts were washed with water, dried and analysed by LC-MS. The mycolic acid methyl esters were visualized with 10 % CuSO4 in 8 % H3PO4 spray followed by charring at 200 °C for analytical purposes.
LC-MS assay.
LC-MS was performed on an Agilent 6220 high resolution time of flight mass spectrometer equipped with a MultiMode source selected to be in the dual atmospheric pressure chemical ionization/electrospray ionization mode. The HPLC is equipped with an Agilent 1200 binary pump. In LC-MS experiments, a Waters XBridge (C18, 2.1×150 mm) with 5 μM particle size was used. For analysis of free mycolates (negative mode), mycolate esters (positive mode) and intact TDM (positive mode), the same HPLC conditions were used. The column temperature was 45 °C. The separation was done using a gradient of solvent A (99 % methanol, 1 % 500 mM ammonium acetate) and solvent B (79 % n-propanol, 20 % hexane, 1 % 500 mM ammonium acetate) from 100 % A to 100 % B at a flow rate of 0.32 ml min−1 over a period of 45 min. The drying gas temperature was 300 °C and the vaporizer temperature was set at 200 °C. Typically, 5 μl of a 1 mg ml−1 sample was injected for analysis. The mass spectrum was acquired from m/z 250 to 3200 Da with a frequency of one scan per second. The resulting data were analysed by using Agilent's Mass Hunter program.
RESULTS AND DISCUSSION
LysB hydrolyses the mAGP complex
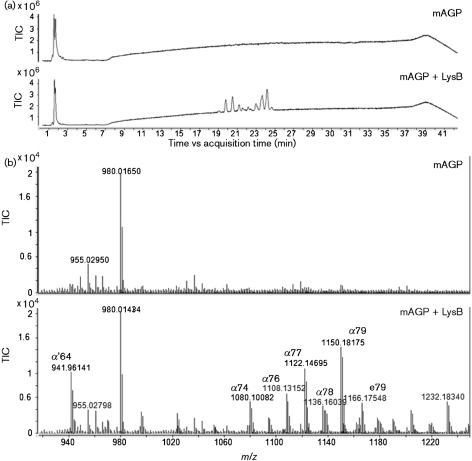
In order to evaluate the ability of LysB to act on mAGP, this complex was obtained from M. smegmatis and incubated with LysB for 24 h. Released mycolic acids were extracted and analysed by LC-MS (negative ion mode). As shown in Fig. 1, free mycolic acids were observed in samples treated with LysB, while in the control sample (where LysB is absent) no mycolic acids were detected. The dominant mycolic acids were those expected from M. smegmatis – α- and α′-mycolates – which contain no oxygen beyond the hydroxyl group in the meromycolate moiety, as well as epoxy mycolates containing an additional oxygen in the meromycolate moiety (Barry et al., 1998; Marrakchi et al., 2008). Since, in the mAGP complex, the non-reducing termini pentaarabinosyl motifs in the arabinogalactan are esterified to mycolic acids, the observation of the release of mycolic residues demonstrates that LysB acts on this bond and thus disrupts the inner leaflet of the mycobacterial outer membrane (Hoffmann et al., 2008; Zuber et al., 2008). This result is consistent with the esterase activity of Ms6 LysB, previously reported by our group (Gil et al., 2008), and with the report of a mycolyl arabinogalactan esterase activity for the homologous LysB found in mycobacteriophage D29 (Payne et al., 2009). Despite the fact that these homologous proteins share a low identity with the amino acid sequence, they have in common the conserved motif GXSXG of lipolytic enzymes (Gil et al., 2008, Hatfull et al., 2006). This suggests a common function and thus a common substrate for them. The esterase activity against the mycobacterial outer membrane is parallel to the lambda Rz/Rz1 homologues of Gram-negative bacteria, which, as proposed by Berry et al. (2008), mediate the final step of host lysis by fusing the inner and outer membranes. Although different from a Gram-negative outer membrane, the existence of an outer membrane layer in the mycobacterial envelope has long been postulated (Minnikin, 1982). Despite the fact that its arrangement is still under debate, recent work has clearly shown its existence (Hoffmann et al., 2008; Zuber et al., 2008). The existence of additional lysis genes in mycobacteriophage genomes, coding for proteins able to break the outer membrane linkage to the cell wall skeleton, enables the mycobacterial phage to efficiently escape from their lysed host.
