Abstract
Functionally distinct chromatin domains are delineated by distinct posttranslational modifications of histones, and in some organisms by differences in DNA methylation. Proper establishment and maintenance of chromatin domains is critical but not well understood. We previously demonstrated that heterochromatin in the filamentous fungus Neurospora crassa is marked by cytosine methylation directed by trimethylated Lysine 9 on histone H3 (H3K9me3). H3K9me3 is the product of the DIM-5 Lysine methyltransferase and is recognized by a protein complex containing heterochromatin protein-1 and the DIM-2 DNA methyltransferase. To identify additional components that control the establishment and function of DNA methylation and heterochromatin, we built a strain harboring two selectable reporter genes that are silenced by DNA methylation and employed this strain to select for mutants that are defective in DNA methylation (dim). We report a previously unidentified gene (dim-7) that is essential for H3K9me3 and DNA methylation. DIM-7 homologs are found only in fungi and are highly divergent. We found that DIM-7 interacts with DIM-5 in vivo and demonstrated that a conserved domain near the N terminus of DIM-7 is required for its stability. In addition, we found that DIM-7 is essential for recruitment of DIM-5 to form heterochromatin.
Keywords: DNA methylation, heterochromatin, KMT, histone methylation, Neurospora
Heterochromatin was originally defined based on its cytological properties. This densely staining component of the genome remains condensed throughout the cell cycle and is typically rich in repeated DNA (1). Proper regulation of heterochromatin is critical, both because heterochromatin formation can silence genes and because heterochromatin is essential for normal centromere function (1). Heterochromatin in Neurospora crassa shares molecular features with heterochromatin in higher eukaryotes, including A:T-rich, repetitive DNA, Histone H3 trimethyl Lysine 9 (H3K9me3), Heterochromatin Protein-1 (HP1), and DNA methylation (2, 3). The A:T-rich DNA found in the N. crassa genome is almost exclusively the product of the homology-dependent genome defense mechanism RIP (repeat-induced point mutation), which operates in the sexual phase of the life cycle (4). All DNA methylation in N. crassa is dependent on the DIM-5 H3 Lysine methyltransferase (KMT), which catalyzes trimethylation of H3K9 associated with RIP'd DNA (5). HP1 recognizes and binds H3K9me3 and directs DNA methylation by recruiting the DNA methyltransferase (DNMT), DIM-2, through interaction of the chromo shadow domain of HP1 and PXVXL-like motifs in DIM-2 (6, 7). Although much has been learned about the steps leading to DNA methylation, the mechanisms responsible for controlling DIM-5 remain unclear.
Although the control of H3K9 methylation has not been fully elucidated in any organism, it is currently best understood in the fission yeast, Schizosaccharomyces pombe. Establishment and maintenance of H3K9me at centromeres, telomeres, and the silent mating-type locus of S. pombe requires the KMT complex ClrC, as well as components of the RNA interference machinery and the HP1 homolog, Swi6 (1). Studies with other organisms have implicated small RNA pathways (8–10), DNA methylation (11), DNA binding proteins (12–14), and histone modifications (15) in the control of H3K9 methylation. In contrast to S. pombe, neither the RNAi machinery nor HP1 is required for the establishment or maintenance of H3K9me3 in N. crassa (2, 7, 16).
Biochemical analyses revealed that modification of certain residues on H3 inhibit the activity of H3 K9 KMTs (15). It is therefore possible that DIM-5 scans the entire genome and catalyzes H3K9 methylation only upon “reading” the correct pattern of histone modifications. Recent work demonstrated that H3 S10 dephosphorylation by protein phosphatase-1 is required for normal H3K9 methylation in N. crassa, lending credibility to this hypothesis (17). It is also possible, however, that DIM-5 is recruited to A:T-rich regions of the genome to establish H3K9 methylation. A:T-rich, RIP'd DNA is a potent signal for de novo H3K9 methylation and subsequent DNA methylation (2, 18, 19). Treatment of N. crassa with the drug Distamycin-A, which binds to the minor groove of A:T-rich DNA, leads to a loss of DNA methylation (19), suggesting that an A:T-rich binding factor directs DIM-5 to RIP'd DNA. Functionally relevant proteins that recognize A:T-rich DNA in N. crassa are currently unknown.
