Abstract
Two distinct human herpesvirus 6 (HHV-6) variants infect humans. HHV-6B is the etiologic agent of roseola and is associated with life-threatening neurological diseases, such as encephalitis, as well as organ transplant failure. The epidemiology and disease association for HHV-6A remain ill-defined. Specific anti-HHV-6 drugs do not exist and classic antiherpes drugs have secondary effects that are often problematic for transplant patients. Clinical trials using IFN were also performed with inconclusive results. We investigated the efficacy of type I IFN (α/β) in controlling HHV-6 infection. We report that cells infected with laboratory strains and primary isolates of HHV-6B are resistant to IFN-α/β antiviral actions as a result of improper IFN-stimulated gene (ISGs) expression. In contrast, HHV-6A-infected cells were responsive to IFN-α/β with pronounced antiviral effects observed. Type II IFN (γ)-signaling was unaltered in cells infected by either variant. The HHV-6B immediate-early 1 (IE1) physically interacts with STAT2 and sequestrates it to the nucleus. As a consequence, IE1B prevents the binding of ISGF3 to IFN-responsive gene promoters, resulting in ISG silencing. In comparison, HHV-6A and its associated IE1 protein displayed marginal ISG inhibitory activity relative to HHV-6B. The ISG inhibitory domain of IE1B mapped to a 41 amino acid region absent from IE1A. Transfer of this IE1B region resulted in a gain of function that conferred ISG inhibitory activity to IE1A. Our work is unique in demonstrating type I IFN signaling defects in HHV-6B-infected cells and highlights a major biological difference between HHV-6 variants.
Keywords: immediate-early 1, interferon-stimulated genes, STAT2
Secreted type 1 IFNs (α and β) bind to a receptor linked with Tyk2 and Jak1 tyrosine kinases (1, 2). Activated Jak1 and Tyk2 in turn phosphorylate STAT1 and STAT2, which assemble with IRF9 to form the IFN-stimulated gene factor 3 (ISGF3) complex that binds to IFN-stimulated response elements (ISRE) and promote transcription (3–6). Products of IFN-stimulated genes (ISGs) play essential roles in antiviral defense.
Human herpesvirus 6 (HHV-6), isolated more than 20 years ago (7), is an increasingly recognized medically relevant pathogen. Two distinct HHV-6 variants (A and B) exist. These viruses have distinct biological properties and are associated with specific pathological conditions. The most common clinically defined disease associated with HHV-6B is roseola (8). HHV-6B reactivation is also common after hematopoietic stem cell transplantation and is responsible for severe complications (9). Much less is known regarding HHV-6A, but results suggest that this variant is more neurotropic than HHV-6B (10).
In the present work, we studied the activation of ISGs during HHV-6 infection. Our results show a striking difference between HHV-6 variants, with HHV-6B effectively dwarfing ISGs activation and HHV-6A having only limited effects. Using RNA interference, we have identified that the immediate-early 1 (IE1) protein of HHV-6B is responsible for this effect. Finally, we demonstrate that IE1 inhibitory actions were mediated through physical interactions and nuclear sequestration of STAT2, leading to impaired assembly and binding of ISGF3 to ISRE elements.
Results
HHV-6B-Infected Cells Are Resistant to the Antiviral Actions of IFN.
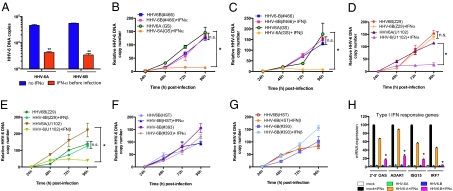
Work performed in the early 1990s suggests that HHV-6 is sensitive to type I IFN antiviral effects (11, 12). We revisited this issue and tested whether the timing of infection relative to the IFN treatment affected the outcome. IFN-α treatment of SupT1 cells before infection was very effective at restricting growth of both HHV-6 variants (Fig. 1A). In contrast, when peripheral blood mononuclear cells (PBMCs) were infected with HHV-6 before IFN-α or IFN-β treatment (1,000 U/mL), the replication of all HHV-6B strains (Z29 and HST) and primary isolates (#393 and #466) tested was unaffected, although that of HHV-6A (GS and U1102) was reduced by 90% (Fig. 1 B–G). Similar effects were observed at lower (10–100 U/mL) IFN doses (Fig. S1 A–D). To test for potential type I IFN signaling defects, SupT1 cells were infected with HHV-6, then stimulated with IFN-α, and ISG expression determined (Fig. 1H). Compared with mock-infected/IFN-α-treated cells, ISG expression in HHV-6B-infected cells was reduced and that of HHV-6A-infected cells was much less affected. SupT1 cells were infected with similar efficiencies by both HHV-6 variants, as determined by IE1 mRNA and p41 expression (Fig. S1 E and F). To determine specificity, type II IFN signaling was also studied. No differences were observed in response to IFN-γ stimulation between mock- and HHV-6-infected cells, regardless of the HHV-6 variant (Fig. S1G). These results indicated that HHV-6B specifically impairs type I IFN signaling.
Fig. 1.