Fig. 1.
LC-MS (negative ion mode) identification of mycolic acids released from mAGP after LysB treatment. (a) Total ion chromatograms from non-treated and treated (LysB) mAGP samples. (b) The negative ion mass spectra (averaged from 19 to 25 min) of non-treated and treated mAGP samples from m/z 920 to 1232. The mAGP sample treated with LysB showed the presence of several peaks, identified as mycolic acids. α, α-mycolates; α′, α′-mycolates; e, epoxy mycolates. The ion at m/z 980 corresponds to a calibration reference compound.
LysB hydrolyses lipids from the extractable lipid fraction
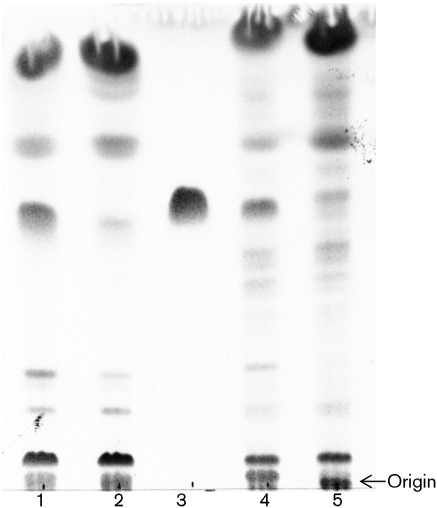
Taking into account that LysB has a broad range of lipolytic activity (Gil et al., 2008), we decided to investigate its ability to hydrolyse other cell wall lipids. Total extractable lipids from M. smegmatis were extracted and treated with LysB. TLC analysis revealed that the lipid profile changes after LysB treatment (Fig. 2, lanes 1 and 2). One of the major lipids disappears from the LysB-treated sample. Interestingly the same result is observed with total lipids from M. tuberculosis H37Ra (Fig. 2, lanes 4 and 5) and M. bovis BCG (data not shown). This indicates that LysB is not specific for M. smegmatis, as it is able to hydrolyse lipids from other mycobacterial species. The lipid hydrolysed from M. smegmatis by LysB was designated Msmeg_1. Its RF value was very similar to that of the Mtb TDM (Fig. 2, lane 3), suggesting that it could be the M. smegmatis version of TDM. The long acyl chains of the mycolic acid residues differ in structure and chain length within TDM and thus, TDMs from different mycobacteria have slightly different RF values (Fujita et al., 2005).
Fig. 2.
Thin layer chromatogram developed with chloroform/methanol/water (20 : 4 : 0.5 by vol.) of total lipid extract from M. smegmatis (lane 1), total lipid extract from M. smegmatis treated with LysB (2), Mtb TDM standard (3), total lipid extract from M. tuberculosis H37Ra (4) and total lipid extract from M. tuberculosis H37Ra treated with LysB (5).