It is notable that DNA methylation inhibits transcription elongation in N. crassa, thereby silencing the expression of methylated sequences (20). Here we report the development of a dual reporter strain harboring two selectable markers that are silenced in a DNA methylation-dependent manner. We used this strain to select for mutants that are defective in DNA methylation, which led to the identification of DIM-7. We demonstrate that DIM-7 is essential for both H3K9 and DNA methylation. In addition, we show that DIM-7 interacts with DIM-5 and is required for recruitment of DIM-5 to A:T-rich, RIP'd DNA to form heterochromatin.
Results
Reporter Strain Construction.
A copy of the bacterial hph gene flanked by RIP'd copies of the N. crassa am gene (amRIP-hphm-amRIP) is reversibly silenced by DNA methylation that spreads from the mutated flanking sequences (21). Unfortunately, this methylated hph allele is ineffective for direct selection of Dim− mutants because of relatively high rates of spontaneous reactivation (∼10−2 to 10−5). We therefore designed a second methylated reporter gene, reasoning that simultaneous reactivation of two methylated genes would be a rare event. We built a construct containing the bacterial bar gene flanked by an inverted duplication of the N. crassa his-3 gene. The bar gene should not be subject to RIP or to excision by homologous recombination between the flanking his sequences, whereas the his sequences should be susceptible to RIP and DNA methylation (21). The relevant sequences were introduced into the his-3 locus of a strain that contains amRIP-hphm-amRIP (N1674) by targeted gene replacement and a primary transformant was crossed to obtain homokaryotic progeny and to trigger RIP. Three strains were identified with methylated copies of hph and bar, and one of these strains (N2984) was crossed again to introduce an inl deletion allele and to recover equivalent strains of both mating types (N2977 and N3311).
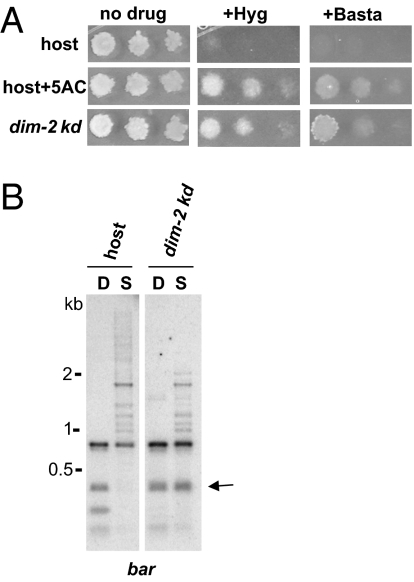
As one test of whether both the hph gene and the bar gene were reversibly silenced by DNA methylation, we grew strain N2977 in the presence or absence of the DNMT inhibitor 5-azacytidine (5AC) and assayed serial dilutions of conidial suspensions for growth on media with and without hygromycin or basta. The reporter strain was able to grow on media containing hygromycin or basta if treated with 5AC, but not otherwise (Fig. 1A), suggesting that both the hph and bar genes were indeed silenced by DNA methylation. We also tested if inactivation of the dim-2 DNMT gene by quelling (i.e., RNA-dependent gene silencing) (22) would similarly reactivate the genes. The reporter strain (N2984) was cotransformed with fragments of dim-2 along with a plasmid containing the selectable Bml gene (23). Cotransformants are efficiently recovered in N. crassa and we expected a high fraction of them (∼50%) to show quelling of dim-2 (22). Indeed, we readily identified several such strains by plating conidia of transformants on hygromycin and basta (Fig. 1A). Southern hybridizations using the methyl-cytosine-sensitive restriction enzyme, Sau3AI, and its methyl-cytosine-insensitive isoschizomer, DpnII, confirmed that reactivation of expression was correlated with reduced DNA methylation of the bar reporter gene (Fig. 1B). For example, the dim-2 knock-down strain N3854 was resistant to basta and showed reduced DNA methylation in bar sequences, unlike its parental strain.
Fig. 1.
DNA methylation-dependent silencing of hphm and barm reporter genes. (A) Suspensions of 104, 103, or 102 conidia of strain N2977, grown in the absence (host) or presence of 5-azacytidine (host+5AC), and a dim-2 knock-down strain (dim-2 kd; strain N3854) were spot-tested on media with or without basta or hygromycin. (B) Genomic DNA from the parental strain (host) and a dim-2 knockdown (dim-2 kd) was digested with the methyl-cytosine-sensitive restriction enzyme Sau3AI or its insensitive isoschizomer, DpnII, and used for Southern hybridizations with a probe corresponding to the bar reporter gene. The arrow indicates loss of DNA methylation in the dim-2 kd strain.