Type I IFN signaling defects in HHV-6B-infected cells. (A) SupT1 cells were treated with IFN-α (1,000 U/mL) for 18 h before infection [multiplicity of infection (m.o.i.) = 0.5] with HHV-6 (GS and Z29). Two days after infection, intracellular HHV-6 DNA copies were measured by quantitative PCR (qPCR). **, P < 0.01 relative to no IFN group. (B–G) PBMCs were infected with HHV-6B laboratory strains (Z29, HST), HHV-6A laboratory strains (U1102, GS), or HHV-6B primary isolates (#393, #466) for 24 h at an m.o.i. of 0.1. The next day, IFN-α (B, D, F) or IFN-β (C, E, G) was added (1,000 U/mL) and viral DNA was isolated every day for 4 d. Viral DNA was assessed by qPCR. Results are expressed as mean (triplicate) ± SDs of HHV-6 DNA copy number after normalization with GAPDH. *, P < 0.05 between HHV-6A +/- IFN. (H) Two-day-old HHV-6A (GS)- or HHV-6B (Z29)-infected SupT1 cells were stimulated with IFN-α (1,000 U/mL) for 8 h, after which type I IFN-responsive ISG mRNA expression was determined by RT-qPCR. Results are expressed as a percentage of the ISG levels in mock-infected and IFN-treated cells and are representative of two experiments performed in triplicates. *, P < 0.05 relative to mock-infected and IFN-treated cells.
These results suggest that an HHV-6B-encoded gene product is abrogating type I IFN antiviral effects. SupT1 cells infected with HHV-6B in the presence of phosphonoacetic acid and stimulated with IFN-α demonstrated reduced ISG mRNA levels, indicating that late viral gene expression was not required for this effect (Fig. S2A). Effectiveness of phosphonoacetic acid treatment was verified by monitoring the lack of late gene (U54) expression (Fig. S2B). Together, these data imply that IE or E gene products are responsible for ISG inhibition.
IE1 Is Responsible for Impaired Type I IFN Signaling in Infected Cells.
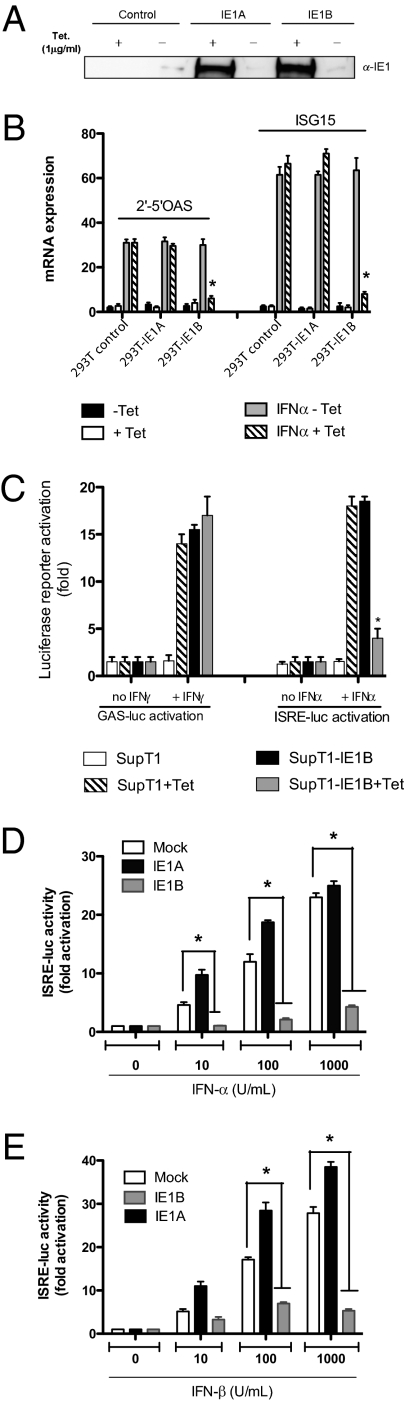
We hypothesized that IE1, being among the least conserved protein (63% amino acid identity) between HHV-6 variants (13, 14), could be responsible for the divergent ISG modulation observed. To study this hypothesis, we made use of IE1 tetracycline (Tet)-inducible cell lines (15). Cells were either untreated or cultured with Tet (1 μg/mL) to induce IE1A (HHV-6A) or IE1B (HHV-6B) expression (Fig. 2A) followed by IFN-α stimulation. Expression of ISGs in IE1B-expressing (+Tet) cells was severely reduced compared with control or IE1A-expressing cells (Fig. 2B). To test whether type II IFN-responsive genes were affected by IE1 expression, we made use of SupT1 IE1-inducible cell lines, which are responsive to both type I and type II IFNs. Activation of a type II IFN-responsive promoter (GAS-Luc reporter) was not affected; however, the activation of the ISRE-Luc reporter, under the control of a type I IFN responsive promoter, was inhibited by IE1B (+Tet) (Fig. 2C). IE1B protein was able to shut down the induction of the ISRE-Luc reporter gene expression upon stimulation with either IFN-α or IFN-β, in striking contrast with IE1A inability to dwarf type I IFN signaling. IE1A failed to inhibit ISRE-Luc activation, even at low IFN doses (Fig. 2 D and E). These results indicate that IE1B but not IE1A is effective at blocking type I IFN signaling cascade with no effect on type II IFN signaling.
Fig. 2.
Effects of HHV-6 IE1 expression on the activation of ISGs. (A) IE1 expression in Tet-inducible 293T control and IE1 cell lines ± Tet (1 μg/mL). (B) Tet-inducible 293T control and IE1 cell lines were treated ± Tet (1 μg/mL) for 24 h followed by stimulation with IFN-α. Total RNA was extracted and type I ISG expression analyzed by RT-qPCR. Results are expressed as mean (triplicate) ± SD. ISG expression after normalization with GAPDH mRNA. Results are representative of three independent experiments. *, P < 0.01 relative to IFN, -Tet samples. (C) Control- and IE1B-inducible SupT1 cells were transfected with GAS-Luc or ISRE-Luc reporters. Twenty-four hours posttransfection, cells were incubated with IFN-γ or IFN-α. Sixteen hours later, luciferase activity was determined. Results are expressed as mean (triplicate) induction (n-fold) of GAS or ISRE luciferase activity ± SD relative to SupT1 cells. Results are representative of three independent experiments. *, P < 0.01 relative to SupT1-IE1B cells. (D and E) HEK-293T cells were transfected with 50 ng of reporter-luciferase ISRE-Luc vector together with IE1A- or IE1B-expression vectors. Twenty-four hours later, cells were stimulated with increasing amount of IFN-α (D) or IFN-β (E) for 16 h and luciferase activity determined. Results are expressed as the level of activation (n-fold) relative to the activity in control, mock-stimulated transfected cells, and are representative of two independent experiments. *, P < 0.05 as determined by using two-tailed t-test.