Msmeg_1 is the M. smegmatis trehalose dimycolate
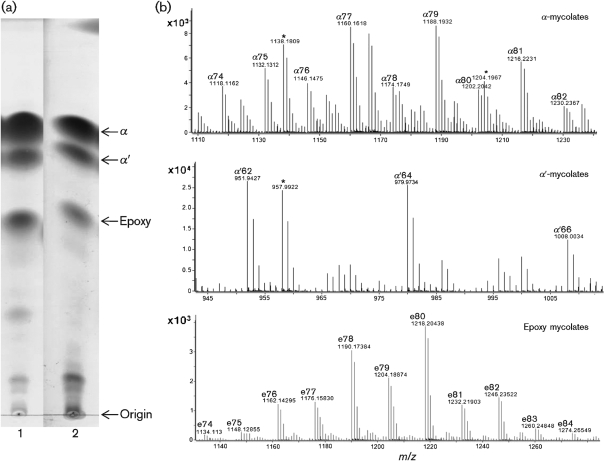
To investigate the composition of Msmeg_1, we produced a large amount of M. smegmatis cell mass, extracted Msmeg_1 and purified it by silica gel chromatography. To search for the presence of mycolic acids, Msmeg_1 was de-esterified with TBAH and concomitantly, the released fatty acids were methyl esterified with methyl iodide and then analysed by using TLC. After a threefold development with n-hexane/ethyl acetate (95 : 5, v/v), we observed three groups of MAMEs from both Msmeg_1 and intact M. smegmatis (Fig. 3a). The three spots observed correspond to α-, α′- and epoxy mycolates. The different types of MAMEs were eluted from the TLC and analysed by LC-MS. This analysis revealed the presence of M+Na ions with m/z values ranging from 951 to 1274, and the major components were identified as shown in Fig. 3(b). This result demonstrates that Msmeg_1 has mycolic acids in its structure. The pseudo-molecular ions obtained show a mass increase of 38 amu compared with those in Fig. 1. This corresponds to the mycolic acids being analysed as methyl esters and thus detected as sodiated methyl esters (positive mode) rather than as non-esterified carboxylate anions (negative mode). In addition, after separation into the mycolate classes, more details can be seen than are apparent in Fig. 1, namely the epoxy mycolates range from 74 to 84 carbon atoms, the α′-mycolates range from 62 to 66 carbon atoms and the α-mycolates range from 74 to 82 carbon atoms. As is the case for the free mycolates (Fig. 1), the exact mass values for components identified as epoxy mycolates are all consistent with the additional oxygen atom in the molecule.
Fig. 3.
Analysis of MAMEs by TLC and MS. (a) TLC fractionation of MAMEs into subclasses: thin-layer chromatogram of MAMEs from whole M. smegmatis (lane 1) and Msmeg_1 (lane 2). The development solvent was n-hexane/ethyl acetate (95 : 5, v/v), used three times. α, α-mycolates; α′, α′-mycolates; e, epoxy mycolates. (b) LC-MS analyses of MAME subclasses extracted from Msmeg_1, carried out in positive ion mode. Top, α-mycolates; middle, α′-mycolates; bottom, epoxy mycolates. The presence or absence of peaks marked with an asterisk (*) was variable and, based on their masses, was probably due to oxidation reactions occurring on the silica gel.
TDM is composed of mycolic acids and a trehalose disaccharide; to confirm the presence of the sugar molecule, we performed alkaline hydrolysis on Msmeg_1. The sugar was extracted and acetylated, and the resulting products were analysed by gas chromatography-MS. In parallel, two standards (MTb TDM and trehalose) were treated exactly as Msmeg_1. Our results showed that the retention time and mass spectra (data not presented) obtained for Msmeg_1 and the MTb TDM standard were the same as the trehalose standard, clearly demonstrating that the sugar in Msmeg_1 is the trehalose. Taken as a whole, the results clearly show that Msmeg_1 is in fact M. smegmatis TDM.
Treatment of purified Msmeg_1 with LysB
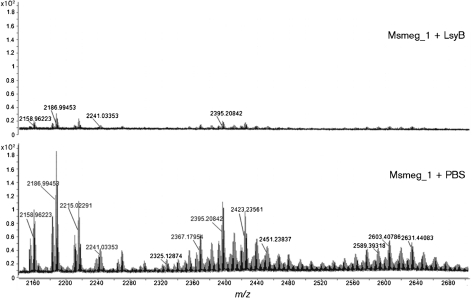
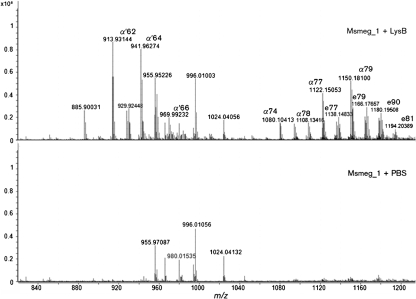
When we analysed pure Msmeg_1, treated and non-treated with LysB, in the positive ion mode by LC-MS, the M+Na pseudo-molecular ions in the region from m/z 2160 to 2700 showed the presence of several peak clusters for the non-treated Msmeg_1, while no such ions were observed for the treated sample (Fig. 4). This result is in accordance with our previous TLC results, in which LysB was able to hydrolyse Msmeg_1. LC-MS analysis (negative mode) for free mycolate in the same samples showed the presence of the expected mycolates in the LysB-treated sample but not in the untreated sample (Fig. 5). These analyses directly showed the action of LysB on Msmeg_1.
