Abstract
The signal recognition particle (SRP) is a ribonucleoprotein complex that targets proteins for secretion in a co-translational manner. While originally thought to be essential in all bacteria, recent data show that the SRP is dispensable in at least some streptococcal species. The SRP from the human pathogen group A Streptococcus (GAS, Streptococcus pyogenes) is predicted to be composed of protein Ffh and 4.5S RNA. Deletion of ffh alters the secretion of several GAS proteins, and leads to a severe reduction in virulence. Here, we report that mutation of the gene encoding 4.5S RNA results in phenotypes both similar to and distinct from that observed following ffh mutation. Similarities include a reduction in secretion of the haemolysin streptolysin O, and attenuation of virulence as assessed by a murine soft tissue infection model. Differences include a reduction in transcript levels for the genes encoding streptolysin O and NAD-glycohydrolase, and the reduced secretion of the SpeB protease. Several differences in transcript abundance between the parental and mutant strain were shown to be dependent on the sensor-kinase-encoding gene covS. Using growth in human saliva as an ex vivo model of upper respiratory tract infection we identified that 4.5S RNA mutation leads to a 10-fold reduction in colony-forming units over time, consistent with the 4.5S RNA contributing to GAS growth and persistence during upper respiratory tract infections. Finally, we determined that the 4.5S RNA was essential for GAS to cause lethal infections in a murine bacteraemia model of infection. The data presented extend our knowledge of the contribution of the SRP to the virulence of an important Gram-positive pathogen.
INTRODUCTION
The human bacterial pathogen group A Streptococcus (GAS; Streptococcus pyogenes) can cause diseases that range from self-limiting pharyngitis and pyoderma to the severely invasive streptococcal toxic shock syndrome and necrotizing fasciitis (Cunningham, 2000). Key to the ability of GAS to cause such disease diversity is the production of numerous freely secreted or cell-surface-associated virulence factors that modulate the host immune response and enhance bacterial survival (Olsen et al., 2009). Recently, progress has been made in the study of the molecular mechanisms governing GAS protein secretion, with several proteins shown to be targeted to a single distinct membrane microdomain termed the ExPortal for secretion (Rosch & Caparon, 2005; Rosch et al., 2007).
The signal recognition particle (SRP) pathway is a universally conserved pathway that delivers secreted and transmembrane proteins to the secretory apparatus (Herskovits et al., 2000). The SRP is a ribonucleotide–protein complex that, in Escherichia coli, is composed of the fifty-four homologue (Ffh) protein and the 4.5S RNA. Ffh recognizes and binds to the secretion signal sequences of secreted or transmembrane proteins as they emerge from the ribosome. In a process that is accelerated by the 4.5S RNA, the Ffh–nascent polypeptide complex binds the membrane receptor filament temperature-sensitive Y (FtsY), which delivers the nascent polypeptide to the Sec translocation machinery (Bradshaw & Walter, 2007). Ffh–FtsY complex formation also stimulates the GTPase activities of both proteins, leading to their dissociation and making them available for additional rounds of protein targeting (Powers & Walter, 1995). Thus, the 4.5S RNA functions as a molecular switch that couples GTP hydrolysis to protein secretion (Bradshaw et al., 2009).
With regard to Gram-positive organisms, the SRP pathway has been best characterized in Bacillus subtilis (Yamane et al., 2004). The B. subtilis SRP pathway is similar to that of E. coli in that B. subtilis has ffh and ftsY homologues, as well as a homologue of the 4.5S RNA (termed the small cytoplasmic RNA, or scRNA). The B. subtilis SRP RNA is larger than the E. coli homologue, however: 271 nt compared to 114 nt (Struck et al., 1989). Despite the size difference, the two RNAs are functionally conserved, with E. coli 4.5S RNA able to complement a B. subtilis scRNA mutant (Nakamura et al., 1992). The SRP pathway is a major contributor to protein export in B. subtilis vegetative cells, and in spore formation (Yamane et al., 2004; Zanen et al., 2006).
While originally thought to be essential in all bacteria, the SRP pathway has recently been shown to be dispensable in at least some streptococcal species. While isogenic 4.5S RNA, ffh and ftsY mutant strains of the cariogenic oral pathogen Streptococcus mutans are viable, they have diminished tolerance to several stresses, including low pH and high salt (Hasona et al., 2005; Kremer et al., 2001). In the absence of stress, growth yield was similar between the parental and SRP mutant S. mutans strains, although the doubling time increased. Consistent with the function of the SRP pathway, the concentration of membrane-associated proteins differed between parental and SRP mutant strains (Hasona et al., 2007). The abundance of 116 gene transcripts also differed between parental and ffh mutant strains, highlighting the extensive transcriptional remodelling required to overcome disruption of the SRP pathway (Hasona et al., 2007).
Characterization of the SRP pathway in GAS has thus far been limited to a study that focused on the ffh gene (Rosch et al., 2008). An ffh mutant GAS strain was successfully constructed, showing that the SRP pathway was not essential in this pathogen. As was observed in S. mutans, the GAS ffh mutant gave growth yields similar to those of the parental strain when grown in nutrient-rich media but with an increased doubling time. Growth of the ffh mutant strain was identified as being glucose-dependent, with a severe reduction in growth rate and yield in media containing low concentrations of glucose (Rosch et al., 2008). In addition to ffh mutation resulting in medium-specific growth defects, the secreted virulence factors NAD-glycohydrolase and streptolysin O were present at lower concentrations in the supernatant of the ffh mutant strain, and the growth-phase expression of the secreted protease SpeB was deregulated. Importantly, the ffh mutant strain was severely attenuated in its ability to cause disease in a zebrafish model of necrotic myositis and a murine model of soft tissue infection (Rosch et al., 2008). Here, we investigate the 4.5S RNA-encoding gene and show that it is also a non-essential gene in GAS. Although the 4.5S RNA gene is not essential, mutants affected in this gene are attenuated for growth in an ex vivo model of upper respiratory tract infection, and severely attenuated for virulence in mouse models of bacteraemia and soft tissue infection. Of potential interest, several phenotypic differences were noted that appear to distinguish ffh and 4.5S RNA mutant strains. These data further our knowledge of the SRP pathway in GAS, and given the essential nature of this pathway in many bacterial species, its study in GAS may generate data pertinent to other pathogens.
METHODS
Bacterial strains and culture conditions.