HHV-6B IE1 Protein Prevents the Formation and Binding of ISGF3 Complex to ISRE Elements in Human Cells.
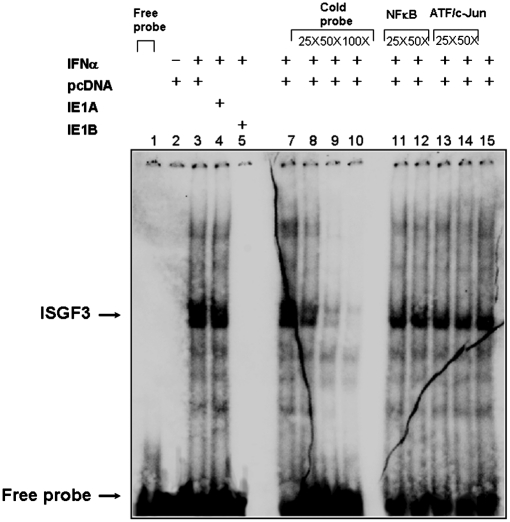
Western blot analyses disclosed that steady-state and phosphorylated protein levels of STAT1, STAT2, and IRF9 remained unchanged, despite IE1A or IE1B expression and before or after IFN-α stimulation (Fig. S3A). DNA-binding activity of the ISGF3 transcription complex in IE1-expressing cells was studied next by EMSA. IFN-α stimulation resulted in pronounced gel shift signals in control (Lane 2) and IE1A expressing cells (Lane 3) (Fig. 3). In contrast, the ISGF3 complex was absent from IE1B expressing cells (Lane 4). The specificity of ISGF3 complex was confirmed using excess homologous (ISG15) (Lanes 7–10) or heterologous (NF-κB) (Lanes 11–12) and (ATF-2/c-Jun) (Lanes 13–14) cold probes. Because of the heterotrimeric nature of the ISGF3 complex and difficulties in inducing a supershift with antibodies directed against ISGF3 components, we performed Western blot of mobility-shift gels, as described (16). Protein-DNA complexes formed with extracts from untreated or superinduced (18 h of IFN-γ, followed by 30 min of IFN-α) A549 cells. The positions of the bands stained by antibodies against ISGF3 subunits on Western blot were determined by alignment with the autoradiographs from control lanes of the mobility-shift gel (Fig. S3B). We concluded that the inhibition of type I IFN signaling in HHV6B-infected cells is caused by a defect in ISGF3 binding to ISRE elements.
Fig. 3.
Effects of IE1 on binding of ISGF3 to ISRE. ISGF3 DNA binding activity in resting and IFN-α-activated control and IE1-expressing cells was analyzed by EMSA. Nuclear extracts were incubated with a 32P-labeled ISRE probe from ISG15 promoter in the presence or absence of competitor probes. (Lane 1) 1, free probe; (Lanes 2–5) binding of ISGF3 complex to the ISG15 probe in the absence or in the presence of IE1; (Lanes 7–10) homologous competition using increasing doses of unlabeled ISG15 probe; (Lanes 11–14) heterologous competition using increasing doses of unlabeled oligonucleotides corresponding to the NFκB (Lanes 11–12) and ATF/c-Jun (Lanes 13–14) binding motifs. The results are representative of three experiments.
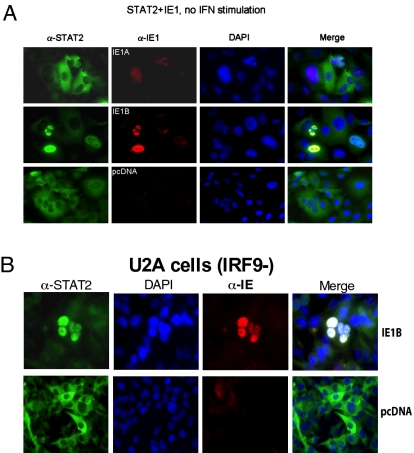
Immunofluorescence microscopy was used next to examine the subcellular localization of ISGF3 components in IE1-expressing cells. STAT1 localized in the cytoplasm of control and IE1-expressing cells (Fig. S4A). Interestingly, STAT2 localized in the nuclei of IE1B-transfected cells, even though no IFN was added (Fig. 4A). When cells were treated with IFN-α for 1 h, STAT1 and STAT2 accumulated in the nuclei of control and IE1A, IE1B cells (Fig S4 B and C). Twenty-four hours after IFN-α activation, STAT1 and STAT2 were redistributed in the cytoplasm of control and IE1A-transfected cells but STAT1 and STAT2 remained in the nuclei of IE1B-expressing cells (Fig. S4 D and E). To confirm and extend these results, we studied endogenous STAT2 localization in U2A cells, a 2fTGH-derivative that lacks IRF9 (17, 18). IE1B expression caused nuclear accumulation of STAT2 in U2A cells (Fig. 4B), suggesting that IE1B affects the constitutive cytoplasmic-nuclear shuttling and sequesters STAT2 in the nucleus in an IRF9-independent manner.