We next asked if the simultaneous reversion frequency of both markers was significantly lower than the reversion frequencies for the individual markers. Indeed, although spontaneous hygromycin- or basta-resistant colonies arose at an average frequency of ∼1 × 10−5, conidia plated on both drugs gave rise to revertants at frequency of 2.7 × 10−7 (Table S1). Although the combined reversion frequency was greater than would be expected if silencing of the markers were completely independent, we expected that it would be adequate to select Dim− mutants.
Selection of Dim Mutants.
In a UV-mutagenesis experiment, ∼500,000 conidia of the dual reporter strain (N2977) were exposed to UV light and challenged with basta. Next, 154 basta-resistant colonies were isolated and tested for hygromycin resistance. About one-third (48/154) grew in the presence of both drugs and Southern hybridization revealed reduced DNA methylation in 47 of these strains. These strains were crossed to the dual reporter strain of the opposite mating type (N3311) to attempt to isolate homokaryotic progeny. Dim− progeny strains were successfully isolated from 10 independent Dim− mutants. Complementation tests demonstrated that 3 of the 10 Dim− isolates comprised a single complementation group defining the dim-7 gene and preliminary mapping localized dim-7 to the right arm of LGV (24).
dim-7 Is Essential for DNA Methylation.
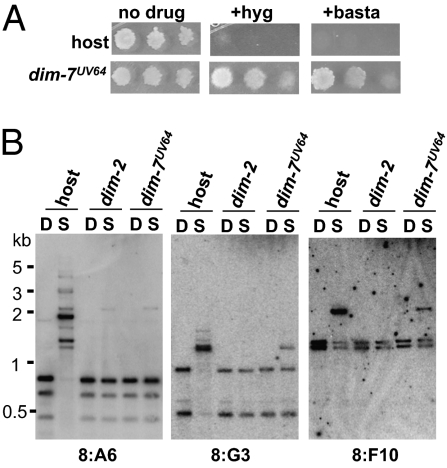
Initial characterization of the dim-7UV64 strain (N3312) revealed that it exhibited robust growth on media containing basta or hygromycin (Fig. 2A). We probed Sau3AI- and DpnII-digested DNA from dim-7UV64, along with methylation-positive (WT) and negative (dim-2) control strains, for known methylated regions and found that DNA methylation was reduced at all regions tested. At the 8:A6 region, dim-7UV64 exhibited an apparent complete loss of DNA methylation, like the dim-2 strain; methylation was significantly reduced in the dim-7UV64 strain at both 8:G3 and 8:F10 (Fig. 2B).
Fig. 2.
The dim-7UV64 strain exhibits reduced DNA methylation. (A) Suspensions of 104, 103, or 102 conidia from strains N2977 (host) and N3312 (dim-7UV64) were spot-tested on media with or without basta or hygromycin, as indicated. (B) Genomic DNA from strain N2977 (host), N1851 (Δdim-2), and N3312 (dim-7UV64) was digested with Sau3AI or DpnII and used for Southern hybridizations with probes corresponding to the methylated 8:A6, 8:G3, or 8:F10 regions (3).