GAS strain MGAS2221 is representative of the highly virulent M1T1 clone responsible for significant morbidity and mortality since the mid-1980s in the USA, Canada and Western Europe (Sumby et al., 2005b). GAS strain 2221covS : : 7bp is a previously described covS mutant derivative of MGAS2221 (Trevino et al., 2009). The genome of strain 2221covS : : 7bp has been analysed by a comparative genome resequencing method and the 7 bp insertion in covS shown to be the only genetic alteration relative to MGAS2221 (Sumby et al., 2006). GAS strains were grown overnight in vitro in Todd–Hewitt broth with 0.2 % yeast extract (THY broth) at 37 °C (5 % CO2).
Construction of a 4.5S RNA isogenic mutant strain.
An isogenic 4.5S RNA mutant strain was created from parental strain MGAS2221 by replacement of the 4.5S RNA gene with a non-polar spectinomycin-resistance cassette via a previously described PCR overlap extension method (Sumby et al., 2008). PCR primers used in the construction of strain 2221Δ4.5S are listed in Supplementary Table S1, available with the online version of this paper. Confirmation of isogenic mutant strain construction was gained via PCR, sequencing and Southern blot analyses. For the Southern analysis, genomic DNAs were digested with ScaI, separated by gel electrophoresis, transferred to membrane, and probed with a region of DNA that was PCR amplified from upstream of the 4.5S RNA gene (using primers 4.5SG and 4.5SH; Table S1).
Complementation of isogenic 4.5S RNA mutant strain 2221Δ4.5S.
Strain 2221Δ4.5S was complemented by introduction of the wild-type 4.5S RNA-encoding gene on the E. coli–GAS shuttle vector pDC123 (Chaffin & Rubens, 1998). Forward and reverse primers used to amplify the 4.5S RNA gene contained BglII and NsiI restriction enzyme sites, respectively (Table S1). PCR products digested with BglII/NsiI were ligated into similarly digested pDC123. All inserts were sequenced to ensure that no spurious mutants had arisen during PCR and cloning.
Construction of a covS/4.5S RNA double mutant strain.
To create a covS/4.5S RNA double mutant strain we made use of strain 2221covS : : 7bp, a previously described derivative of MGAS2221 that has a 7 bp insertion mutation in covS (Trevino et al., 2009). The 4.5S RNA gene was mutated in strain 2221covS : : 7bp, creating the double mutant strain 2221covS : : 7bp/Δ4.5S, as described above for creation of strain 2221Δ4.5S. Confirmation of isogenic mutant strain construction was gained via PCR, sequencing and Southern blot analyses (data not shown).
Western blot analysis of culture supernatant proteins.
GAS strains were grown in THY broth to the exponential (OD600 0.5) and stationary (OD600 1.7) phases of growth. All strains tested gave similar numbers of c.f.u. at these optical densities, and hence no normalization to c.f.u. number was required (data not shown). Culture supernatants were obtained by centrifugation and filtration through a 0.22 μm filter. Proteins were precipitated out of the supernatants via the addition of 3.5 vols ice-cold ethanol and incubating at −20 °C for 4 h. Precipitated proteins were collected via centrifugation at 4000 g for 20 min at 4 °C. Protein pellets were air-dried at room temperature for 20 min before resuspending in 500 μl SDS-PAGE loading buffer. Samples were separated by SDS-PAGE using 12 % Tris/HCl gels, transferred to nitrocellulose membrane, and probed using rabbit primary antibodies to Sic, streptolysin O (SLO), Spd3 and SpeB. The SLO primary antibody was purchased from American Research Products. The Sic, Spd3 and SpeB primary antibodies were generated in-house using either recombinant proteins (Sic and SpeB) or a C-terminal peptide (Spd3), as previously described (Sumby et al., 2005a). Secondary antibodies were HRP-conjugated to enable detection by standard methods using the SuperSignal West Pico Rabbit IgG Detection kit (Pierce). Primary antibodies were used at a 1/8000 dilution, while the secondary antibody was used at a 1/20 000 dilution.
Total RNA isolation.
GAS strains were grown in THY broth to the exponential phase (OD600 ∼0.5) and added to 2 vols RNAprotect (Qiagen). After incubating at room temperature for 5 min, samples were centrifuged for 10 min at 4 °C and 5000 g. Cell pellets were quick frozen in liquid nitrogen and stored at −80 °C until ready for use. Frozen GAS cell pellets were resuspended in 100 μl TE buffer and transferred to 2 ml tubes containing fine glass shards (lysing matrix B tubes, MP Biomedicals). The tubes were placed into a glass bead beater (FastPrep machine, THERMO 101) and processed for 15 s at speed 4. The tubes were centrifuged for 5 s at 14 000 g to reduce foaming and an additional processing in the FastPrep machine was performed following addition of 650 μl buffer RLT (Qiagen). Samples were centrifuged for 30 s at 14 000 g to collect contents and 600 μl was transferred to a 1.5 ml tube containing 900 μl 100 % ethanol. RNA samples were subsequently bound to, washed on and eluted from RNeasy columns (Qiagen) according to the manufacturer's miRNeasy protocol. Contaminating genomic DNA was removed from eluted RNA samples via three 40 min incubations at 37 °C with 2 μl TURBO DNase-free (Applied Biosystems), with DNA removal being verified by PCR. The quality and quantity of isolated RNA was assessed using a 2100 Bioanalyser system (Agilent Technologies).
cDNA synthesis and quantitative RT-PCR.
cDNA was synthesized from total GAS RNA using the reverse transcriptase Superscript III (Invitrogen) according to the manufacturer's instructions. TaqMan quantitative RT-PCR was performed using an ABI 7500 Fast System (Applied Biosystems). Gene transcript levels of mutant and complemented strains were compared to parental strain MGAS2221 using the 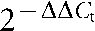 method as described previously (Shelburne et al., 2008). TaqMan primer and probe sequences are listed in Table S1. The constitutively transcribed housekeeping gene proS was used as an internal control, similar to previously published quantitative RT-PCR analyses (Shelburne et al., 2008; Virtaneva et al., 2003).
method as described previously (Shelburne et al., 2008). TaqMan primer and probe sequences are listed in Table S1. The constitutively transcribed housekeeping gene proS was used as an internal control, similar to previously published quantitative RT-PCR analyses (Shelburne et al., 2008; Virtaneva et al., 2003).
Growth of GAS in human saliva.
Human subjects research was conducted in accordance with the rules and regulations set forth by the TMHRI Institutional Review Board. Human saliva was collected from donor individuals, pooled, cleared by centrifugation, and filter-sterilized as previously described (Shelburne et al., 2005b). To compare the ability of GAS strains to grow in human saliva we first grew overnight cultures in THY broth. The following morning each overnight culture was diluted 1 : 100 with sterile PBS, followed by a second 1/100 dilution into 10 ml aliquots of filter-sterilized human saliva (hence a 1/10 000 final dilution of the overnight culture into human saliva). GAS/saliva solutions were incubated at 37 °C with 5 % CO2. Samples (100 μl) of the GAS/saliva solutions were recovered over time, serial dilutions performed, and GAS c.f.u. enumerated following overnight incubation on blood agar plates (37 °C; 5 % CO2). The experiment was repeated three times and mean values are reported.