Fig. 4.
LC-MS analysis (positive ion mode) of purified Msmeg_1 treated with LysB and non-LysB-treated enzyme-treated Msmeg_1. The non-treated Msmeg_1 spectrum revealed the presence of several ion clusters between m/z 2158 and 2647. The isotope peaks containing 12C exclusively are labelled.
Fig. 5.
LC-MS (negative mode) identification of mycolic acids from released Msmeg_1 after LysB treatment (top panel). The bottom panel is a control with no LysB treatment. The mycolates, thus identified, were consistent with the more detailed analysis of TDM shown in Fig. 3. α, α-mycolates; α′, α′-mycolates; e, epoxy mycolates. The ion at m/z 980 is from the calibration standard; the ions at m/z 955, 996 and 1024 (found in both the control and the sample) are of unknown origin, and can also be seen in Fig. 1.
The mass spectrum of the intact Msmeg_1 (Fig. 4) shows abundant M+Na ions in three clusters with the dominant m/z values of 2187, 2395 and 2603 (fully 12C isotope mass). These correspond to the combinations of α′/α′, α′/α, and a mixture of α/α and α/epoxy mycolates esterified to trehalose. In Table 1, the most probable combination of mycolic acids present in each molecular species was determined based on the weights of mycolyl residues known to be present from our mycolate analysis (Figs 3 and 5). Similar heterogeneity in intact TDM molecules from other mycobacterial species has been found previously (Fujita et al., 2005).
Table 1.
The most probable combination of mycolic acids present in TDM of different molecular masses isolated from M. smegmatis
| Mass no. of TDM (M+Na)* | M. smegmatis mycolic acid combination† |
|---|---|
| 2158.96 (C124 Δ2) | α′62 : α′62 |
| 2186.99 (C126 Δ2) | α′62 : α′64 |
| 2215.02 (C128 Δ2) | α′62 : α′66; α′64 : α′64 |
| 2325.12 (C136 Δ3) | α′62 : α74 |
| 2353.15 (C138 Δ3) | α′62 : α76; α′64 : α74 |
| 2367.17 (C139 Δ3) | α′62 : α77; α′64 : α75 |
| 2395.20 (C141 Δ3) | α′62 : α79; α′64 : α77; α′66 : α75 |
| 2423.23 (C143 Δ3) | α′62 : α81; α′64 : α79; α′66 : α77 |
| 2575.37 (C154 Δ4) | α74 : α80; α75 : α79; α76 : α78; α77 : α77 |
| 2589.39 (C155 Δ4) | α74 : α81; α75 : α80; α76 : α79; α77 : α78 |
| 2603.40 (C156 Δ4) | α74 : α82; α75 : α81; α76 : α80; α77 : α79; α78 : α78 |
| 2617.41 (C157 Δ4) | α75 : α82; α76 : α81; α77 : α80; α78 : α79 |
| 2631.44 (C158 Δ4) | α76 : α82; α77 : α81; α78 : α80; α79 : α79 |
| 2661.43 (C159 epoxy Δ3) | α82 : e77; α81 : e78; α80 : e79; α79 : e80; α78 : e81; α77 : e82; α76 : e83; α75 : e84 |
| 2675.43 (C160 epoxy Δ3) | α82 : e78; α81 : e79; α80 : e80; α79 : e81; α78 : e82; α76 : e84; α77 : e83; α76 : e84 |
*The C number refers to the number of carbons in the mycolate chains (does not include the 12 carbons of trehalose). The Δ number indicates the total number of double bonds.
†α, α-mycolates; α′, α′-mycolates; e, epoxy mycolates.