Fig. 4.
Subcellular localization of STAT1, STAT2 and IE1. (A) A549 cells and (B) U2A cells (IRF9-) were nucleofected with pcDNA, IE1A, or IE1B expression vectors. Forty-eight hours posttransfection cells were analyzed for STAT2 (green) or IE1 (red) expression. Nuclei were stained with DAPI (blue). The localization of the proteins relative to the nucleus is presents in the merged pictures. Results are representative of three experiments.
Physical Interaction Between IE1 and ISGF3 Components.
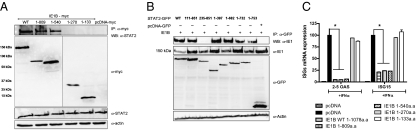
We could successfully pull down STAT2 by immunoprecipitating IE1 from HHV-6B-infected cells, suggesting that these two proteins physically interact (Fig S5 A and B). Similar results were obtained in 2fTGH, U3A (STAT1−) and U2A (IRF9−) cells, indicating that IE1B/STAT2 interaction was independent of STAT1 or IRF9 (Fig S5C). To map the IE1B/STAT2 interacting regions, we expressed and tested myc-tagged mutants of IE1B (Fig. S5D) for interactions with STAT2. WT, 1–809 and 1–540 IE1B mutants interacted with STAT2, although the 1–270 and 1–133 mutants failed to do so, suggesting that the IE1 region binding to STAT2 lies between amino acids 270 and 540 (Fig. 5A). To map the STAT2 regions binding to IE1B, we used GFP-STAT2 mutants (19) (Fig. S5E). IE1 is able to interact with WT and all STAT2 mutants, with the exception of the ΔN235 mutant (Fig. 5B). At first, this would suggest that IE1B binds STAT2 within the 111 to 235 amino acid region, but results from Banninger and Reich (19) indicate that the ΔN235 STAT2 mutant, which lacks the coiled-coil domain responsible for binding to IRF9, is cytoplasmic and does not shuttle to the nucleus. Considering that IE1B is a nuclear protein (20), physical interactions between the ΔN235 and IE1B would not be possible under the current experimental settings. Taking this into consideration, we concluded that IE1 binds STAT2 within the 111 to 397 region.
Fig. 5.
Mapping of IE1B and STAT2 interacting domains. (A) WT and IE1B mutants were expressed in 293T cells followed 48 h later by α-myc IP and Western blot for STAT2. The three lower panels show expression of IE1B mutants, STAT2, and actin in whole-cell extracts. (B) GFP-STAT2 constructs were cotransfected with IE1B (WT) in 293T cells and 48 h later cells were harvested and subjected to IP using α-GFP antibody followed by Western blot for IE1. The three lower panels show expression of IE1B, STAT2 (GFP-tag), and actin in whole-cell extracts. (C) 293T cells were transfected with WT or IE1B mutant vectors, followed 24 h later by IFN-α stimulation. After 18 h, 2' to 5' OAS and ISG15 mRNA expression was measured by RT-qPCR. Results are expressed as a percentage of the ISG mRNA expression relative to control vector-transfected and IFN-treated cells after normalization of samples with GAPDH mRNA expression. Results are representative of three experiments. *, P < 0.01.
We next determined whether binding of IE1B to STAT2 correlated with ISG inhibition. WT and IE1B mutants were expressed into 293T cells followed by IFN-α stimulation. The 1–809 and 1–540 mutants were capable of suppressing ISG activation as efficiently as WT IE1, unlike mutants 1–270 and 1–133, which had lost their inhibitory potential (Fig. 5C). We concluded that a direct correlation existed between the ability of IE1B to bind STAT2 and the ability of IE1B to inhibit ISGs activation.
IE1 Is Responsible for the Impaired Type I IFN Signaling in HHV-6B–Infected Cells.
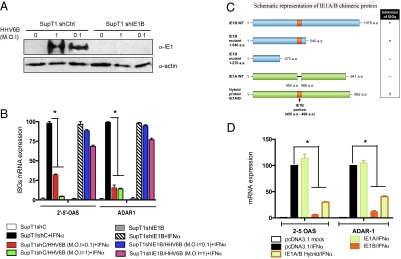
We made use of stable shIE1B knockdown SupT1 cells, which express constitutive shRNA specific for the IE1B transcript (SupT1-shIE1B), to suppress IE1 expression during HHV-6 infection. As shown in Fig. 6A, near total knockdown of IE1B expression is observed in SupT1-shIE1B cells following infection with two HHV-6 infectious doses. In uninfected cells (SupT1shC and SupT1-shIE1B), IFN-α efficiently stimulated the expression of ISGs (Fig. 6B). Control cells (shC) infected with HHV-6B displayed reduced ISGs activation; however, those in which IE1B was knockdown (shIE1B), ISGs activation levels were comparable to those of uninfected cells. These results sug-gest that the IE1 from HHV-6B specifically counteracts type I IFN signaling pathway during infection.
Fig. 6.
