Abstract
Histone deacetylases (HDAC) 1 and 2 are highly similar enzymes that help regulate chromatin structure as the core catalytic components of corepressor complexes. Although tissue-specific deletion of HDAC1 and HDAC2 has demonstrated functional redundancy, germ-line deletion of HDAC1 in the mouse causes early embryonic lethality, whereas HDAC2 does not. To address the unique requirement for HDAC1 in early embryogenesis we have generated conditional knockout embryonic stem (ES) cells in which HDAC1 or HDAC2 genes can be inactivated. Deletion of HDAC1, but not HDAC2, causes a significant reduction in the HDAC activity of Sin3A, NuRD, and CoREST corepressor complexes. This reduced corepressor activity results in a specific 1.6-fold increase in histone H3 K56 acetylation (H3K56Ac), thus providing genetic evidence that H3K56Ac is a substrate of HDAC1. In culture, ES cell proliferation was unaffected by loss of either HDAC1 or HDAC2. Rather, we find that loss of HDAC1 affects ES cell differentiation. ES cells lacking either HDAC1 or HDAC2 were capable of forming embryoid bodies (EBs), which stimulates differentiation into the three primary germ layers. However, HDAC1-deficient EBs were significantly smaller, showed spontaneous rhythmic contraction, and increased expression of both cardiomyocyte and neuronal markers. In summary, our genetic study of HDAC1 and HDAC2 in ES cells, which mimic the embryonic epiblast, has identified a unique requirement for HDAC1 in the optimal activity of HDAC1/2 corepressor complexes and cell fate determination during differentiation.
Keywords: corepressor, acetylation, deacetylation, chromatin
Class I histone deacetylases (HDAC1, -2, -3, and -8) are highly conserved enzymes present in the nucleus of all cells, where they help modulate levels of gene expression (1). The best characterized substrates of HDACs are the N-terminal tails of the core histones H2A, H2B, H3 and H4. Deacetylation of histone tails results in a “tightening” of chromatin, due to the electrostatic potential of unacetylated lysine residues to promote internucleosomal interactions (2, 3) and the loss of a binding site for components of the transcriptional machinery containing a bromodomain, e.g., TAFII1 (4).
Among the class I HDACs, HDAC1 and HDAC2 are the most similar (83% amino acid identity), sharing an almost identical catalytic core domain and a conserved C-terminal tail (5). In mammalian cells HDAC1 and HDAC2 interact together (6) to form the catalytic core of a number of higher-order complexes including Sin3A, NuRD, CoREST, and NODE (reviewed in ref. 1). These complexes are then targeted to chromatin by sequence-specific (often cell-specific) transcription factors to repress transcription in cooperation with other chromatin modifiers, such as the lysine-specific demethylase, LSD1, in the CoREST complex (7–9). As part of these multiprotein complexes, the activity of HDAC1 and HDAC2 has been implicated in the regulation of cell cycle progression by tumor suppressors (10–12), differentiation (13, 14), cellular aging (15), and cancer (16). Indeed, a variety of HDAC inhibitors that target both HDAC1 and -2 are currently being tested in the clinic as potential anticancer therapeutics (17).
Mouse genetics has demonstrated an essential role in embryogenesis for HDAC1 (18, 19) and many components of HDAC1/2 complexes, including Sin3A (20), SDS3 (21), MBD3 (22), and LSD1 (23, 24). Germ-line deletion of HDAC1 results in early embryonic lethality around embryonic day (e)10.5, although aberrant development occurs as early as e7.5. In contrast to these early embryonic phenotypes, constitutive HDAC2 knockout mice survive embryogenesis and either die shortly after birth in one model (19) or survive to adulthood in others (25–27), albeit at reduced Mendelian frequencies. In a number of cell types, deletion of both HDAC1 and HDAC2 is required to generate a phenotype (14, 19, 28). This result suggests that the activity of HDAC1 and HDAC2 is mostly redundant, with the requirement for both HDAC1 and HDAC2 occurring only at certain key developmental periods, such as gastrulation. To investigate the essential role of HDAC1 during early embryogenesis we have compared the biochemical, proliferative, and differentiation properties of HDAC1 and HDAC2 conditional knockout embryonic stem (ES) cells. ES cells are the in vitro counterpart of epiblast cells of the early postimplantation embryo and their differentiation mimics many of the changes in gene expression associated with embryonic development (29). We find that loss of HDAC1, but not HDAC2, reduces the level of HDAC activity associated with HDAC1/2 complexes and leads to the enhanced differentiation of embryoid bodies.
Results
Generation of Conditional Knockout ES Cell Lines for HDAC1 and HDAC2.
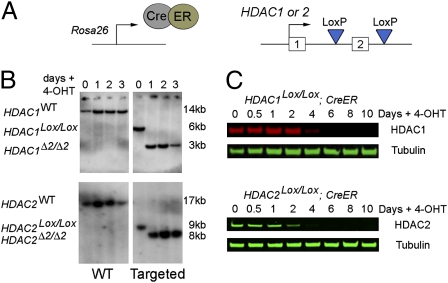
Beginning with an E14 ES cell line expressing an inducible Cre/Estrogen Receptor (CreER) construct from the endogenous ROSA26 locus (30), we used homologous recombination to produce HDAC1Lox/Lox; CreER and HDAC2Lox/Lox; CreER cell lines in which exon 2 of each gene is flanked by LoxP sites (see Fig. 1A and Figs. S1 and S2 for detailed methods). Induction of Cre activity by addition of 4-hydroxy tamoxifen (OHT) to the growth media resulted in complete recombination of each allele and deletion of exon 2 (HDAC1Δ2/Δ2 or HDAC2 Δ2/Δ2) within 24 h (Fig.1B). Loss of exon 2 disrupts the ORF of both HDAC1 and HDAC2 such that a premature stop codon is introduced in exon 3. Following deletion of exon 2 a further 4–5 days of culture are required before HDAC1 and HDAC2 protein levels are reduced below 10% of those of control cells (Fig. 1C), suggesting that a combination of a long-lived mRNA and recruitment into a stable protein complex results in relatively slow protein turnover. In the subsequent experiments, HDAC1Lox/Lox; CreER and HDAC2Lox/Lox; CreER cells 10–24 days postrecombination (hence referred to as HDAC1Δ2/Δ2 and HDAC2Δ2/Δ2) were used to assess their biochemical and growth properties.