Thus, TDM from M. smegmatis has now been shown to be a substrate for LysB. In addition, structural details beyond the early initial characterization of TDM in M. smegmatis (Mompon et al., 1978) have been obtained. These include the composition of the mycolic acid components (especially the epoxy mycolates) and the determination of which pairs of mycolates are combined on the trehalose disaccharide (Table 1).
Treatment of intact M. smegmatis with LysB
Enzymes that attack the mycobacterial cell envelope from the outside are needed as a novel therapeutic method to kill mycobacteria. Such enzymes can also be used as reagents to gain access to the contents of the mycobacterial cell for DNA and RNA analysis, and also for immunohistochemical analysis. Currently, lysozyme has some efficacy for the non-therapeutic purposes but is not as efficient as would be desired and must be used at very high concentrations. Its lack of robust activity makes it an impractical reagent to develop on its own for therapeutic purposes. Although the relatively high molecular mass of LysB (predicted molecular mass of 38.3 kDa) raised the concern that it would not be able to gain access to the mycolate bonds, we did test its ability to inhibit the growth of M. smegmatis. We found that, in the presence, but not in the absence, of Tween 80, LysB effectively killed M. smegmatis. Further investigation revealed that this was probably not due to the detergent itself but due to the toxicity of oleic acid released from the Tween 80 by LysB (F. Gil, unpublished observation). Therefore, although we have demonstrated that LysB is able to hydrolyse the mycolate–arabinogalactan bond and the mycolate–trehalose bond when Ms6 LysB is added from the outside, it is unable to disrupt the outer membrane sufficiently to kill the mycobacteria.
Conclusions
In the present work, we demonstrate that the mycobacteriophage Ms6 LysB lipolytic enzyme is able to hydrolyse ester bonds that are necessary for the integrity of the mycobacterial envelope. In particular, the arabinosyl head groups of the inner leaflet of the outer membrane (ultimately attaching the inner leaflet to peptidoglycan) are cleaved from the mycolates. Importantly, TDM is also de-esterified by the enzyme and, although the exact location of TDM in the cell envelope in not precisely defined, it is likely to be present in the outer leaflet of the outer membrane. It should be noted that the gene encoding LysB is localized in the lytic cassette of Ms6, downstream of the endolysin gene, supporting a role in mycobacterial lysis (Garcia et al., 2002). Thus, we propose that LysB activity, together with LysA acting on the peptidoglycan (unpublished data), results in a more efficient breakage of the cell wall, allowing a robust dissemination of the new phage particles. This is in agreement with a report (Payne et al., 2009) in which a deletion of lysB in the mycobacteriophage Giles genome results in a reduction of the mycobacteriophage plaque size, indicating that Giles LysB, although not essential for plaque formation, is required for efficient phage release. Unfortunately, LysB is unable to hinder the growth of M. smegmatis when added to growing cultures. Of note is the fact that LysB hydrolyses the M. smegmatis TDM, a glycolipid present in the lipid outer layer of mycobacteria, which has been associated with virulence in pathogenic bacteria. Our results suggest that TDM from the slow-growing mycobacteria are also hydrolysed by LysB, indicating that homologous proteins from other mycobacteriophages will also hydrolyse this glycolipid. It has already been shown that mycobacteria can survive after removal of the TDM, but viability of these bacteria inside macrophages is significantly reduced (Indrigo et al., 2002).
Acknowledgments
This work was supported by funds provided by Fundação para a Ciência e Tecnologia (FCT) PTDC/SAU-FCF/73017/2006. F. G. and M. J. C. are recipients of fellowships from FCT (SFRH/BD/29167/2006 and SFRH/BD/24452/2005, respectively). Support for M. R. M. and A. E. G. was provided by NIH AI33706. We thank Donald Dick (Department of Chemistry, Colorado State University) for the mass spectrometric analysis and Victoria Jones for reading the manuscript.