Knockdown of IE1 expression in HHV-6-infected cells restores type I IFN-induced transcriptional activation of ISGs. (A) Expression of IE1 in HHV-6-infected SupT1 cells stably expressing control (shCtrl) or IE1B specific shRNA (shIE1B). (B) Uninfected and 3-day-old infected cells were treated with exogenous IFN-α for 16 h and analyzed for 2' to 5' OAS and ADAR1 mRNA expression by RT-qPCR. Results are expressed as percentages of the ISG mRNA expression relative to mock-infected and IFN-stimulated SupT1-shC cells. Results are representative of two experiments. (C) Stick diagram of IE1 proteins. Orange rectangle depicts the 41 amino acid ISG inhibitory domain of IE1B subcloned into IE1A protein. Resulting chimera consists of 41 amino acid IE1B portion (orange) and WT 941 amino acid IE1A backbone (green). (D) Inhibition of IFN-α-induced ISG expression by chimeric IE1A/IE1B protein. 293T cells were transfected with control, IE1A, IE1B, and IE1A/IE1B hybrid expressing vectors. Twenty-four hours later, cells were stimulated with IFN-α, after which expression of ISG was assessed by RT-qPCR. Results are expressed as mean (triplicate) ± SD percentage of the ISG mRNA expression relative to pcDNA-transfected and IFN-treated cells after normalization of samples with GAPDH expression. Results are representative of three experiments. *, P < 0.01.
IE1A/IE1B Chimera Protein Is Capable of Inhibiting Type I IFN Signaling.
Global alignment of IE1 sequences from HHV-6 variants disclosed the presence of a 41 amino acid insertion within the 270–540 region of IE1B (amino acids 455–496), a region that is crucial for ISGs inhibition (Fig. 6C). We generated an IE1A hybrid protein carrying the 41 amino acid insertion from IE1B and tested whether IE1B's ISGs inhibitory activity could be transferred to IE1A. 293T cells were transfected with the IE1A/B hybrid expression vector and stimulated with IFN-α. As shown (Fig. 6D), the level of ISGs following IFN-α treatment is comparable in control and IE1A-expressing cells. In the presence of IE1B or IE1A/B hybrid protein, the expression levels of ISGs were considerably diminished. These results indicate that the transfer of the 41 amino acid segment from IE1B to IE1A protein results in a gain of function for IE1A, making this protein efficient at curtailing ISGs activation following IFN-α treatment.
Discussion
Human herpesviruses are excellent examples of well-adapted pathogens that can establish long-term relationships and persist within their hosts. Of them, HHV-6B colonizes nearly 100% of the human population, making it one of the most successful pathogens existing. In contrast, despite sharing >90% identity with HHV-6B, the A variant is encountered much less frequently in North America, Europe, and Japan (9, 21–26).
Previous reports have indicated that HHV-6 is sensitive to IFNs. In these studies, cells were either treated with IFN before infection or cultured for extended periods of time (10 d) before assessing IFN-α/β anti-HHV-6 effects (11, 12). Under these circumstances, the antiviral machinery triggered by IFN-α/β is induced before viral entry and can efficiently interfere with infection processes. Our data (Fig.1A) confirm that IFN-α pretreatment is effective at preventing infection of both HHV-6 variants. However, different results are obtained when cells are first infected with HHV-6 before stimulation with type I IFN. Under such circumstances, the antiviral effects of IFN-α/β on HHV-6B are almost completely negated, suggesting that this variant encodes proteins alleviating the antiviral effects of IFN. Under identical conditions, the growth of HHV6A was severely impaired, indicating that HHV-6A is less efficient at avoiding the antiviral effects of IFN. Our results indicate that HHV-6B- but not HHV-6A-infected cells respond improperly to IFN-α stimulation, with reduced expression of ISGs that are involved in the establishment of viral interference.
One of the most divergent proteins between HHV-6 variants, with amino acid identities ranging between 62 and 71% (depending on which viral strains are compared), is the IE1 protein (13, 14, 20). IE1 is a phosphorylated and sumoylated nuclear protein that is expressed within 1 h of infection (20, 27). The functions of IE1 during infection are currently unknown, but in recent work we reported that IE1 is efficient at preventing IFN-β gene transcription, indicating that it likely plays important roles for successful infection initiation and establishment of persistence (15). We provide evidence that IE1 from HHV-6B but not HHV-6A is very efficient at inhibiting ISG activation in response to type I IFNs.
We addressed the mechanism by which IE1B might prevent ISGs activation in response to IFN stimulation. The early activation steps (Jak kinases phosphorylation) were unaffected by IE1B; however, subsequent formation and binding of ISGF3 to ISRE elements were affected. The improper assembly of the ISGF3 complex likely originates from the ability of IE1B to bind to the coiled-coil motif of STAT2, a region previously identified as the IRF9 binding site (28). By binding to STAT2, IE1B would alter the association of IRF9 with STAT2 and the binding to ISRE elements, because in the absence of IRF9, STAT2 does not possess DNA-binding properties. The binding of IE1B to STAT2 also causes nuclear accumulation of STAT2, even though cells were not stimulated with IFN-α. Previous work indicated that under resting conditions STAT2 shuttles constantly from the cytoplasm to the nucleus (19, 29). The nuclear export of STAT2, being more efficient than its import, results in a predominance of STAT2 in the cytoplasm. However, in IE1B-expressing cells, the shuttling of STAT2 is affected. After reaching the nucleus, STAT2 binds to IE1B, preventing its export to the cytoplasm and causing its nuclear accumulation. Whether cofactors are involved in the nuclear shuttling of STAT2 is somewhat controversial. Banninger and Reich suggested that the cytoplasmic/nuclear shuttling of STAT2 was dependent on IRF9 (19). In contrast, Frahm et al. state that under resting conditions, the nuclear import of STAT2 is independent of IRF9 (29). The reasons for this divergence are at present unclear. Using the U2A cells (IRF9−), we observed that IE1B could physically interact with STAT2 and cause nuclear retention of STAT2, suggesting that under resting conditions, STAT2 can shuttle independently of IRF9.