Mouse infection assays.
Animal research was conducted in accordance with the rules and regulations set forth by the TMHRI Institutional Animal Care & Use Committee. Mice were infected with GAS grown to mid-exponential phase (OD600 ∼0.5). In our bacteraemia model of infection, female CD-1 mice were injected intraperitoneally with 250 μl of a 1×108 c.f.u. ml−1 suspension of GAS in PBS. Kaplan–Meier survival curves were generated and analysed for statistical significance using the logrank test. In our soft tissue infection model, female Crl:SKH10hrBR mice were injected subcutaneously with 100 μl of a 1×108 c.f.u. ml−1 suspension of GAS in PBS. The volume and ulceration of lesions were determined on days 1, 2, 3, 4, 5, 7, 9 and 11 post-infection, as described by Lukomski et al. (2000). Generalized estimating equations (GEEs) and Fisher's exact test were used to test for significant differences in the volume and ulceration of lesions, respectively. Groups of 20 mice per GAS strain were used for both infection models.
RESULTS
Identification and analysis of 4.5S RNA transcripts
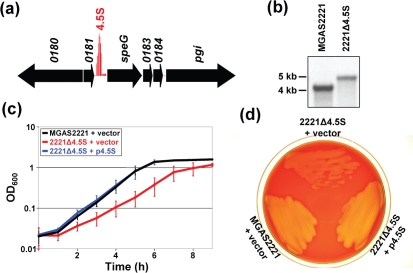
Recently, we used a custom Affymetrix tiling microarray to perform a genome-wide search for small regulatory RNAs in the serotype M1 GAS strain MGAS2221 (Perez et al., 2009). During this analysis we identified a highly transcribed RNA located upstream of, and in the same orientation as, the exotoxin-encoding gene speG (Fig. 1a). Nucleotide blast analysis identified the transcript as being the highly conserved 4.5S RNA component of the SRP. Consistent with the important function predicted for the 4.5S RNA it is constitutively transcribed at high levels during growth in nutrient-rich media (Perez et al., 2009).
Fig. 1.
Mutation of the 4.5S RNA detrimentally affects GAS growth rate in liquid and on solid media. (a) Our previous tiling microarray analysis identified the 4.5S RNA as a highly transcribed RNA located upstream of, and in the same orientation as, the exotoxin-encoding gene speG (Perez et al., 2009). Genes are represented by black arrows facing the direction of transcription. Red vertical lines represent signal intensities from probes tiled within intergenic regions. (b) Southern blot confirming gene replacement in the isogenic 4.5S RNA mutant strain 2221Δ4.5S. The hybridizing fragment size for the parental and mutant strains should be 4.1 kb and 5 kb, respectively. (c) Growth curve comparisons of parental strain MGAS2221 containing empty vector and isogenic 4.5S RNA mutant strain 2221Δ4.5S containing either empty vector or the 4.5S RNA-encoding derivative p4.5S. Growth was in THY broth; mean OD600 readings from duplicate samples grown on three separate occasions are plotted (error bars represent±sd). (d) Mutation of the 4.5S RNA reduces colony size and haemolysis on blood agar plates. The parental, mutant and complemented mutant strains were streaked onto an agar plate containing 5 % sheep blood and incubated overnight at 37 °C.
Construction and characterization of a 4.5S RNA mutant strain
While the composition of the SRP has yet to be determined for GAS, or for any streptococcal species, it is hypothesized to consist of protein Ffh and 4.5S RNA. Recently, it was determined that the ffh gene is not essential in GAS (Rosch et al., 2008). To identify whether the same was true for the RNA component of the SRP particle we set out to create an isogenic 4.5S RNA mutant derivative of GAS isolate MGAS2221. Mutant strain construction was successful as confirmed by PCR, sequence and Southern blot analyses (Fig. 1b and data not shown). Thus, the 4.5S RNA is not an essential gene in GAS. Similar to results observed following ffh mutation (Rosch et al., 2008), mutation of the gene encoding 4.5S RNA did not appreciably affect growth yield compared to the parental strain following growth in THY broth, but did increase the doubling time from 52 min for MGAS2221 to 75 min for 2221Δ4.5S (Fig. 1c). GAS chain length was not appreciably altered by 4.5S RNA mutation (unpublished data). Strain 2221Δ4.5S also gave smaller colonies, and subsequent areas of haemolysis, on blood agar plates than parental strain MGAS2221 (Fig. 1d and Supplementary Fig. S1). The slower growth of strain 2221Δ4.5S could be overcome by complementation with the wild-type 4.5S RNA gene (plasmid p4.5S in Fig. 1c, d).
Effect of 4.5S RNA mutation on GAS protein secretion
To identify whether strain 2221Δ4.5S had an altered protein secretion pattern compared to MGAS2221 we performed a series of Western blot analyses. Strains MGAS2221 and 2221Δ4.5S containing empty vector, and strain 2221Δ4.5S containing the complementing plasmid p4.5S, were grown to the exponential and stationary phases of growth, and secreted proteins were isolated and Western blots performed. For the four secreted GAS virulence factors investigated, streptolysin O (SLO) (Timmer et al., 2009), protease SpeB (Egesten et al., 2009), streptococcal inhibitor of complement (Sic) (Binks et al., 2005) and S. pyogenes DNase 3 (Spd3) (Sumby et al., 2005a), the isogenic mutant strain containing empty vector secreted lower amounts of these proteins than both the parental and complemented mutant strains (Fig. 2). Therefore, the 4.5S RNA is required for the efficient secretion of at least a subset of GAS proteins.
Fig. 2.