Fig. 1.
Generation of HDAC1 and HDAC2 conditional knockout ES cell lines. (A) An E14 ES cell line constitutively expressing a Cre/Estrogen Receptor (CreER) fusion from the ROSA26 locus was used to generate homozygous conditional knockout alleles for HDAC1 and HDAC2. Both copies of exon 2 were flanked by LoxP sites, using consecutive rounds of gene targeting. (B) Southern blots showing wild-type and exon 2 targeted HDAC1Lox/Lox and HDAC2Lox/Lox loci. Addition of 4-hydroxy tamoxifen (OHT) activates the CreER fusion and induces deletion of exon2 (HDAC1Δ2/Δ2, HDAC2Δ2/Δ2) in ES cells; >95% recombination is observed after 24 h. (C) Quantitative Western blot shows ligand-inducible deletion of HDAC1 and -2 protein in whole-cell extracts from HDAC1Lox/Lox and HDAC2Lox/Lox ES cells, respectively. Cells were cultured for up to 10 days (0–3 days in the presence of OHT). α-Tubulin was used to normalize protein loading. Blots were visualized and quantitated using a LiCOR scanner. Data are representative of three independently tested clones for each genotype.
Reduced HDAC Activity Associated with HDAC1/2 Complexes in the Absence of HDAC1.
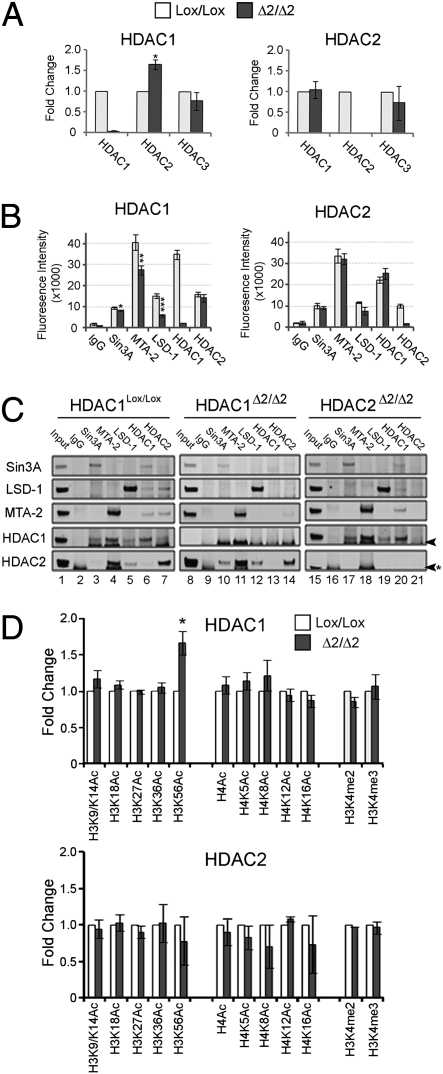
Consistent with previous reports (18, 31), deletion of HDAC1 results in an increased level of HDAC2 protein (Fig. 2A). However, no increase in HDAC2 mRNA is detected, suggesting that increased protein levels may occur from changes in HDAC2 translation and/or degradation in the absence of HDAC1. Interestingly, there is no change in HDAC1 protein levels in the absence of HDAC2 (Fig. 2A). Most, if not all cellular HDAC1 and -2 is associated with higher-order protein complexes in the nucleus (1). We used coimmunoprecipitation of individual components of the Sin3A, NuRD, and CoREST complexes to assess the associated HDAC activity and their integrity in cells lacking either HDAC1 or HDAC2. In the absence of HDAC1 we observe a decrease in the deacetylase activity associated with each of the Sin3A, NuRD, and CoREST complexes, with the largest reduction in the CoREST complex (Fig. 2B). This decrease in activity occurs despite the fact that we detect increased amounts of HDAC2 in each of these same complexes (Fig. 2C, compare HDAC2 amounts in lanes 3–5 with those in lanes 10–12). The increased incorporation of HDAC2 into HDAC1/2 complexes in the absence of HDAC1 may account for the increased abundance of HDAC2 protein (Fig. 2A Left). In contrast, there is no significant alteration in the amount of deacetylase activity associated with the three HDAC1/2 complexes in the absence of HDAC2 (Fig. 2B). These data argue that loss of HDAC1, but not HDAC2, causes a reduction in the HDAC activity of HDAC1/2-containing complexes. However, in the absence of either enzyme, deacetylase activity is still measurable and a physical association still remains, suggesting that the integrity of each complex is retained.
Fig. 2.