We mapped the ISGs inhibitory domain between amino acids 270 and 540 of IE1B. We could correlate the inhibitory potential of the IE1B mutants to their STAT2 binding properties, suggesting that ISG inhibition is a direct consequence of IE1B binding to STAT2. The fact that IE1 from the HHV-6A variant proved ineffective at suppressing ISGs is of interest. IE1B is 1,078 amino acids long, compared with 941 for IE1A. Two insertions of 41 and 75 amino acids in IE1B account for the bulk of the difference in length between the IE1 proteins of both HHV-6 variants (20). Interestingly, one of these two insertions is located within the 270 to 540 region of IE1B that is responsible for ISG inhibition (20). The lack of such regions within IE1A could explain its inability to suppress ISG expression. Using an IE1A hybrid carrying the 41 amino acid insertion from IE1B (amino acids 455–496), we could transfer the ISGs inhibitory effects to IE1A, suggesting that this region represents the inhibitory domain.
To address the possibility that viral proteins other than IE1 may contribute to the type I IFN signaling defects observed in HHV-6-infected cells, we made use of RNA interference technology. We were able to demonstrate that in the absence of IE1B expression, cells regain their ability to respond to IFN-α stimulation and display normal ISG expression levels. These results suggest that IE1B is responsible for the majority of the inhibitory effects. Because the impact of IE1 knockdown on the HHV-6 life cycle is currently unknown, we cannot exclude potential effects from other viral proteins.
The diversity of molecular mechanisms evolved by viruses to block IFN signaling is enormous. Interestingly, HCMV IE1, which shares no sequence homology with HHV-6 IE1, affects the binding of ISGF3 to ISG promoters (30), possibly through binding of STAT2 via a 54 amino acid acidic motif (pI 3.2) located near the COOH terminus (31). SUMOylation of K450 within the acidic domain could reverse ISG inhibition by HCMV IE1 (31). Analysis of the 41 amino acid region of HHV-6 IE1B responsible for ISG inhibition reveals no such acidic motif (pI 6.8). Furthermore, our results indicate that the K802R SUMOylation-negative mutant of IE1 (20) is as effective as WT IE1 at suppressing ISG expression, indicating that HHV-6 and CMV have evolved divergent inhibitory mechanisms.
In summary, this study is unique in reporting that HHV-6B abrogates IFN-α/β signaling. Considering that type I IFNs are often used to treat viral infections, our results could have medical implications. Work by Hong et al. teaches us that in a cohort of multiple sclerosis patients, a direct correlation existed between the HHV-6 plasmatic viral loads and the duration of IFN-α treatment suggesting potential resistance of HHV-6 to short term IFN therapy (32). This situation would be somewhat analogous to our experiments showing resistance of HHV-6–infected cell to IFN-α/β. In addition, work by Gracia-Montojo et al. indicated that in a cohort of multiple sclerosis patients treated with IFN-β, the prevalence of plasmatic HHV-6 DNA was not statistically different from those from untreated multiple sclerosis patients (33). These results suggest limited efficacy of IFN-β as anti-HHV-6 therapy and that prolonged IFN administration is needed to obtain an antiviral effect. These results are in agreement with our findings that HHV-6-infected cells are resistant to type I IFN therapy. Further work is needed to determine the relative usefulness of IFN as a therapy for HHV-6.
Materials and Methods
Cell Culture, Virus Production, and Infection.
SupT1 cells were cultured in RPMI medium 1640 (Sigma-Aldrich) supplemented with 10% FBS (FBS). The 293T cell line (ATCC), 2fTGH and 2fTGH-derived cell lines U2A, U3A, and U6A (generously provided by G. Stark, Lerner Research Institute, Cleveland, OH) were grown in DMEM (Sigma-Aldrich). A549 cells (ATCC) were cultured in F12 media (Invitrogen) supplemented with 10% FBS. The HHV-6A GS, U1102, and HHV-6 Z29 HST strains were propagated and concentrated as described (20). Primary isolates of HHV6B subtype were obtained from the HHV-6 Foundation, and were propagated in PHA-P (3 μg/mL) activated cord blood mononuclear cells (provided by HHV-6 Foundation). The HHV-6 primary isolates infectivity titer in Molt-3 cells was determined 24 h after infection, as previously described (15).
Cells were treated with human IFNα (HUIFN-α2a), human IFNβ (HUIFN-β 1b), (PBL Interferon Source), or IFNγ (Cedarlane) added to maintenance medium at different concentration: 10 to 1,000 U/mL (IFN-α or IFN-β) and 50 ng/mL (IFNγ).
PBMC Isolation and Infection.
PBMCs were obtained from healthy donors after the centrifugation of venous blood over Ficoll-Paque PLUS gradient (GE Healthcare) followed by three washes with PBS. Obtained PBMC were incubated in the presence of PHA-P (1.5 μg/mL) for 3 d, after which HHV6A and HHV6B infections were performed in RPMI medium containing 10% FBS, as described (20).
Plasmids, Transient Transfection, Reporter-Gene Assays.
IE1 expression plasmids were previously described (15, 27). ISRE-Luc and GAS-Luc reporter plasmids were purchased from Clontech. GFP-STAT2 constructs (19) were kindly provided by Nancy Reich, Stony Brook University, New York. 293T cells grown in 12-well plates to 50 to 70% confluence were transfected using the calcium phosphate precipitation procedures (15). Thirty-six hours posttransfection, cells were stimulated with IFNα, IFNβ (10, 100, 1,000, and 1,500 U/mL), or IFNγ (50 ng/mL) and incubated for 8 and 16 h, respectively. Luciferase activity was measured as described (34).