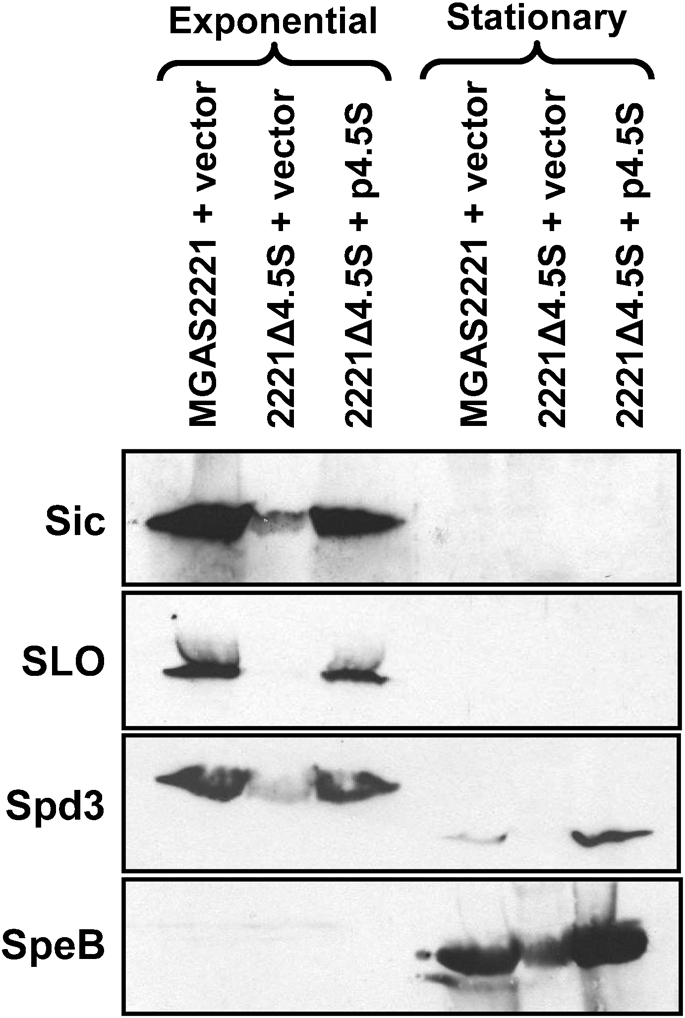
Mutation of the 4.5S RNA alters the GAS secretome. Secreted proteins from GAS strains MGAS2221 (+vector), 2221Δ4.5S (+vector) and 2221Δ4.5S (+p4.5S) were ethanol precipitated and subjected to Western blot analyses. Exponential-phase samples were recovered at an OD600 of 0.5, while stationary-phase samples were recovered at an OD600 of 1.7. All Western blots gave signals at the expected locations given the molecular masses of the target proteins [Sic (35 kDa), SLO (64 kDa), Spd3 (30 kDa) and SpeB (43 kDa)].
Effect of 4.5S RNA mutation on GAS transcript abundance
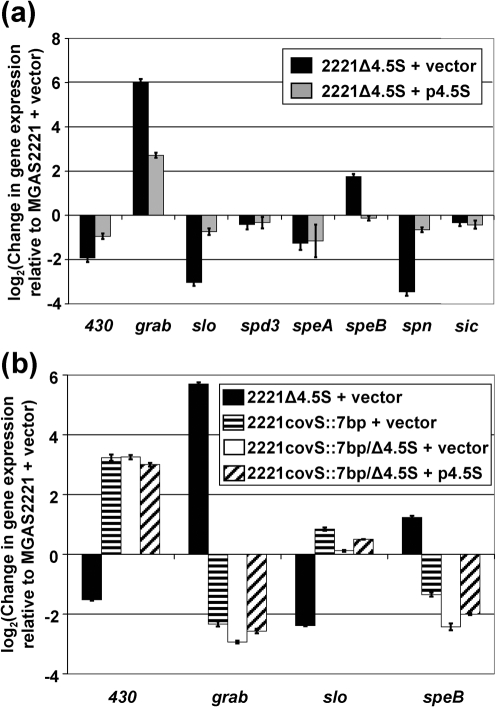
Given that strain 2221Δ4.5S secreted reduced amounts of Sic, SLO, Spd3 and SpeB proteins we tested whether there was a concomitant reduction in the abundance of mRNA transcripts encoding these proteins. To compare transcript abundance in MGAS2221 and 2221Δ4.5S we performed quantitative RT-PCR. Strains were grown to the exponential phase in THY broth, RNA was isolated, cDNA was synthesized, and quantitative RT-PCR was performed (Fig. 3a). Only the relative abundance of slo transcripts matched that observed at the level of protein secretion, namely a reduced concentration in the mutant strain relative to the parental strain. The levels of spd3 and sic transcripts were essentially unaltered, indicating that the reduction in secreted Spd3 and Sic was not the result of decreased transcription of the encoding genes. The relative concentration of speB transcripts increased about fourfold in strain 2221Δ4.5S, the opposite of the phenotype observed at the protein level (Figs 2 and 3a). The discordance between SpeB protein levels and speB transcript levels may be due to the quantitative RT-PCR data being gained from exponential-phase samples, while the Western data only gave signal from the stationary-phase samples.
Fig. 3.
Mutation of the 4.5S RNA alters the GAS transcriptome in a covS-dependent manner. Taqman quantitative RT-PCR was used to compare the exponential-phase transcript levels of several virulence-factor-encoding genes. (a) To assess whether mutation of the 4.5S RNA alters gene transcript abundance we analysed transcripts from parental strain MGAS2221 (+vector), 4.5S RNA mutant strain 2221Δ4.5S (+vector) and complemented strain 2221Δ4.5S (+p4.5S). (b) To assess whether modulation of transcript abundance following 4.5S RNA mutation is CovS-dependent we analysed transcripts from MGAS2221 (+vector), 2221Δ4.5S (+vector), 2221covS : : 7bp (+vector), 2221covS : : 7bp/Δ4.5S (+vector) and 2221covS : : 7bp/Δ4.5S (+p4.5S). Quantitative RT-PCR data were normalized to the transcript levels of the commonly used housekeeping gene proS (Graham et al., 2006; Reid et al., 2003; Shelburne et al., 2008; Sumby et al., 2005a). 430, spy0430. Each experiment was performed in triplicate and means±sd are plotted.
Four additional gene transcripts were also analysed by quantitative RT-PCR, grab, encoding protein G-related α2-macroglobulin-binding protein (Nyberg et al., 2004), speA, encoding the superantigen S. pyogenes exotoxin A (Russell & Sriskandan, 2008), spn, encoding S. pyogenes NAD-glycohydrolase (Meehl et al., 2005), and spy0430, encoding a putative membrane protein (Trevino et al., 2009). grab transcripts were present at higher abundance in strain 2221Δ4.5S, while spy0430, speA and spn transcripts were present at decreased abundance in strain 2221Δ4.5S, relative to parental strain MGAS2221. In each case, transcript levels approached wild-type levels following introduction of plasmid p4.5S into strain 2221Δ4.5S (Fig. 3a). The data are consistent with the global remodelling of the GAS transcriptome in response to loss of the SRP pathway, similar to that observed following SRP disruption in S. mutans (Hasona et al., 2007).