Loss of HDAC1 results in decreased deacetylase activity associated with HDAC1/2 complexes and an increase H3K56Ac. (A) Quantitative Western blot of HDAC1, -2, and -3 obtained from nuclear extracts of the indicated cell types. Protein levels were quantitated using a LiCOR scanner and normalized to the level of α-tubulin. Data represent independent experiments, using three independent clones. (*, P < 0.05; Student's t test). Mean values (n = 3) ± SEM are plotted. (B) Specific antisera to the indicated proteins were used to coimmunoprecipitate Sin3A, NuRD (αMTA-2), and CoREST (α-LSD1) complexes from HDAC1Lox/Lox, HDAC1Δ2/Δ2, HDAC2Lox/Lox, and HDAC2Δ2/Δ2 ES cells. The amount of associated deacetylase activity was measured using a commercially available kit (*, P < 0.05; **, P < 0.01; ***, P < 0.001; Student's t test). Mean values (n = 3) ± SEM are plotted. (C) The remaining material from the coimmunoprecipitation was run on an SDS/PAGE gel and then analyzed by Western blotting using the indicated antibodies. Arrow indicates a nonspecific band. (D) The acetylation status of core histones was detected using quantitative Western blotting. Histones were acid extracted from HDAC1Lox/Lox, HDAC1Δ2/Δ2, HDAC2Lox/Lox, and HDAC2Δ2/Δ2 clones. The signal of specific acetylated lysines was normalized to the total amount of H3 or H4 as appropriate and quantitated using a LiCOR scanner (*, P < 0.01; Student's t test). Mean values (n = 3) ± SEM are plotted. Data represent independent experiments using three different clones of each genotype.
We next examined the global acetylation levels of histones H3 and H4 in the absence of either HDAC1 or HDAC2 (Fig. 2D). We detected a slight increase in the acetylation status of H3K9/14, H4K5, and H4K8 in the absence of HDAC1. However, the only significant difference in HDAC1Δ2/Δ2 cells was a 1.6-fold increase in the level of H3K56 acetylation (H3K56Ac), a modification associated with DNA damage (32, 33), nucleosome assembly (32), and the activity of stem cell factors (34) in higher eukaryotes. This demonstration that H3K56Ac is a substrate for HDAC1 has not been previously characterized. Global histone acetylation levels were unchanged by loss of HDAC2 (Fig. 2D Lower), consistent with an unchanged level of deacetylase activity associated with HDAC1/2 complexes in cells lacking HDAC2 protein. Treatment of HDAC1Lox/Lox ES cells with trichostatin A (TSA) for 12 h produces a 7-fold increase in H3K56Ac levels, compared to the 1.6-fold change associated with loss of HDAC1, suggesting that additional class I and class II HDACs participate in its regulation (Fig. S3A). In contrast to H3K56Ac, which occurs in the histone core, the acetylation status of lysines within the tails of histones H3 and H4 (H3K9/K14 and H4K5/K8/K12/K16) was changed by <1.5-fold in response to TSA. However, TSA treatment produces a robust 5-fold increase in acetylation of these same residues in mouse embryo fibroblasts (MEFs) (Fig. S3B), suggesting that histone tails are hyperacetylated in ES cells, and thus potentially masks a detectable increase in histone acetylation in HDAC1Δ2/Δ2 cells.
Proliferation and Differentiation Capacity of ES Cells Is Not Inhibited by Loss of HDAC1 or HDAC2.
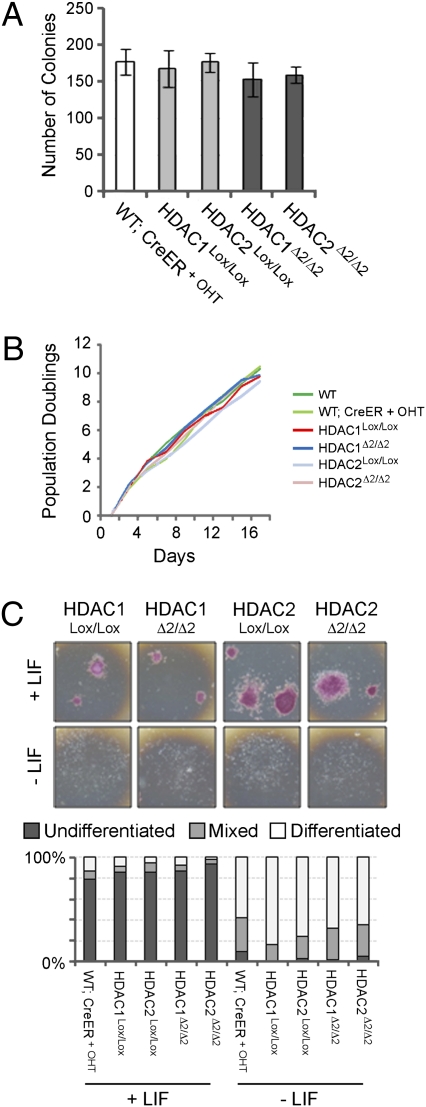
Because HDAC1 has been implicated in cell cycle progression (18, 35, 36), we compared the growth ability of HDAC1Δ2/Δ2 and HDAC2Δ2/Δ2 cells to that of control cells. We found that there was no effect of deleting HDAC1 or HDAC2 on colony formation when plating ES cells at low density (Fig. 3A) or on their population doubling time (Fig. 3B). Loss of MBD3, a central component of the NuRD complex (37), or treatment with HDAC inhibitors (38) has been demonstrated to inhibit ES cell differentiation. We therefore assessed the capacity of HDAC1Δ2/Δ2 and HDAC2Δ2/Δ2 cells to differentiate when grown in the absence of leukemia inhibitory factor (LIF) compared to controls (Fig. 3C). The indicated ES cells were grown in the presence and absence of LIF for 6 days and then assayed for alkaline phosphatase activity, a stem cell marker. WT-CreER cells treated with 4-OHT, untreated homozygous targeted cells (HDAC1Lox/Lox and HDAC2Lox/Lox), and homozygous deleted cells (HDAC1Δ2/Δ2 and HDAC2Δ2/Δ2) lacking HDAC1 and HDAC2, respectively, all showed comparable levels of differentiation. These data demonstrate that the growth characteristics of ES cells lacking either HDAC1 or HDAC2 are similar to those of wild-type cells.