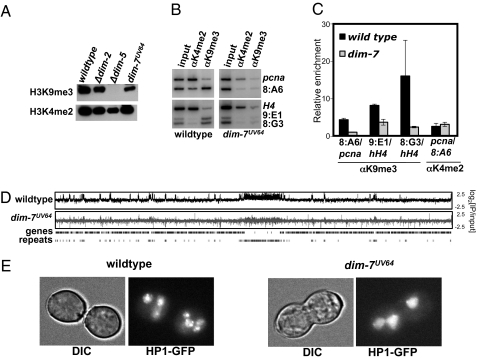
To determine if DIM-7 is required for H3K9 methylation, which is essential for DNA methylation in N. crassa (5), we performed Western analyses on histones isolated from WT, dim-2, dim-5, and dim-7UV64 strains using antibodies specific for H3K9me3. The dim-7UV64 strain showed reduced H3K9me3 (Fig. 3A). We next performed ChIP assays to examine H3K9me3 levels at three methylated regions, 8:A6, 9:E1, and 8:F10, and found a striking reduction in H3K9me3 signals at all three loci (Fig. 3 B and C). To examine the generality of the reduction, we performed ChIP-chip analyses using a high-density LGVII microarray (2). A marked reduction in H3K9me3 was found in virtually all methylated regions of the dim-7UV64 strain, consistent with traditional ChIP experiments (Fig. 3D and Fig. S1). Similarly, cytological examination of an HP1-GFP transgene showed an apparent loss of heterochromatin in the dim-7UV64 strain (Fig. 3E). Whereas HP1-GFP is typically localized to three or four nuclear foci (6), the dim-7 mutant displayed diffuse nuclear localization, although a single focus of HP1-GFP was observed in some nuclei. These results are comparable to those observed previously for the N. crassa dim-5 strain, which lacks H3K9me3 (6).
Fig. 3.
H3K9me3 levels are globally reduced in the dim-7UV64 mutant. (A) Histones were extracted from WT (N150), Δdim-2 (N1851), Δdim-5 (N3074), and dim-7UV64 (N3312) strains and subjected to Western blotting using antibodies to H3K9me3 or H3K4me2. (B) For WT and the dim-7UV64 strain, radio-labeled products obtained by multiplex PCR using using whole-cell extracts (input) or the indicated immunoprecipitate fraction as a template are shown. PCR products corresponding to heterochromatin (8:A6, 9:E1, and 8:G3) (3) or control euchromatin regions (H4 and pcna) are indicated at the right of the panel. (C) The average enrichment of each PCR product relative to the indicated euchromatin internal control is shown graphically for three independent experiments. (D) The distribution of H3K9me3 on LGVII in WT and dim-7UV64. Enrichment values (IP/input) for ChIP-chip experiments using antibodies to H3K9me3 are shown as log2 values. (E) The distribution of HP1-GFP is shown in multinucleate conidia for WT and the dim-7UV64 strain (DIC, differential interference contrast).
Identification of dim-7.
Genetic mapping localized dim-7 to a 431-kb region on LGVR (24), but we were unable identify the gene by complementation with cosmid clones covering the region. Fortunately, in a separate study of proteins associated with DIM-5, a candidate gene was identified. Of 69 candidate DIM-5-interacting proteins identified by mass spectrometry, we obtained the most complete peptide coverage for a protein encoded by NCU04152 (31.7% coverage) (Fig. S2), a gene that resides within the LGV region harboring dim-7. We therefore isolated and sequenced NCU04152 from the dim-7UV64 strain (N3312) and identified a 125-bp deletion at the 3′ end of the gene. The deletion removed 34 codons at the predicted 3′ end of the coding sequence, including the stop codon, and should result in translation of the 3′ UTR to produce an extension of 35 amino acids at the C terminus of the mutant protein.
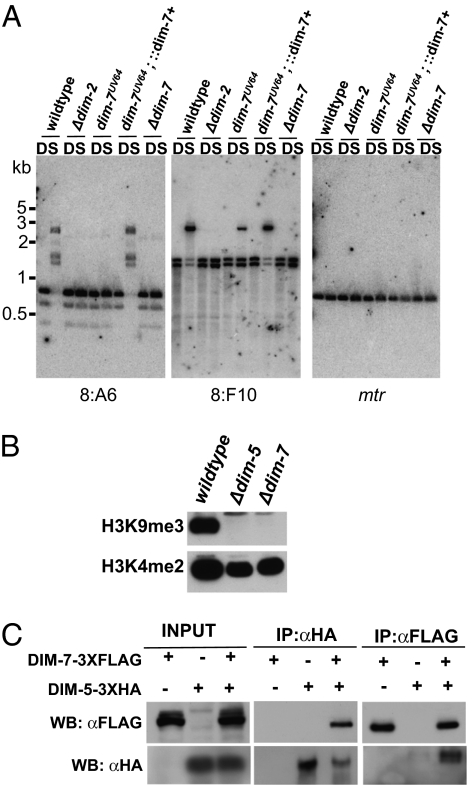
To confirm that this mutation is responsible for the Dim− phenotype, we amplified NCU04152 from a WT strain by PCR and introduced it into a dim-7UV64 strain by cotransformation with a plasmid conferring benomyl-resistence. Five of six benomyl-resistant transformants exhibited increased DNA methylation relative to the dim-7UV64 host strain. Restoration of DNA methylation is shown for a representative transformant in Fig. 4A. We conclude that the deletion within the 3′ end of NCU04152 is responsible for the defects in H3K9 and DNA methylation observed in the dim-7UV64 strain. Two additional alleles of dim-7 were sequenced; curiously, both alleles displayed distinct single nucleotide missense mutations within the stop codon. In particular, dim-7UV30 and dim-7UV68 contained TAA (stop) to GAA (Glutamate) and TAA (stop) to TTA (Leucine) changes, respectively, which should extend the protein by 16 amino acids. All three mutant alleles are predicted to encode nearly full-length proteins and therefore may not result in complete loss of function.