Role of the CovS sensor kinase in remodelling transcript abundance following 4.5S RNA mutation
The changes in transcript abundance following 4.5S RNA mutation are the reverse of that observed following mutation of covS (control of virulence Sensor; also known as csrS), which encodes the membrane-spanning sensor kinase component of the CovR/S two-component system (Bernish & van de Rijn, 1999; Engleberg et al., 2001; Federle et al., 1999; Gryllos et al., 2008; Levin & Wessels, 1998; Trevino et al., 2009). To test whether the transcriptional changes that occur following 4.5S RNA mutation are dependent on a functional covS gene we created a covS/4.5S RNA double mutant strain and compared transcript levels relative to a covS single mutant strain (Fig. 3b). For the four genes analysed by quantitative RT-PCR the increase (grab and speB) or decrease (spy0430 and slo) in transcript abundance following 4.5S RNA mutation was covS-dependent.
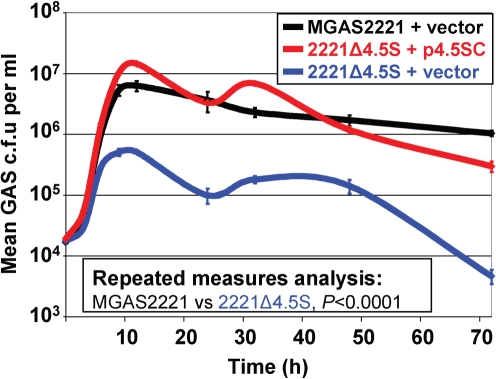
Contribution of the 4.5S RNA to GAS growth and persistence in human saliva
The main portal of GAS transmission is the upper respiratory tract (Hamburger & Robertson, 1948; Shelburne et al., 2005a), and dissemination commonly occurs through dispersal of aerosolized saliva (Hamburger & Robertson, 1948; Katzenell et al., 2001). To investigate the contribution of the 4.5S RNA to the ability of GAS to cause upper respiratory tract infections we used growth and persistence in human saliva as a surrogate for growth and persistence in the upper respiratory tract, as previously described (Shelburne et al., 2005b; Trevino et al., 2009). GAS were inoculated into filter-sterilized aliquots of human saliva and over time the number of GAS c.f.u. per ml of saliva was assayed (Fig. 4). While growth was comparable between all strains during the first 5 h, after this time growth of strain 2221Δ4.5S containing empty vector slowed considerably, such that between 9 and 72 h ∼10-fold higher concentrations of GAS c.f.u. were recovered from the parental and complemented mutant strain. As with growth in THY broth, GAS chain length was not appreciably altered by 4.5S RNA mutation (unpublished data). The data are consistent with the 4.5S RNA contributing to the ability of GAS to grow and persist in human saliva.
Fig. 4.
Mutation of the 4.5S RNA reduces the ability of GAS to grow and persist in human saliva. GAS strains were grown in human saliva and samples recovered over time for enumeration of GAS c.f.u.. C.f.u. were determined after serially diluting, plating on blood agar plates, and incubating at 37 °C overnight. The experiment was repeated three times and means±sem are plotted.
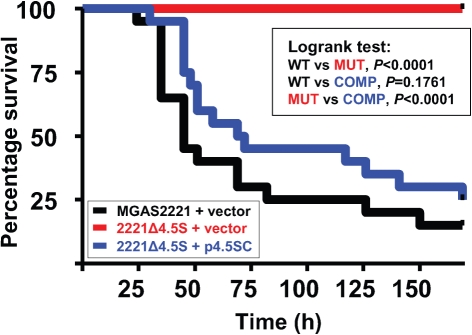
Use of a mouse bacteraemia model of infection to determine the contribution of the 4.5S RNA to GAS virulence
To test whether the 4.5S RNA was required for GAS to cause disease we compared parental, mutant and complemented mutant strains in a well-described mouse bacteraemia model of infection (Sumby et al., 2006). Groups of 20 mice were infected intraperitoneally with 2.5×107 GAS c.f.u., and animal survival was monitored over time. Importantly, all animals infected with mutant strain 2221Δ4.5S survived infection, whereas only 15 % of animals infected with parental strain MGAS2221, and 25 % of animals infected with complemented strain 2221Δ4.5S p4.5S, survived (Fig. 5). Thus, our data show that the 4.5S RNA is critical for GAS to cause lethal infections in a murine bacteraemia model of infection.
Fig. 5.
Mutation of the 4.5S RNA severely attenuates GAS pathogenesis in a mouse bacteraemia model of infection. Shown is a Kaplan–Meier survival plot of groups of 20 mice infected via intraperitoneal injection with 2.5×107 c.f.u. of parental strain MGAS2221 (+vector; WT, black), isogenic mutant 2221Δ4.5S (+vector; MUT, red), or complemented mutant 2221Δ4.5S (+p4.5S; COMP, blue). Significance was tested using the logrank test.
Use of a mouse soft tissue infection model to determine the contribution of the 4.5S RNA to GAS virulence
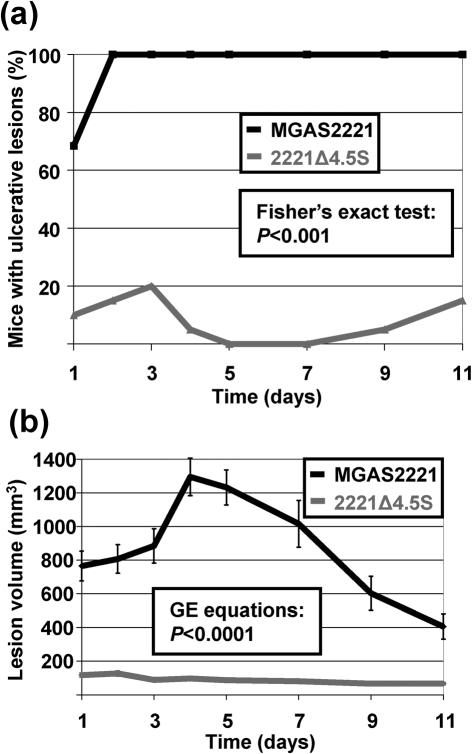
As Rosch et al. (2008) utilized a mouse soft tissue infection model to compare an ffh mutant to its parental strain, we sought to use the same mouse model to identify (a) whether there were phenotypic differences between ffh and 4.5S RNA mutant strains in this animal model when compared to their parental strain, and (b) whether the 4.5S RNA contributes to GAS virulence during soft tissue infection. Groups of 20 mice were infected subcutaneously with 1×107 c.f.u., and the skin lesions that formed over time were monitored. All MGAS2221-infected mice developed ulcerative lesions by day 2 of the infection (Fig. 6a). In contrast, no more than 20 % of animals infected with strain 2221Δ4.5S developed ulcerative lesions at any time during the experiment. In addition to differences in the rate of ulcerative lesion formation, animals also differed significantly in the size of the lesions that formed, with significantly greater lesion volumes observed in animals infected with wild-type GAS (Fig. 6b). Thus, similar to the protein component of the SRP, the 4.5S RNA significantly enhances virulence in a murine soft tissue infection model.