Fig. 3.
Conditional deletion of HDAC1 or -2 does not inhibit the growth or differentiation capacity of ES cells. (A) Colony formation assay: mean values and SDs of cells of the indicated genotype plated at low density and cultured for 8 days, before counting. Mean values (n = 3) ± SEM are plotted. (B) The population doubling times of the indicated cell types were calculated by counting and then replating cells every 2 days for an 18-day period. (C) Differentiation potential. ES cells of the type indicated were plated at low density in the presence (+) or absence (−) of LIF and cultured for 6 days before staining for the presence of alkaline phosphatase. Colonies were scored as undifferentiated (purple), differentiated (white), or mixed. Data represent three independent experiments using two individual clones of each genotype.
Loss of HDAC1, but not HDAC2, Causes Enhanced Differentiation of Embryoid Bodies.
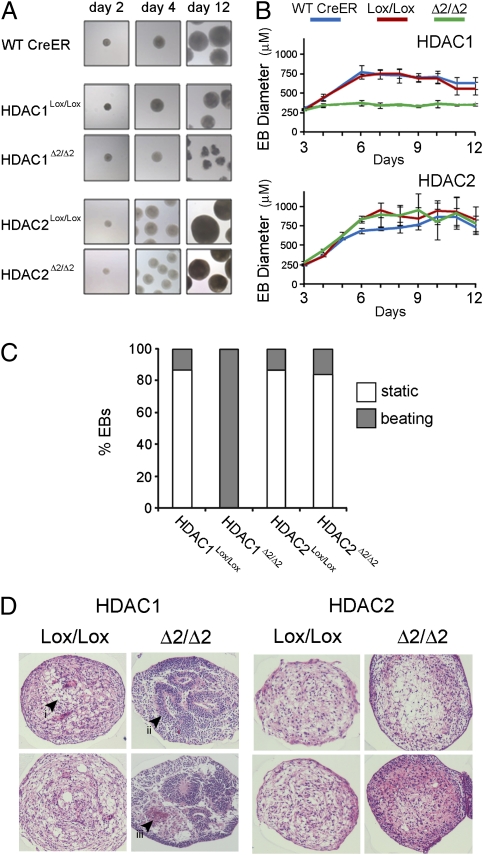
To examine the differentiation process of HDAC knockout cells in greater detail HDAC1Δ2/Δ2 and HDAC2Δ2/Δ2 ES cells were used to generate embryoid bodies (EB), which recapitulate many of the changes associated with gastrulation, including generation of the three primary germ layers, ectoderm, endoderm, and mesoderm. It is presumably during gastrulation, or just before, that HDAC1 null embryos begin to accrue developmental defects that are apparent phenotypically at e7.5 and beyond (18, 19). ES cells lacking either HDAC1 or HDAC2 were able to form EBs over a 2-day period, using the hanging drop method (Fig. 4A, day 2). Continued culture revealed that EBs lacking HDAC1 become irregular, rather than uniformly spherical, and are much reduced in size compared to EBs derived from control cells (Fig. 4A, day 12, and 4B). By contrast, EBs lacking HDAC2 (HDAC2Δ2/Δ2) were uniformly spherical and grew to a similar size to wild-type and HDAC2Lox/Lox controls. At day 8, 100% of HDAC1Δ2/Δ2 EBs had developed a spontaneous rhythmic contraction phenotype, compared to <10% for HDAC1Lox/Lox controls (Fig. 4C), indicative of differentiation into cardiomyocytes and pacemaker cells. Only a limited number of HDAC2Δ2/Δ2 EBs were observed to spontaneously contract. The reduction in HDAC1Δ2/Δ2 EB size, their irregular shape, and spontaneous contraction suggested that increased differentiation was occurring. To visualize the differentiation and examine the cell types produced, day 12 EBs of the indicated genotype were embedded in paraffin, sectioned laterally, and then stained with hematoxylin and eosin (Fig. 4D). Histological examination of HDAC2Δ2/Δ2 and undeleted control EBs (HDAC1Lox/Lox and HDAC2Lox/Lox) revealed a relatively uniform organization consisting mainly of mesenchyme surrounded by ground substrate. HDAC1Δ2/Δ2 EBs, in contrast, have a large number of organized cellular structures, including epithelial rosettes with associated basement membrane and many regions of cell death, indicative of profound differentiation (Fig. 4D, compare Left and Right).
Fig. 4.
Loss of HDAC1 enhances embryoid body differentiation. (A) Representative examples of EBs at the indicated time points. (B) Mean size and SDs of EBs during a 12-day experiment. Different genotypes are indicated. Mean values (n = 3) ± SEM are plotted. (C) Percentage of EBs with a rhythmic beating phenotype. Individual genotypes are indicated. (D) Hematoxylin and eosin-stained sections of day 12 EBs of the indicated genotype. (i) Regions of mesenchyme, unstained areas demark ground substance; (ii) structured region of organized epithelium (epithelial rosette) on basement membrane; (iii) region of cell death and apoptosis. Both ii and iii are characteristic of EBs derived from HDAC1Δ2/Δ2 ES cells. Data are representative of three independently tested clones for each genotype.