Fig. 4.
DIM-7 is essential for DNA and H3K9 methylation and interacts with DIM-5. (A) Genomic DNA from WT (N150), Δdim-2 (N1851), dim-7UV64 (N3312), dim-7UV64;::dim-7+(N3868), and Δdim-7 (N3855) was digested using Sau3AI or DpnII and used for Southern hybridizations with probes corresponding to the methylated 8:A6 or 8:F10 regions (3). The blots were also probed with mtr, an unmethylated gene, to confirm that the digests were complete. (B) Histones were extracted from WT (N150), Δdim-5 (N3074), and Δdim-7 (N3855) and subjected to Western blotting using antibodies to H3K9me3 or H3K4me2. (C) Immunoprecipiation experiments were performed using extracts from strains expressing DIM-7–3XFLAG, DIM-5–3XHA, or both (indicated by + or −). The input fraction, the α-HA immunoprecipitate fraction (IP:αHA), and the α-FLAG immunoprecipitate fraction (IP:αFLAG) were subjected to Western blotting and probed with the α-FLAG or α-HA antibodies as indicated (WB).
To determine if dim-7 is dispensable for viability, and if so is essential for H3K9 and DNA methylation, we attempted to generate a homokaryotic dim-7 knockout strain. We obtained a heterokaryon strain in which dim-7 had been replaced with an hph cassette (Δdim-7) from the Neurospora genome project (25), crossed this strain to a Sad-1 strain to prevent meiotic silencing by unpaired DNA (26), and isolated hygromycin-resistant progeny. Hygromycin-resistant progeny exhibited severe growth defects but were viable, indicating that dim-7 is not essential. We then examined DNA methylation in the homokaryotic null mutant and found that it showed no DNA methylation, in contrast to the case with the three alleles isolated in our selection (Fig. 4A). Similarly, Western blots revealed that the Δdim-7 strain displayed an apparent complete loss of H3K9me3, like dim-5 strains (Fig. 4B).
DIM-7 Interacts with DIM-5 and Requires a Conserved Domain Near the DIM-7 N Terminus.
The fact that mass spectrometry of DIM-5-associated proteins revealed DIM-7 suggested that these proteins interact, either directly or indirectly. To explore the possibility that DIM-7 is a bona fide binding partner of DIM-5, we created a 3XFLAG, C-terminal tagged version of DIM-7 by using a “knock-in” approach and visualized the fusion protein by Western blotting (27). The DIM-7–3XFLAG protein showed an apparent molecular weight of ∼80 kDa, consistent with its predicted size. The dim-7–3xflag strain was then crossed to a strain containing a dim-5–3xha construct to generate a strain containing tagged versions of both DIM-7 and DIM-5. We examined DNA methylation levels in these strains to verify that the fusion proteins were functional (Fig. S3A) and performed coimmunoprecipitation experiments to assay for interaction. Western blots of the input fraction and of the immunoprecipitate obtained with the anti-FLAG antibodies revealed that both DIM-7–3XFLAG and DIM-5–3XHA were present in both the input and the immunoprecipitate (Fig. 4C). We next performed the reciprocal immunoprecipitation with antibodies to HA. Similarly, both DIM-7–3XFLAG and DIM-5–3XHA were detected in the input and immunoprecipitate. These data confirm that DIM-7 and DIM-5 interact in vivo. We failed to observe a direct interaction between DIM-5 and DIM-7 in vitro or using the yeast two-hybrid assay, raising the possibility that the proteins interact via an intermediary in a protein complex.