Fig. 6.
Mutation of the 4.5S RNA severely attenuates GAS pathogenesis in a mouse model of soft tissue infection. Groups of 20 mice were infected subcutaneously with 1×107 GAS c.f.u. and monitored over time. (a) Ulceration of the skin lesion that formed at the infection site occurred at a significantly lower rate in animals infected with mutant strain 2221Δ4.5S compared to those infected with parental strain MGAS2221. (b) Lesion size was significantly larger in animals infected with parental strain MGAS2221 than those infected with strain 2221Δ4.5S. Error bars represent±sem.
DISCUSSION
The 4.5S RNA accelerates binding between Ffh and its membrane-associated receptor FtsY, targeting Ffh-bound nascent polypeptides for secretion by the co-translational pathway (Bradshaw et al., 2009). Here, we present data showing that the 4.5S RNA-encoding gene is not essential in GAS and by extension, and in support of a study that investigated the protein component of the SRP (Rosch et al., 2008), that the SRP pathway is not essential. While the 4.5S RNA gene is not essential, mutation of this gene results in pleiotropic effects, including an approximately 50 % increase in doubling time during growth in nutrient-rich liquid media (Fig. 1c); a reduction in the concentration of secreted virulence factors Sic, SLO, Spd3 and SpeB (Fig. 2); altered gene transcript levels (Fig. 3a); a significant reduction in c.f.u. during growth in human saliva (an ex vivo upper respiratory tract infection model; Fig. 4); and a reduction in virulence as assessed by mouse models of bacteraemia and soft tissue infection (Figs 5 and 6). Given these diverse phenotypes it is apparent that 4.5S RNA function is central to many processes in GAS.
Mutation of the gene encoding the protein component of the SRP pathway (ffh) reduced secretion of the potent haemolysin SLO (Rosch et al., 2008). It was therefore not surprising that a similar reduction in SLO secretion was observed following mutation of the 4.5S RNA gene (Fig. 2). The fact that the immune evasion protein Sic, the DNase Spd3 and the cysteine protease SpeB were also identified at a lower concentration in supernatants of 2221Δ4.5S compared to MGAS2221 is consistent with 4.5S mutation detrimentally affecting the secretion of at least a subset of secreted proteins (Fig. 2). A reduction in SpeB secretion was not observed following mutation of ffh. Rather, SpeB levels increased following ffh mutation, at least in part due to the earlier growth phase expression of SpeB (Rosch et al., 2008). It should be noted that as a direct comparison between parental, ffh and 4.5S RNA mutant strains was not performed, it is possible that subtle differences in the experimental conditions between our dataset and that of Rosch and colleagues account for the observed differences.
We observed that six out of the eight genes tested by quantitative RT-PCR differed more than twofold between the parental and isogenic 4.5S RNA mutant strain (Fig. 3a). Furthermore, for the four genes tested, we identified that the direction of change in transcript abundance following 4.5S RNA mutation was dependent on a functional covS gene (Fig. 3b). Thus, covS is epistatic to the 4.5S RNA with respect to the remodelling of transcript abundance of the tested genes. These data are consistent with disruption of the SRP pathway leading to the constitutive activation of CovS or the activation/inhibition of a CovS-regulated regulator.
While the initial growth of strain 2221Δ4.5S in human saliva mirrored that of the parental and complemented mutant strains, by the 6 h time point growth slowed significantly (Fig. 4). Thus, by 9 h there was an order of magnitude fewer c.f.u. in the 2221Δ4.5S-inoculated saliva samples. The 2221Δ4.5S growth curve in saliva mirrors that observed with a malE mutant strain (Shelburne et al., 2006). The malE gene encodes a cell-surface maltodextrin-binding lipoprotein that contributes to the ability of GAS to utilize complex carbohydrates. Whether MalE requires the SRP pathway for secretion, thus explaining the growth defect of the 4.5S RNA mutant strain in human saliva, or whether the attenuated secretion of other proteins is responsible for the saliva growth phenotype (e.g. phosphotransferase system components, membrane-localized importers and/or exporters), remains to be investigated.
Our data show that the 4.5S RNA is essential for GAS virulence in a mouse bacteraemia model of infection (Fig. 5). In addition, we show that the 4.5S RNA plays a major role in the ability of GAS to cause disease in a mouse soft tissue infection model (Fig. 6), which is consistent with data from the study of the protein component of the SRP (Rosch et al., 2008). Due to the severity of the virulence defect of strain 2221Δ4.5S we believe that the reduced secretion of no single virulence factor is responsible. Rather, a combination of the reduced secretion of multiple virulence factors (Fig. 2), an altered transcriptome (Fig. 3a) and a reduced growth rate (Figs 1c and 4) all contribute to the virulence attenuation of 2221Δ4.5S.
The fact that GAS survives disruption of the SRP pathway implies that an alternative mechanism exists for the secretion of at least a subset of SRP-secreted proteins. Many bacteria use the twin-arginine translocation (Tat) pathway to transport folded proteins across the membrane (De Buck et al., 2008). However, this pathway is unlikely to be present in GAS due to the absence of homologues of conserved components of this pathway (Dilks et al., 2003). A possible alternative to the GAS SRP pathway may involve the YidC/Oxa1/Alb3 family of proteins (Pohlschroder et al., 2005). Members of the YidC/Oxa1/Alb3 family of proteins are involved in the insertion of proteins into the membranes of bacteria, mitochondria and chloroplasts, respectively. The mechanism by which these proteins promote insertion of substrate proteins into membranes is unknown. Similar to most Gram-positive bacteria, GAS encode two YidC proteins, YidC1 and YidC2. While not studied in GAS, YidC1 and YidC2 have been studied in S. mutans, where deletion of yidC1 caused only a minor growth phenotype while deletion of yidC2 resulted in a major stress phenotype similar to an SRP mutant strain (Hasona et al., 2005). Disruption of both yidC2 and components of the SRP pathway gave S. mutans strains that were severely impaired in growth and stress tolerance. Thus, the S. mutans YidC2 and SRP pathways appear to serve separate yet overlapping roles in protein export (Funes et al., 2009). Given the importance of secreted and cell-surface-associated proteins in GAS virulence, further study of the SRP and YidC pathways may open new avenues for development of novel treatment and/or preventive regimes based upon the inhibition of protein secretion.