Increased Expression of Cardiomyocyte-, Muscle-, and Neuronal-Specific Markers in Embryoid Bodies Lacking HDAC1.
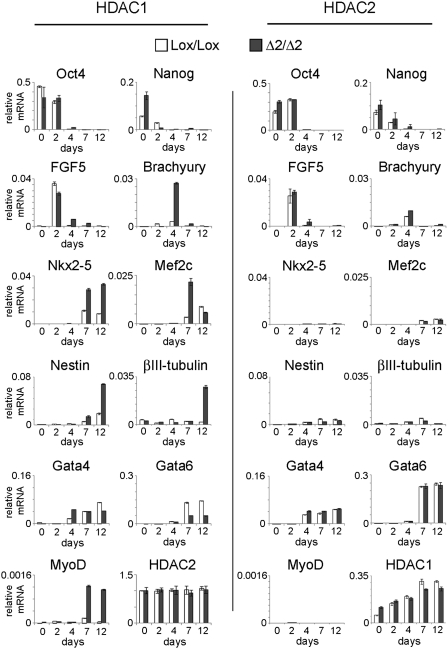
To identify the cell types present during wild-type and mutant EB differentiation, we collected RNA at time points from day 0 (cycling ES cells) to day 12 (mature EBs) and then performed quantitative (q)RT-PCR for lineage-specific markers (Fig. 5). Consistent with the process of ES cell differentiation, the pluripotency markers Oct4 and Nanog are repressed in both HDAC1Δ2/Δ2 and HDAC2Δ2/Δ2 EBs. We also observe that HDAC1Δ2/Δ2 and HDAC2Δ2/Δ2 EBs show a similar induction (day 2), and then repression (day 7), of FGF5, suggesting that loss of either HDAC does not adversely affect primitive ectoderm formation, a critical intermediate in the formation of the three primary germ layers. A key regulator of mesodermal specification is the T-box transcription factor, brachyury. Induction of brachyury during EB differentiation is enhanced in EBs lacking HDAC1 compared with HDAC2 deleted cells and undeleted controls. Consistent with increased brachyury levels and the spontaneous “beating” phenotype of HDAC1Δ2/Δ2 EBs, we monitor increased expression of the cardiomyocyte-specific markers Nkx2-5 and Mef2c, in addition to the skeletal muscle marker, MyoD. The neuronal cell markers, nestin and βIII-tubulin, are similarly elevated in day 12 HDAC1Δ2/Δ2 EBs. However, Gata-6, a marker of endodermal differentiation, is reduced by loss of HDAC1, indicating preferential differentiation toward mesodermal and ectodermal lineages at the expense of endoderm under these differentiation conditions (Fig. 5).
Fig. 5.
Loss of HDAC1 causes precocious differentiation into mesodermal and ectodermal lineages in EBs. Quantitative RT-PCR data for genes characteristic of undifferentiated stem cells (Oct-4, Nanog), endoderm (Gata4, Gata6), primitive ectoderm (Fgf5), mesoderm (Brachyury, Nkx2-5, Mef2c, and MyoD) and ectoderm (Nestin, βIII-tubulin) was performed as indicated on mRNA collected at days 0, 2, 4, 7, and 12 during EB differentiation. Mean values (n = 3) ± SEM are plotted. Values indicate expression of the specific gene relative to GAPDH measured using Universal ProbeLibrary hydrolysis probes. Data are representative of two independently tested clones for each genotype.
Discussion
HDAC1, but Not HDAC2, Regulates H3K56Ac and Is Required for Optimal Deacetylase Activity of HDAC1/2 Complexes in ES Cells.
To assess the embryonic requirement for HDAC1, in relation to HDAC2, we generated ES cells in which each gene can be inactivated conditionally. We find that loss of HDAC1, but not HDAC2, reduces the amount of deacetylase activity associated with HDAC1/2 complexes, particularly NuRD and CoREST (Fig. 2B), despite a compensatory increase in HDAC2 protein and its incorporation into these complexes. Lagger and colleagues (18) noted a similar decrease in Sin3A- and NuRD-associated HDAC activity in their HDAC1−/− ES cell system. Perhaps the most obvious explanation for a reduction in the deacetylase activity of HDAC1/2 complexes in the absence of HDAC1 is that HDAC1 is a slightly more effective deacetylase than HDAC2 in ES cells. It remains to repeat these experiments in somatic cells, particularly those cell types where HDAC1 and -2 are functionally redundant, to ask if this dependence for HDAC1 is embryonic specific. If so, then this result may explain in part the postgastrulation lethality of HDAC1−/− mice (18, 19) and the ES cell differentiation phenotype (discussed below).
Loss of HDAC1 alone results in increased H3K56Ac in ES cells (Fig. 2D). This solitary detected change in lysine acetylation may reflect that H3K56Ac is a relatively rare modification in mammalian cells (≈1% of histone H3) (32, 34). In contrast, histone tails appear to be hyperacetylated in ES cells (compare TSA-induced acetylation in ES cells versus MEFs, Fig. S3), potentially masking the detection of increased acetylation due to the loss of HDAC1. It has been shown previously that H3K56Ac is a substrate for the NAD-dependent deacetylases, SIRT1 and SIRT2 (32); we demonstrate here that H3K56Ac levels are also regulated by HDAC1 (Fig. 2D), in addition to other zinc-dependent HDACs (Fig. S3). However, it remains formally possible that loss of HDAC1 might increase H3K56Ac levels via an indirect mechanism. Potentially, elevated CBP/p300 levels, which acetylate H3K56 in mammalian cells, or, conversely, down-regulation of Sirt1 (32) could cause the increase in H3K56Ac levels. We are currently performing experiments with purified HDAC1/2 complexes from ES cells to address whether H3K56Ac is a direct target for deacetylation by HDAC1.