BLAST searches using the predicted DIM-7 protein identified similar proteins in filamentous fungi but failed to identify similar proteins outside of this group and searches of the SMART or PFAM domain databases failed to detect any conserved domains within the predicted N. crassa DIM-7 sequence (28, 29). In a further attempt to identify potential homologs of DIM-7, we performed PSI-BLAST using the predicted N. crassa DIM-7 sequence. Multiple iterations of PSI-BLAST revealed limited similarity between DIM-7 and a predicted Raf2p homolog from Schizosaccharomyces japonicus (XP_002173332), suggesting that DIM-7 is a homolog of S. pombe Raf2p, which has been identified as a partner of Clr4 in the ClrC complex (30). We performed sequence alignments with several DIM-7 homologs, plus S. pombe Raf2p, and found that a C2H2 zinc finger domain and a domain near the N terminus were conserved among DIM-7 homologs from filamentous fungi and S. pombe Raf2p, whereas most of the protein was not well conserved (Fig. S4).
To determine if either of the conserved domains are required for interaction with DIM-5 or for DNA methylation, we introduced a DIM-7–3XFLAG fusion construct, or three mutant versions of the FLAG-tagged protein, at the his-3 locus of a Δdim-7 strain (Fig. S4). Deletion or mutation of the C2H2 domain resulted in slightly reduced expression but did not abolish interaction with DIM-5 or DNA methylation (Fig. S3 B and C). In contrast, we failed to detect any DIM-7–3XFLAG harboring a deletion of the conserved N-terminal domain, suggesting that this domain is necessary for stability of the protein. Attempts to immunoprecipitate the altered protein failed to yield detectable DIM-7–3XFLAG or DIM-5, and this construct failed to complement the DNA methylation defect of the parental strain (Fig. S3 B and C).
DIM-7 Is Required for Recruitment of DIM-5 to Heterochromatin.
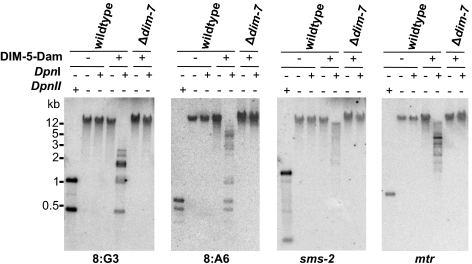
Western blots revealed that DIM-7 is not required for expression or stability of DIM-5 (Fig. S3C), but it seemed possible that DIM-7 is required to direct DIM-5 to heterochromatin domains. ChIP assays to look for DIM-5–3XFLAG or DIM-7–3XFLAG at several heterochromatic regions failed to detect significant enrichment of the tagged proteins. We therefore tested the DamID method, which utilizes Escherichia coli DNA adenine methyltransferase (Dam) to probe chromatin (31), to examine the genomic localization of DIM-5, reasoning that this method may be more sensitive than ChIP. We created a DIM-5-Dam translational fusion and introduced this construct into a dim-5 strain. The fusion protein partially complemented DNA methylation, indicating that it was partially functional (Fig. S3D). We next introduced this construct into Δdim-7 and dim+ strains and tested for Dam activity by treating genomic DNA with DpnI, which specifically cuts GATC sites containing methylated adenine bases, but does not digest unmethylated GATC sites. The digested DNA was fractionated by electrophoresis and probed for the heterochromatin regions 8:G3 and 8:A6, as well as for the euchromatic genes mtr and sms-1 (Fig. 5). In the wild-type background, the heterochromatin probes detected low molecular-weight fragments corresponding to completely digested DNA and some intermediate molecular-weight fragments corresponding to partially digested DNA. In contrast, only high molecular-weight DNA was detected in the Δdim-7 background. Importantly, probes for sms-2 and mtr hybridized to high molecular-weight DNA, corresponding to largely undigested DNA in both the WT and dim-7 strains. These results were reproducible in replicate experiments and for independently isolated dim+ and Δdim-7 strains. Thus, these data suggest that DIM-5 is specifically recruited to heterochromatin regions in the wild-type background and that this recruitment depends on DIM-7.
Fig. 5.