Abbreviations
LC-MS, liquid chromatography-mass spectrometry
mAGP, mycolyl-arabinogalactan peptidoglycan
MAMEs, mycolic acids methyl esters
Mtb TDM, trehalose 6,6′-dimycolate from M. tuberculosis
TBAH, tetrabutylammonium hydroxide
TDM, trehalose 6,6′-dimycolate
TLC, thin-layer chromatography
References
- Barry, C. E., III, Lee, R. E., Mdluli, K., Sampson, A. E., Schroeder, B. G., Slayden, R. A. & Yuan, Y. (1998). Mycolic acids: structure, biosynthesis and physiological functions. Prog Lipid Res 37, 143–179. [DOI] [PubMed] [Google Scholar]
- Berry, J., Summer, E. J., Struck, D. K. & Young, R. (2008). The final step in the phage infection cycle: the Rz and Rz1 lysis proteins link the inner and outer membranes. Mol Microbiol 70, 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhamidi, S., Scherman, M. S., Rithner, C. D., Prenni, J. E., Chatterjee, D., Khoo, K.-H. & McNeil, M. (2008). The identification and location of succinyl residues and the characterization of the interior arabinan region allow for a model of the complete primary structure of Mycobacterium tuberculosis mycolyl arabinogalactan. J Biol Chem 283, 12992–13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick, D. C. & Brennan, P. J. (2008). Biosynthesis of the arabinogalactan–peptidoglycan complex of Mycobacterium tuberculosis. In The Mycobacterial Cell Envelope, pp. 25–40. Edited by M. Daffé & J.-M. Reyrat. Washington, DC: American Society for Microbiology.
- Daffé, M. (2008). The global architecture of the mycobacterial cell envelope. In The Mycobacterial Cell Envelope, pp. 3–11. Edited by M. Daffé & J.-M. Reyrat. Washington, DC: American Society for Microbiology.
- Daffé, M. & Draper, P. (1998). The envelope layers of mycobacteria with reference to their pathogenicity. Adv Microb Physiol 39, 131–203. [DOI] [PubMed] [Google Scholar]
- Fischetti, V. A. (2001). Phage antibacterials make a comeback. Nat Biotechnol 19, 734–735. [DOI] [PubMed] [Google Scholar]
- Fischetti, V. A. (2005). Bacteriophage lytic enzymes: novel anti-infectives. Trends Microbiol 13, 491–496. [DOI] [PubMed] [Google Scholar]
- Fujita, Y., Naka, T., McNeil, M. R. & Yano, I. (2005). Intact molecular characterization of cord factor (trehalose 6,6′-dimycolate) from nine species of mycobacteria by MALDI-TOF mass spectrometry. Microbiology 151, 3403–3416. [DOI] [PubMed] [Google Scholar]
- Garcia, M., Pimentel, M. & Moniz-Pereira, J. (2002). Expression of mycobacteriophage Ms6 lysis genes is driven by two sigma (70)-like promoters and is dependent on a transcription termination signal present in the leader RNA. J Bacteriol 184, 3034–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil, F., Catalao, M. J., Moniz-Pereira, J., Leandro, P., McNeil, M. & Pimentel, M. (2008). The lytic cassette of mycobacteriophage Ms6 encodes an enzyme with lipolytic activity. Microbiology 154, 1364–1371. [DOI] [PubMed] [Google Scholar]
- Hatfull, G. F., Pedulla, M. L., Jacobs-Sera, D., Cichon, P. M., Foley, A., Ford, M. E., Gonda, R. M., Houtz, J. M., Hryckowian, A. J. & other authors (2006). Exploring the mycobacteriophage metaproteome: phage genomics as an educational platform. PLoS Genet 2, e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, C., Leis, A., Niederweis, M., Plitzko, J. M. & Engelhardt, H. (2008). Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc Natl Acad Sci U S A 105, 3963–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indrigo, J., Hunter, R. L., Jr & Actor, J. K. (2002). Influence of trehalose 6,6′-dimycolate (TDM) during mycobacterial infection of bone marrow macrophages. Microbiology 148, 1991–1998. [DOI] [PubMed] [Google Scholar]
- Liu, J., Barry, C. E., III, Besra, G. S. & Nikaido, H. (1996). Mycolic acid structure determines the fluidity of the mycobacterial cell wall. J Biol Chem 271, 29545–29551. [DOI] [PubMed] [Google Scholar]
- Loeffler, J. M., Nelson, D. & Fischetti, V. A. (2001). Rapid killing of Streptococcus pneumoniae with bacteriophage cell wall hydrolase. Science 294, 2170–2172. [DOI] [PubMed] [Google Scholar]
- Marrakchi, H., Bardou, F., Lanéele, M. & Daffé, M. (2008). A comprehensive overview of mycolic acid structure and biosynthesis. In The Mycobacterial Cell Envelope, pp. 41–62. Edited by M. Daffé & J.-M. Reyrat. Washington, DC: American Society for Microbiology.