Loss of HDAC1 or HDAC2 Does Not Affect the Cell Cycle in ES Cells.
HDAC1 and HDAC2 have been linked to cell cycle regulation by their association with the tumor suppressor, Rb (10–12) and transcriptional repression of the CDK inhibitor, p21 (18, 31, 36). However, we found that there was no change in the growth potential of either HDAC1Δ2/Δ2 or HDAC2Δ2/Δ2 ES cells (Fig. 3 A and B), nor did we observe any change in the level of p21 mRNA (although its expression is increased by treatment with TSA, Fig. S4). Phenotypically therefore, our cells differ from the HDAC1−/− ES cells used by Lagger et al. (18), which may reflect the difference in their derivation, either by sequential gene targeting followed by conditional deletion or from blastocysts bearing a constitutive deletion, respectively. Although p21 is the prototypical HDAC1 target gene, it is interesting to note that siRNA-mediated knockdown of HDAC1 produces a cell type-specific up-regulation of p21 (36). Rather than affecting the cell cycle, we find that deletion of HDAC1 causes an increase in the amount and variety of cell types produced during ES cell differentiation.
HDAC1 Is Required for the Controlled Reprogramming of ES Cells upon Differentiation.
Unlike deletion of the NuRD component MBD3 (37), loss of either HDAC1 or HDAC2 did not affect the potential of ES cells to exit the pluripotent stem cell program upon LIF withdrawal (Fig. 3C). Inactivation of Oct4 occurs concomitantly with deacetylation of its promoter region shortly after differentiation begins (39). However, the ability to switch off Oct4 (and Nanog) expression is unperturbed by loss of either HDAC1 or -2. Early differentiation of ES cells into primitive ectoderm, key to formation of the three primary germ layers, was also unaffected, as suggested by the presence of Oct4 and spike of FGF5 expression in day 2 EBs (Fig. 5). From days 4 to 12 HDAC1Δ2/Δ2 EBs are distinct from either undeleted controls or HDAC2Δ2/Δ2 EBs, by their reduced size, irregular shape, substantially increased contractility, and superior, more defined cellular organization. Further analysis of EB transcriptional profiles indicates that HDAC1Δ2/Δ2 EBs exhibit preferential differentiation into mesoderm (particularly cardiomyocytes, Nkx2-5, and Mef2c) and latterly neuroectoderm cell lineages (nestin and βIII-tubulin) possibly at the expense of endoderm (Gata4 and Gata6). In contrast, transcriptional differences have yet to be detected in HDAC2Δ2/Δ2 EBs, as might be expected because we observe no change in the deacetylase activity of corepressor complexes in the absence of HDAC2. Thus it appears that HDAC1 acts to moderate the degree, nature, and pace of ES cell differentiation. These data concur with a number of studies in which HDAC inhibitors have been applied to pluripotent cells. Treatment of day 7 EBs with TSA promotes cardiomyocyte differentiation (40), as measured by an increase in Nkx2-5 expression, which may be a direct target of HDAC1 (13). Similarly, the addition of HDAC inhibitors to neuronal precursors results in a selective increase in neurons, but not oligodendrocytes or astrocytes (41–43), which argues in favor of a specific, rather than general role for HDACs in differentiation.
ES cells contain a number of distinct HDAC1/2-containing complexes including Sin3A, NuRD, CoREST (Fig. 2C), and a recently described complex termed the Nanog and Oct-4-associated deacetylase (NODE) complex (44). The NODE complex is similar in composition to NuRD, but lacks MBD3 and Rbbp7. Interestingly, perturbation of the NuRD complex by deletion of MBD3 causes a defect in ES cell differentiation, concomitant with a reduced Mta2-HDAC1 interaction (37). Conversely, disruption of NODE causes increased differentiation, associated with the activation of endodermal-specific markers (GATA6 and Foxa2) (44). An shRNA-mediated knockdown of HDAC2 in the same study revealed no effect on differentiation, consistent with our genetic deletion of HDAC2 (Fig. 3C). Therefore, two distinct HDAC1/2-containing complexes appear to have opposing functions in regard to ES cell fate and lineage commitment. However, it is not clear if a distinct NODE complex occurs in EBs, where loss of HDAC1 causes precocious differentiation. ES cell pluripotency and cell fate determination are profoundly affected by MAPK, Wnt, and TGFβ signaling pathways (45). It is interesting to note therefore that HDAC1 and -2 have been shown to be downstream effectors of Wnt signaling during cardiomyocyte (13) and oligodendrocyte (46) differentiation. A more prudent application of our system using defined (serum-free) media, over prolonged periods, is currently underway to decipher the roles of HDAC1 and -2 in lineage-specific differentiation programs.
We have shown that HDAC1 is necessary for the optimal deacetylase activity of HDAC1/2 corepressor complexes and regulates the acetylation status of H3K56Ac. This change in the biochemical activity of HDAC1Δ2/Δ2 cells correlates with a precocious differentiation phenotype. The increased differentiation potential of these HDAC1Δ2/Δ2 ES (epiblast-like) cells may provide plausible in vitro evidence as to why HDAC2−/− embryos survive at least until the perinatal stage and insight into why HDAC1−/− embryos die shortly after gastrulation. The ability to regulate HDAC1 and HDAC2 genetically in a pluripotent system should allow us to identify the target genes and nonhistone targets, which regulate key developmental pathways.