Protein–Protein Interaction Assays, Immunobloting.
For protein immunoprecipitation, transfected cells were lysed in RIPA buffer. Whole-cell extracts were immunoprecipitated with anti-IE1 sera (20) and protein G-coated Sepharose beads (Amersham). Proteins were separated by SDS/PAGE, transferred to nitrocellulose membranes, and detected with anti-IE1 (20), anti-IRF9 (ISGF3γ), anti STAT1 p84/p91 (E-23), anti-p-STAT1 (Tyr-701), anti-STAT2 (H-190), anti-p-STAT2 (Tyr-690) (Santa Cruz Biotechnology), and antiactin (C2) antibodies (Jackson ImmunoResearch Laboratories).
Electrophoretic Mobility-Shift Assay.
Nuclear extracts were prepared from “superinduced” (18 h of rhIFN-γ, followed by 30 min of IFN-α) IE1A, IE1B, and control-vector nucleofected A549 cells. Samples were assayed for ISGF3 binding in gel-shift analysis using a 32P-labeled double-stranded oligonucleotide corresponding to the ISRE of the IFN-inducible ISG15 gene (5′GGCTTCAGTTTCGGTTTCCCTTTCCCGAG-3′). For competition assays, excess of cold probe were added to the mixture. After migration, gels were dried and exposed to imaging plates (FujiFilm Canada).
RNA Interference, Generation of shIE1 Stable, and Inducible Cell Lines.
shRNA oligos used for targeting IE1 are listed in Table S1. After annealing, these shRNA oligos were cloned into pTER+ expression vector (35). SupT1 cells stably expressing the tetracycline repressor from the pcDNA6/Tr plasmid (Invitrogen) were electroporated (250V, 960 μF, 25 ms) with pTER-shControl, pTER-shIE1A, and pTER-shIE1B plasmids and selected with Zeocin (85 μg/mL).
Immunofluorescence.
A549 cells and derivatives were nucleofected with 5 μg of control, IE1A-, or IE1B-expressing vector and processed for immunofluorescence, as described (15).
Real-Time Quantitative PCR.
Measurements of gene expression was performed as described (15). Primer pairs and probes are listed in Table S1. All samples were normalized using GAPDH gene expression. Intracellular HHV-6 genomic copy numbers were determined as described (15).
Statistical Analysis.
Experimental groups were compared using unpaired two-tailed t test. Results were considered significantly different when P < 0.05.
Supplementary Material
Acknowledgments
We acknowledge the valuable ongoing collaboration with the HHV-6 Foundation. This work was funded in part by Grant MOP-89706 from the Canadian Institutes of Health Research and a senior scholarship from Fonds de la Recherche en Santé Québec (to L.F.)
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909951107/DCSupplemental.
References
- 1.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 2.Honda K, Yanai H, Takaoka A, Taniguchi T. Regulation of the type I IFN induction: A current view. Int Immunol. 2005;17:1367–1378. doi: 10.1093/intimm/dxh318. [DOI] [PubMed] [Google Scholar]
- 3.Marié I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miettinen M, Sareneva T, Julkunen I, Matikainen S. IFNs activate toll-like receptor gene expression in viral infections. Genes Immun. 2001;2:349–355. doi: 10.1038/sj.gene.6363791. [DOI] [PubMed] [Google Scholar]
- 5.Sato M, et al. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 1998;441:106–110. doi: 10.1016/s0014-5793(98)01514-2. [DOI] [PubMed] [Google Scholar]
- 6.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 7.Salahuddin SZ, et al. Isolation of a new virus, HBLV, in patients with lympho-proliferative disorders. Science. 1986;234:596–601. doi: 10.1126/science.2876520. [DOI] [PubMed] [Google Scholar]
- 8.Yamanishi K, et al. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet. 1988;1:1065–1067. doi: 10.1016/s0140-6736(88)91893-4. [DOI] [PubMed] [Google Scholar]
- 9.Zerr DM, et al. Clinical outcomes of human herpesvirus 6 reactivation after hematopoietic stem cell transplantation. Clin Infect Dis. 2005;40:932–940. doi: 10.1086/428060. [DOI] [PubMed] [Google Scholar]
- 10.Hall CB, et al. Persistence of human herpesvirus 6 according to site and variant: Possible greater neurotropism of variant A. Clin Infect Dis. 1998;26:132–137. doi: 10.1086/516280. [DOI] [PubMed] [Google Scholar]
- 11.Kikuta H, et al. Interferon induction by human herpesvirus 6 in human mononuclear cells. J Infect Dis. 1990;162:35–38. doi: 10.1093/infdis/162.1.35. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi K, et al. Interferon and natural killer cell activity in patients with exanthem subitum. Pediatr Infect Dis J. 1992;11:369–373. doi: 10.1097/00006454-199205000-00006. [DOI] [PubMed] [Google Scholar]
- 13.Dominguez G, et al. Human herpesvirus 6B genome sequence: Coding content and comparison with human herpesvirus 6A. J Virol. 1999;73:8040–8052. doi: 10.1128/jvi.73.10.8040-8052.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isegawa Y, et al. Comparison of the complete DNA sequences of human herpesvirus 6 variants A and B. J Virol. 1999;73:8053–8063. doi: 10.1128/jvi.73.10.8053-8063.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaworska J, Gravel A, Fink K, Grandvaux N, Flamand L. Inhibition of transcription of the beta interferon gene by the human herpesvirus 6 immediate-early 1 protein. J Virol. 2007;81:5737–5748. doi: 10.1128/JVI.02443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seegert D, et al. A novel interferon-alpha-regulated, DNA-binding protein participates in the regulation of the IFP53/tryptophanyl-tRNA synthetase gene. J Biol Chem. 1994;269:8590–8595. [PubMed] [Google Scholar]