Acknowledgments
This research was funded in part by award number R21AI078159 from the National Institute of Allergy and Infectious Diseases (to P. S.), and a Research Scholar Award from The Methodist Hospital Research Institute (to P. S.). We thank Kathryn Pflughoeft (UT Health Science Center – Houston) for critical reading of this manuscript.
Abbreviations
GAS, group A Streptococcus (Streptococcus pyogenes)
SLO, streptolysin O
SRP, signal recognition particle
Footnotes
A supplementary table of primers and a supplementary figure are available with the online version of this paper.
References
- Bernish, B. & van de Rijn, I. (1999). Characterization of a two-component system in Streptococcus pyogenes which is involved in regulation of hyaluronic acid production. J Biol Chem 274, 4786–4793. [DOI] [PubMed] [Google Scholar]
- Binks, M. J., Fernie-King, B. A., Seilly, D. J., Lachmann, P. J. & Sriprakash, K. S. (2005). Attribution of the various inhibitory actions of the streptococcal inhibitor of complement (SIC) to regions within the molecule. J Biol Chem 280, 20120–20125. [DOI] [PubMed] [Google Scholar]
- Bradshaw, N. & Walter, P. (2007). The signal recognition particle (SRP) RNA links conformational changes in the SRP to protein targeting. Mol Biol Cell 18, 2728–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw, N., Neher, S. B., Booth, D. S. & Walter, P. (2009). Signal sequences activate the catalytic switch of SRP RNA. Science 323, 127–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffin, D. O. & Rubens, C. E. (1998). Blue/white screening of recombinant plasmids in Gram-positive bacteria by interruption of alkaline phosphatase gene (phoZ) expression. Gene 219, 91–99. [DOI] [PubMed] [Google Scholar]
- Cunningham, M. W. (2000). Pathogenesis of group A streptococcal infections. Clin Microbiol Rev 13, 470–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Buck, E., Lammertyn, E. & Anné, J. (2008). The importance of the twin-arginine translocation pathway for bacterial virulence. Trends Microbiol 16, 442–453. [DOI] [PubMed] [Google Scholar]
- Dilks, K., Rose, R. W., Hartmann, E. & Pohlschroder, M. (2003). Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J Bacteriol 185, 1478–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egesten, A., Olin, A. I., Linge, H. M., Yadav, M., Morgelin, M., Karlsson, A. & Collin, M. (2009). SpeB of Streptococcus pyogenes differentially modulates antibacterial and receptor activating properties of human chemokines. PLoS One 4, e4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engleberg, N. C., Heath, A., Miller, A., Rivera, C. & DiRita, V. J. (2001). Spontaneous mutations in the CsrRS two-component regulatory system of Streptococcus pyogenes result in enhanced virulence in a murine model of skin and soft tissue infection. J Infect Dis 183, 1043–1054. [DOI] [PubMed] [Google Scholar]
- Federle, M. J., McIver, K. S. & Scott, J. R. (1999). A response regulator that represses transcription of several virulence operons in the group A streptococcus. J Bacteriol 181, 3649–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funes, S., Hasona, A., Bauerschmitt, H., Grubbauer, C., Kauff, F., Collins, R., Crowley, P. J., Palmer, S. R., Brady, L. J. & Herrmann, J. M. (2009). Independent gene duplications of the YidC/Oxa/Alb3 family enabled a specialized cotranslational function. Proc Natl Acad Sci U S A 106, 6656–6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, M. R., Virtaneva, K., Porcella, S. F., Gardner, D. J., Long, R. D., Welty, D. M., Barry, W. T., Johnson, C. A. & Parkins, L. D. (2006). Analysis of the transcriptome of group A Streptococcus in mouse soft tissue infection. Am J Pathol 169, 927–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryllos, I., Tran-Winkler, H. J., Cheng, M. F., Chung, H., Bolcome, R., III, Lu, W., Lehrer, R. I. & Wessels, M. R. (2008). Induction of group A Streptococcus virulence by a human antimicrobial peptide. Proc Natl Acad Sci U S A 105, 16755–16760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger, M., Jr & Robertson, O. H. (1948). Expulsion of group A hemolytic streptococci in droplets and droplet nuclei by sneezing, coughing and talking. Am J Med 4, 690–701. [DOI] [PubMed] [Google Scholar]
- Hasona, A., Crowley, P. J., Levesque, C. M., Mair, R. W., Cvitkovitch, D. G., Bleiweis, A. S. & Brady, L. J. (2005). Streptococcal viability and diminished stress tolerance in mutants lacking the signal recognition particle pathway or YidC2. Proc Natl Acad Sci U S A 102, 17466–17471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasona, A., Zuobi-Hasona, K., Crowley, P. J., Abranches, J., Ruelf, M. A., Bleiweis, A. S. & Brady, L. J. (2007). Membrane composition changes and physiological adaptation by Streptococcus mutans signal recognition particle pathway mutants. J Bacteriol 189, 1219–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskovits, A. A., Bochkareva, E. S. & Bibi, E. (2000). New prospects in studying the bacterial signal recognition particle pathway. Mol Microbiol 38, 927–939. [DOI] [PubMed] [Google Scholar]
- Katzenell, U., Shemer, J. & Bar-Dayan, Y. (2001). Streptococcal contamination of food: an unusual cause of epidemic pharyngitis. Epidemiol Infect 127, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer, B. H., van der Kraan, M., Crowley, P. J., Hamilton, I. R., Brady, L. J. & Bleiweis, A. S. (2001). Characterization of the sat operon in Streptococcus mutans: evidence for a role of Ffh in acid tolerance. J Bacteriol 183, 2543–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, J. C. & Wessels, M. R. (1998). Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol Microbiol 30, 209–219. [DOI] [PubMed] [Google Scholar]
- Lukomski, S., Nakashima, K., Abdi, I., Cipriano, V. J., Ireland, R. M., Reid, S. D., Adams, G. G. & Musser, J. M. (2000). Identification and characterization of the scl gene encoding a group A Streptococcus extracellular protein virulence factor with similarity to human collagen. Infect Immun 68, 6542–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehl, M. A., Pinkner, J. S., Anderson, P. J., Hultgren, S. J. & Caparon, M. G. (2005). A novel endogenous inhibitor of the secreted streptococcal NAD-glycohydrolase. PLoS Pathog 1, e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, K., Imai, Y., Nakamura, A. & Yamane, K. (1992). Small cytoplasmic RNA of Bacillus subtilis: functional relationship with human signal recognition particle 7S RNA and Escherichia coli 4.5S RNA. J Bacteriol 174, 2185–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyberg, P., Rasmussen, M. & Bjorck, L. (2004). α2-Macroglobulin-proteinase complexes protect Streptococcus pyogenes from killing by the antimicrobial peptide LL-37. J Biol Chem 279, 52820–52823. [DOI] [PubMed] [Google Scholar]
- Olsen, R. J., Shelburne, S. A. & Musser, J. M. (2009). Molecular mechanisms underlying group A streptococcal pathogenesis. Cell Microbiol 11, 1–12. [DOI] [PubMed] [Google Scholar]
- Perez, N., Trevino, J., Liu, Z., Ho, S. C. M., Babitzke, P. & Sumby, P. (2009). A genome-wide analysis of small regulatory RNAs in the human pathogen group A Streptococcus. PLoS One 4, e7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlschroder, M., Hartmann, E., Hand, N. J., Dilks, K. & Haddad, A. (2005). Diversity and evolution of protein translocation. Annu Rev Microbiol 59, 91–111. [DOI] [PubMed] [Google Scholar]
- Powers, T. & Walter, P. (1995). Reciprocal stimulation of GTP hydrolysis by two directly interacting GTPases. Science 269, 1422–1424. [DOI] [PubMed] [Google Scholar]
- Reid, S. D., Virtaneva, K. & Musser, J. M. (2003). Group A Streptococcus vaccine research: historical synopsis and new insights. In Bacterial Vaccines, pp. 155–173. Edited by R. W. Ellis & B. R. Brodeur. Georgetown, TX: Landes Bioscience.