Materials and Methods
Generation of HDAC1 and HDAC2 Knockout ES Cell Lines and ES Cell Culture.
E14 ES cells, expressing a CreER fusion protein from the ROSA26 locus (30), were used to generate HDAC1Lox/Lox; CreER and HDAC2Lox/Lox; CreER cell lines by consecutive rounds of gene targeting. For detailed description of targeting vectors and gene targeting strategies see SI Materials and Methods and Figs. S1 and S2. ES cell lines were maintained on gelatinized plates in standard ES cell medium consisting of Knockout DMEM (Invitrogen), 15% FCS, 1× glutamine/penicillin/streptomycin (Invitrogen), 100 μM β-mercaptoethanol (Sigma-Aldrich), and LIF.
Population Doubling, Colony Formation, and Alkaline Phosphatase Assays.
ES cell lines of 1.5 × 106 cells were seeded in triplicate in six-well plates, trypsinized, and counted and replated every 2–3 days for a period of 18 days. Colony formation was assessed by seeding 7 × 102 cells in triplicate in six-well plates followed by culture for 8 days and counted. For alkaline phosphatase assays cells were plated at 7 × 102 cells per well in six-well plates in the presence of LIF. After overnight culture, cells were cultured in the presence or absence of LIF for 6 days and then stained with a commercial alkaline phosphatase assay kit (Millipore).
Protein and Enzymatic Analysis.
To confirm deletion of HDAC1 and HDAC2, cells were lysed with hypotonic buffer: 10 mM Hepes (pH 7.9), 10 mM KCl, 0.1 mM EDTA, 10 mM DTT, and 1× protease inhibitor mixture (Sigma). After centrifugation at 1,500 × g for 5 min nuclear extracts were prepared from pellets with 20 mM Hepes (pH 7.9), 0.4 M NaCl, 1 mM EDTA, 25% glycerol, and 1× protease inhibitor mixture. Ten micrograms of nuclear extract was separated using SDS/PAGE and membranes were probed with the appropriate antibodies (Table S1). Acid extraction of histones was performed as previously described (47). Five micrograms of extract was loaded in each lane and membranes were probed using a panel of antibodies raised against a number of histone modifications (for antibodies used to probe membranes see Table S1). The Odyssey Infrared Imaging System was used to quantify protein signal using the appropriate IRDye conjugated secondary antibodies (LiCOR Biosciences). For immunoprecipitation, 70 μg of nuclear extract was incubated overnight at 4 °C with antibody-coated protein-G agarose beads (see Table S1 for appropriate antibodies). After four washes in nuclear extract buffer, beads were split into two aliquots. One aliquot was used to assess the enzymatic activity of the immunoprecipitates using a commercially available deacetylase assay (Active Motif); the remaining aliquot was resolved by SDS/PAGE and probed with antibodies raised against known components of the immunoprecipitated complexes.
In Vitro Differentiation.
ES cell differentiation was induced by culturing ES cells as EBs according to the hanging drop method for 2 days. Drops of differentiation medium (i.e., standard ES cell medium without LIF) containing 7 × 102 ES cells were plated onto the lids of culture dishes. After 48 h, EBs were transferred to culture plates coated with a hydrophobic coating, Sylgard 184 (Dow Corning), and cultured in suspension under constant rotation for a further 10 days.
RNA Isolation, Reverse Transcription, and Quantitative Real-Time PCR.
Total RNA was isolated using a standard TRIzol (Invitrogen) protocol. Two micrograms of total RNA was reverse transcribed using Q-Script one-step Supermix (Quanta Biosciences) and the resulting cDNA was diluted with an equal amount of DEPC treated H2O. Multiplex assays were designed using the Universal ProbeLibrary Assay Design Centre (www.roche-applied-science.com; see Table S2 for primers and probes). For each reaction, 2 μl of diluted cDNA was used in all subsequent multiplex qRT-PCR reactions using the Light Cycler Probes Master Mix (Roche) as per the manufacturer's instructions. Reactions were carried out on a Roche Light Cycler 480 under the following conditions: initial denaturation for 10 min at 94 °C followed by 40 cycles of 10 sec at 94 °C, 20 sec at 55 °C, and 5 sec at 72 °C.
Histology and Immunocytochemistry.
Day 12 EBs were fixed with 4% paraformaldehyde and embedded in paraffin blocks. Five-micrometer sections were then stained with hematoxylin and eosin.
Supplementary Material
Acknowledgments
We thank Ian Eperon, John Schwabe, and Salvador Macip for useful discussions and critical reading of the manuscript. We thank Jenny Edwards, Emma Stringer, Felix Beck, and Kees Straatman for technical assistance. E14 ES cells and ROSA26-CreER targeting vector were kindly provided by David Adams. This work was supported by a Medical Research Council studentship (to C.T.F.) and a Career Development Award (G0600135) (to S.M.C.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000478107/-/DCSupplemental.