- 17.Li X, Leung S, Qureshi S, Darnell JE, Jr, Stark GR. Formation of STAT1-STAT2 heterodimers and their role in the activation of IRF-1 gene transcription by interferon-alpha. J Biol Chem. 1996;271:5790–5794. doi: 10.1074/jbc.271.10.5790. [DOI] [PubMed] [Google Scholar]
- 18.Pellegrini S, John J, Shearer M, Kerr IM, Stark GR. Use of a selectable marker regulated by alpha interferon to obtain mutations in the signaling pathway. Mol Cell Biol. 1989;9:4605–4612. doi: 10.1128/mcb.9.11.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banninger G, Reich NC. STAT2 nuclear trafficking. J Biol Chem. 2004;279:39199–39206. doi: 10.1074/jbc.M400815200. [DOI] [PubMed] [Google Scholar]
- 20.Gravel A, Gosselin J, Flamand L. Human herpesvirus 6 immediate-early 1 protein is a sumoylated nuclear phosphoprotein colocalizing with promyelocytic leukemia protein-associated nuclear bodies. J Biol Chem. 2002;277:19679–19687. doi: 10.1074/jbc.M200836200. [DOI] [PubMed] [Google Scholar]
- 21.Dewhurst S, McIntyre K, Schnabel K, Hall CB. Human herpesvirus 6 (HHV-6) variant B accounts for the majority of symptomatic primary HHV-6 infections in a population of U.S. infants. J Clin Microbiol. 1993;31:416–418. doi: 10.1128/jcm.31.2.416-418.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall CB, et al. Characteristics and acquisition of human herpesvirus (HHV) 7 infections in relation to infection with HHV-6. J Infect Dis. 2006;193:1063–1069. doi: 10.1086/503434. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka-Taya K, et al. Seroepidemiological study of human herpesvirus-6 and -7 in children of different ages and detection of these two viruses in throat swabs by polymerase chain reaction. J Med Virol. 1996;48:88–94. doi: 10.1002/(SICI)1096-9071(199601)48:1<88::AID-JMV14>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 24.van Loon NM, et al. Direct sequence analysis of human herpesvirus 6 (HHV-6) sequences from infants and comparison of HHV-6 sequences from mother/infant pairs. Clin Infect Dis. 1995;21:1017–1019. doi: 10.1093/clinids/21.4.1017. [DOI] [PubMed] [Google Scholar]
- 25.Ward KN, Thiruchelvam AD, Couto-Parada X. Unexpected occasional persistence of high levels of HHV-6 DNA in sera: detection of variants A and B. J Med Virol. 2005;76:563–570. doi: 10.1002/jmv.20399. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto T, Mukai T, Kondo K, Yamanishi K. Variation of DNA sequence in immediate-early gene of human herpesvirus 6 and variant identification by PCR. J Clin Microbiol. 1994;32:473–476. doi: 10.1128/jcm.32.2.473-476.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gravel A, Dion V, Cloutier N, Gosselin J, Flamand L. Characterization of human herpesvirus 6 variant B immediate-early 1 protein modifications by small ubiquitin-related modifiers. J Gen Virol. 2004;85:1319–1328. doi: 10.1099/vir.0.19610-0. [DOI] [PubMed] [Google Scholar]
- 28.Martinez-Moczygemba M, Gutch MJ, French DL, Reich NC. Distinct STAT structure promotes interaction of STAT2 with the p48 subunit of the interferon-alpha-stimulated transcription factor ISGF3. J Biol Chem. 1997;272:20070–20076. doi: 10.1074/jbc.272.32.20070. [DOI] [PubMed] [Google Scholar]
- 29.Frahm T, Hauser H, Köster M. IFN-type-I-mediated signaling is regulated by modulation of STAT2 nuclear export. J Cell Sci. 2006;119:1092–1104. doi: 10.1242/jcs.02822. [DOI] [PubMed] [Google Scholar]
- 30.Paulus C, Krauss S, Nevels M. A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling. Proc Natl Acad Sci USA. 2006;103:3840–3845. doi: 10.1073/pnas.0600007103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huh YH, et al. Binding STAT2 by the acidic domain of human cytomegalovirus IE1 promotes viral growth and is negatively regulated by SUMO. J Virol. 2008;82:10444–10454. doi: 10.1128/JVI.00833-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hong J, Tejada-Simon MV, Rivera VM, Zang YC, Zhang JZ. Anti-viral properties of interferon beta treatment in patients with multiple sclerosis. Mult Scler. 2002;8:237–242. doi: 10.1191/1352458502ms794oa. [DOI] [PubMed] [Google Scholar]
- 33.Garcia-Montojo M, De Las Heras V, Bartolome M, Arroyo R, Alvarez-Lafuente R. Interferon beta treatment: bioavailability and antiviral activity in multiple sclerosis patients. J Neurovirol. 2007;13:504–512. doi: 10.1080/13550280701649670. [DOI] [PubMed] [Google Scholar]
- 34.Gravel A, Tomoiu A, Cloutier N, Gosselin J, Flamand L. Characterization of the immediate-early 2 protein of human herpesvirus 6, a promiscuous transcriptional activator. Virology. 2003;308:340–353. doi: 10.1016/s0042-6822(03)00007-2. [DOI] [PubMed] [Google Scholar]
- 35.van de Wetering M, et al. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 2003;4:609–615. doi: 10.1038/sj.embor.embor865. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.