- Rosch, J. W. & Caparon, M. G. (2005). The ExPortal: an organelle dedicated to the biogenesis of secreted proteins in Streptococcus pyogenes. Mol Microbiol 58, 959–968. [DOI] [PubMed] [Google Scholar]
- Rosch, J. W., Hsu, F. F. & Caparon, M. G. (2007). Anionic lipids enriched at the ExPortal of Streptococcus pyogenes. J Bacteriol 189, 801–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosch, J. W., Vega, L. A., Beyer, J. M., Lin, A. & Caparon, M. G. (2008). The signal recognition particle pathway is required for virulence in Streptococcus pyogenes. Infect Immun 76, 2612–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, H. H. & Sriskandan, S. (2008). Superantigens SPEA and SMEZ do not affect secretome expression in Streptococcus pyogenes. Microb Pathog 44, 537–543. [DOI] [PubMed] [Google Scholar]
- Shelburne, S. A., III, Granville, C., Tokuyama, M., Sitkiewicz, I., Patel, P. & Musser, J. M. (2005a). Growth characteristics of and virulence factor production by group A Streptococcus during cultivation in human saliva. Infect Immun 73, 4723–4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne, S. A., III, Sumby, P., Sitkiewicz, I., Granville, C., DeLeo, F. R. & Musser, J. M. (2005b). Central role of a bacterial two-component gene regulatory system of previously unknown function in pathogen persistence in human saliva. Proc Natl Acad Sci U S A 102, 16037–16042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne, S. A., III, Sumby, P., Sitkiewicz, I., Okorafor, N., Granville, C., Patel, P., Voyich, J., Hull, R., DeLeo, F. R. & Musser, J. M. (2006). Maltodextrin utilization plays a key role in the ability of group A Streptococcus to colonize the oropharynx. Infect Immun 74, 4605–4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne, S. A., III, Keith, D., Horstmann, N., Sumby, P., Davenport, M. T., Graviss, E. A., Brennan, R. G. & Musser, J. M. (2008). A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc Natl Acad Sci U S A 105, 1698–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struck, J. C., Hartmann, R. K., Toschka, H. Y. & Erdmann, V. A. (1989). Transcription and processing of Bacillus subtilis small cytoplasmic RNA. Mol Gen Genet 215, 478–482. [DOI] [PubMed] [Google Scholar]
- Sumby, P., Barbian, K. D., Gardner, D. J., Whitney, A. R., Welty, D. M., Long, R. D., Bailey, J. R., Parnell, M. J., Hoe, N. P. & other authors (2005a). Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proc Natl Acad Sci U S A 102, 1679–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumby, P., Porcella, S. F., Madrigal, A. G., Barbian, K. D., Virtaneva, K., Ricklefs, S. M., Sturdevant, D. E., Graham, M. R., Vuopio-Varkila, J. & other authors (2005b). Evolutionary origin and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J Infect Dis 192, 771–782. [DOI] [PubMed] [Google Scholar]
- Sumby, P., Whitney, A. R., Graviss, E. A., DeLeo, F. R. & Musser, J. M. (2006). Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog 2, e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumby, P., Zhang, S., Whitney, A. R., Falugi, F., Grandi, G., Graviss, E. A., Deleo, F. R. & Musser, J. M. (2008). A chemokine-degrading extracellular protease made by group A Streptococcus alters pathogenesis by enhancing evasion of the innate immune response. Infect Immun 76, 978–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmer, A. M., Timmer, J. C., Pence, M. A., Hsu, L. C., Ghochani, M., Frey, T. G., Karin, M., Salvesen, G. S. & Nizet, V. (2009). Streptolysin O promotes group A Streptococcus immune evasion by accelerated macrophage apoptosis. J Biol Chem 284, 862–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevino, J., Perez, N., Ramirez-Pena, E., Liu, Z., Shelburne, S. A., III, Musser, J. M. & Sumby, P. (2009). CovS simultaneously activates and inhibits the CovR-mediated repression of distinct subsets of group A Streptococcus virulence factor-encoding genes. Infect Immun 77, 3141–3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtaneva, K., Graham, M. R., Porcella, S. F., Hoe, N. P., Su, H., Graviss, E. A., Gardner, T. J., Allison, J. E., Lemon, W. J. & other authors (2003). Group A Streptococcus gene expression in humans and cynomolgus macaques with acute pharyngitis. Infect Immun 71, 2199–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane, K., Bunai, K. & Kakeshita, H. (2004). Protein traffic for secretion and related machinery of Bacillus subtilis. Biosci Biotechnol Biochem 68, 2007–2023. [DOI] [PubMed] [Google Scholar]
- Zanen, G., Antelmann, H., Meima, R., Jongbloed, J. D., Kolkman, M., Hecker, M., van Dijl, J. M. & Quax, W. J. (2006). Proteomic dissection of potential signal recognition particle dependence in protein secretion by Bacillus subtilis. Proteomics 6, 3636–3648. [DOI] [PubMed] [Google Scholar]