References
- 1.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: From bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luger K, Richmond TJ. The histone tails of the nucleosome. Curr Opin Genet Dev. 1998;8:140–146. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 3.Robinson PJ, et al. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J Mol Biol. 2008;381:816–825. doi: 10.1016/j.jmb.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 5.Brunmeir R, Lagger S, Seiser C. Histone deacetylase HDAC1/HDAC2-controlled embryonic development and cell differentiation. Int J Dev Biol. 2009;53:275–289. doi: 10.1387/ijdb.082649rb. [DOI] [PubMed] [Google Scholar]
- 6.Taplick J, et al. Homo-oligomerisation and nuclear localisation of mouse histone deacetylase 1. J Mol Biol. 2001;308:27–38. doi: 10.1006/jmbi.2001.4569. [DOI] [PubMed] [Google Scholar]
- 7.Humphrey GW, et al. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J Biol Chem. 2001;276:6817–6824. doi: 10.1074/jbc.M007372200. [DOI] [PubMed] [Google Scholar]
- 8.You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST–human histone deacetylase complex. Proc Natl Acad Sci USA. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hakimi MA, et al. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc Natl Acad Sci USA. 2002;99:7420–7425. doi: 10.1073/pnas.112008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brehm A, et al. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 11.Luo RX, Postigo AA, Dean DC. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 12.Magnaghi-Jaulin L, et al. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 13.Liu Z, et al. WNT signaling promotes Nkx2.5 expression and early cardiomyogenesis via downregulation of Hdac1. Biochim Biophys Acta. 2009;1793:300–311. doi: 10.1016/j.bbamcr.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 14.Montgomery RL, Hsieh J, Barbosa AC, Richardson JA, Olson EN. Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc Natl Acad Sci USA. 2009;106:7876–7881. doi: 10.1073/pnas.0902750106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pegoraro G, et al. Ageing-related chromatin defects through loss of the NURD complex. Nat Cell Biol. 2009;11:1261–1267. doi: 10.1038/ncb1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ropero S, et al. A truncating mutation of HDAC2 in human cancers confers resistance to histone deacetylase inhibition. Nat Genet. 2006;38:566–569. doi: 10.1038/ng1773. [DOI] [PubMed] [Google Scholar]
- 17.Marks PA, Xu WS. Histone deacetylase inhibitors: Potential in cancer therapy. J Cell Biochem. 2009;107:600–608. doi: 10.1002/jcb.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagger G, et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montgomery RL, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cowley SM, et al. The mSin3A chromatin-modifying complex is essential for embryogenesis and T-cell development. Mol Cell Biol. 2005;25:6990–7004. doi: 10.1128/MCB.25.16.6990-7004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.David G, Turner GM, Yao Y, Protopopov A, DePinho RA. mSin3-associated protein, mSds3, is essential for pericentric heterochromatin formation and chromosome segregation in mammalian cells. Genes Dev. 2003;17:2396–2405. doi: 10.1101/gad.1109403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15:710–723. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 25.Guan JS, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trivedi CM, et al. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat Med. 2007;13:324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 27.Zimmermann S, et al. Reduced body size and decreased intestinal tumor rates in HDAC2-mutant mice. Cancer Res. 2007;67:9047–9054. doi: 10.1158/0008-5472.CAN-07-0312. [DOI] [PubMed] [Google Scholar]
- 28.Haberland M, Johnson A, Mokalled MH, Montgomery RL, Olson EN. Genetic dissection of histone deacetylase requirement in tumor cells. Proc Natl Acad Sci USA. 2009;106:7751–7755. doi: 10.1073/pnas.0903139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith AG. Embryo-derived stem cells: Of mice and men. Annu Rev Cell Dev Biol. 2001;17:435–462. doi: 10.1146/annurev.cellbio.17.1.435. [DOI] [PubMed] [Google Scholar]
- 30.Vooijs M, Jonkers J, Berns A. A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep. 2001;2:292–297. doi: 10.1093/embo-reports/kve064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zupkovitz G, et al. Negative and positive regulation of gene expression by mouse histone deacetylase 1. Mol Cell Biol. 2006;26:7913–7928. doi: 10.1128/MCB.01220-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tjeertes JV, Miller KM, Jackson SP. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 2009;28:1878–1889. doi: 10.1038/emboj.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie W, et al. Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol Cell. 2009;33:417–427. doi: 10.1016/j.molcel.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glaser KB, et al. Role of class I and class II histone deacetylases in carcinoma cells using siRNA. Biochem Biophys Res Commun. 2003;310:529–536. doi: 10.1016/j.bbrc.2003.09.043. [DOI] [PubMed] [Google Scholar]
- 36.Senese S, et al. Role for histone deacetylase 1 in human tumor cell proliferation. Mol Cell Biol. 2007;27:4784–4795. doi: 10.1128/MCB.00494-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaji K, et al. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- 38.Lee JH, Hart SR, Skalnik DG. Histone deacetylase activity is required for embryonic stem cell differentiation. Genesis. 2004;38:32–38. doi: 10.1002/gene.10250. [DOI] [PubMed] [Google Scholar]
- 39.Feldman N, et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- 40.Kawamura T, et al. Acetylation of GATA-4 is involved in the differentiation of embryonic stem cells into cardiac myocytes. J Biol Chem. 2005;280:19682–19688. doi: 10.1074/jbc.M412428200. [DOI] [PubMed] [Google Scholar]
- 41.Balasubramaniyan V, et al. Effects of histone deacetylation inhibition on neuronal differentiation of embryonic mouse neural stem cells. Neuroscience. 2006;143:939–951. doi: 10.1016/j.neuroscience.2006.08.082. [DOI] [PubMed] [Google Scholar]
- 42.Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci USA. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siebzehnrubl FA, et al. Histone deacetylase inhibitors increase neuronal differentiation in adult forebrain precursor cells. Exp Brain Res. 2007;176:672–678. doi: 10.1007/s00221-006-0831-x. [DOI] [PubMed] [Google Scholar]
- 44.Liang J, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 45.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: Lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 46.Ye F, et al. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci. 2009;12:829–838. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.