- Minnikin, D. E. (1982). Lipids: complex lipids, their chemistry, biosynthesis and roles. In The Biology of Mycobacteria:, Physiology, Identification and Classification, vol. 1, pp. 95–184. Edited by C. Ratledge & J. L. Stanford. London: Academic Press.
- Mompon, B., Federici, C., Toubiana, R. & Lederer, E. (1978). Isolation and structural determination of a “cord-factor” (trehalose 6,6′ dimycolate) from Mycobacterium smegmatis. Chem Phys Lipids 21, 97–101. [DOI] [PubMed] [Google Scholar]
- Payne, K., Sun, Q., Sacchettini, J. & Hatfull, G. F. (2009). Mycobacteriophage lysin B is a novel mycolylarabinogalactan esterase. Mol Microbiol 73, 367–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phetsuksiri, B., Baulard, A. R., Cooper, A. M., Minnikin, D. E., Douglas, J. D., Besra, G. S. & Brennan, P. J. (1999). Antimycobacterial activities of isoxyl and new derivatives through the inhibition of mycolic acid synthesis. Antimicrob Agents Chemother 43, 1042–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portugal, I., Anes, E. & Moniz-Pereira, J. (1989). Temperate mycobacteriophage from M. smegmatis. Acta Leprol 7, 243–244. [PubMed] [Google Scholar]
- Rao, V., Fujiwara, N., Porcelli, S. A. & Glickman, M. S. (2005). Mycobacterium tuberculosis controls host innate immune activation through cyclopropane modification of a glycolipid effector molecule. J Exp Med 201, 535–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuch, R., Nelson, D. & Fischetti, V. A. (2002). A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 418, 884–889. [DOI] [PubMed] [Google Scholar]
- Snapper, S. B., Melton, R. E., Mustafa, S., Kieser, T. & Jacobs, W. R., Jr (1990). Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol 4, 1911–1919. [DOI] [PubMed] [Google Scholar]
- Summer, E. J., Berry, J., Tran, T. A., Niu, L., Struck, D. K. & Young, R. (2007). Rz/Rz1 lysis gene equivalents in phages of Gram-negative hosts. J Mol Biol 373, 1098–1112. [DOI] [PubMed] [Google Scholar]
- Yoong, P., Schuch, R., Nelson, D. & Fischetti, V. A. (2004). Identification of a broadly active phage lytic enzyme with lethal activity against antibiotic-resistant Enterococcus faecalis and Enterococcus faecium. J Bacteriol 186, 4808–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, R. (2005). Phage lysis. In Phages: Their Role in Bacterial Pathogenesis and Biotechnology, pp. 92–127. Edited by M. K. Waldor, D. I. Friedman & S. L. Adhya. Washington, DC: American Society for Microbiology.
- Young, I., Wang, I. & Roof, W. D. (2000). Phages will out: strategies of host cell lysis. Trends Microbiol 8, 120–128. [DOI] [PubMed] [Google Scholar]
- Zimmer, M., Vukov, N., Scherer, S. & Loessner, M. J. (2002). The murein hydrolase of the bacteriophage phi3626 dual lysis system is active against all tested Clostridium perfringens strains. Appl Environ Microbiol 68, 5311–5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber, B., Chami, M., Houssin, C., Dubochet, J., Griffiths, G. & Daffé, M. (2008). Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J Bacteriol 190, 5672–5680. [DOI] [PMC free article] [PubMed] [Google Scholar]