DIM-7 is required for recruitment of DIM-5 to heterochromatin domains. Genomic DNA from WT (N150), which does not express DIM-5-Dam, as well as dim+ (N3864) and Δdim-7 (N3865) strains expressing DIM-5-Dam were incubated with or without DpnI, which cuts GATC only when the adenine is methylated. As an indicator of completely digested DNA, genomic DNA from the WT strain was incubated with the cytosine methylation-insensitive enzyme DpnII. Digested DNA was used for Southern hybridizations with probes corresponding to the heterochromatic 8:A6 and 8:G3 regions and the euchromatic mtr and sms-2 genes.
Discussion
Constitutive heterochromatin, which is typically associated with silenced, repetitive DNA and frequently with DNA methylation, was once regarded as the “garbage dump” of the genome. It is becoming increasingly clear, however, that heterochromatin makes important contributions to organisms by participating in essential processes, such as genome defense and centromere function (1). Moreover, misregulation of heterochromatin and DNA methylation is a hallmark of human cancers (32). Despite its importance, the mechanisms that direct heterochromatin formation are largely unknown in most eukaryotes. Similarly, the control of DNA methylation is incompletely understood in animals, plants, and fungi. To uncover genes that control heterochromatin and DNA methylation in the model fungus N. crassa, we developed a genetic selection to identify mutants that show reactivation of two silent reporter genes. Implementation of this selection scheme led to the identification of 48 mutant strains. It is significant that DNA methylation is not essential in N. crassa, which allows for isolation of mutants that are completely defective for DNA methylation. Despite this, we failed to obtain mutant progeny from crosses of 38 of 48 mutant strains. The failure to isolate Dim− progeny from many of these strains may indicate that the genes responsible for the Dim− phenotype are essential. Significantly, we failed to isolate mutations in dim-2, hpo, or dim-5, indicating that our mutagenesis was not saturating; therefore, it is likely that selection of additional mutants will continue to uncover novel genes in the DNA methylation pathway.
Of the 10 mutant strains that produced viable progeny, 3 harbored mutations in dim-7. It is notable that complementary genetic and biochemical analyses led to the identification of the dim-7 gene. We previously mapped the dim-7 mutation to a small region containing 89 genes, but we were unable to identify dim-7 by complementation, and purification of DIM-5 in a separate study led to the identification of 69 putative interacting proteins, many of which represent nonspecific interacting proteins (i.e., false positives). We were ultimately able to identify dim-7 because it was found concurrently using both genetic and biochemical approaches. Similarly, our DIM-5 affinity purification identified other proteins, including CUL4 and DDB1, which are required for DNA methylation and were recently identified genetically by us and others (33).
We demonstrated that DIM-7 and DIM-5 interact in vivo, but we failed to observe direct interaction of these proteins in vitro or using the yeast two-hybrid assay. Although it is difficult to interpret these negative results, they may indicate that the DIM-7-DIM-5 interaction requires posttranslational modification of one or both proteins. Alternatively, additional cofactors may mediate interaction of these proteins. Candidates can be found among proteins identified by proteomic analysis of DIM-5-associated proteins. An important future goal is to determine how DIM-7 functions within a putative DIM-5 complex to control H3K9 methylation. We note that DIM-7 possesses limited similarity with Raf2p, a member of the S. pombe ClrC complex (30). Indeed, ClustalW alignments revealed conservation within an N-terminal domain and a C-terminal C2H2 zinc finger domain, suggesting that DIM-7 may be homologous to Raf2p. Given that the zinc finger domains of DIM-7 and Raf2p are conserved, it is surprising that this region of the protein is dispensable for DIM-7 function. This may reflect a difference in the mechanism of heterochromatin formation between S. pombe and N. crassa (i.e., RNAi-dependent vs. RNAi-independent heterochromatin formation). Nevertheless, our data suggest that N. crassa DIM-5 may require a complex of interacting proteins to perform H3K9 methylation, similar to its S. pombe homolog, Clr4.
Clr4 includes a chromo domain, which is important for its association with heterochromatin (34), whereas DIM-5 lacks a chromo domain. This finding could explain why we were unable to demonstrate association of DIM-5 with heterochromatin using ChIP assays, and suggests that DIM-5 associates only transiently with chromatin. Reasoning that even transient chromatin association of an expressed DIM-5-Dam fusion protein may be sufficient to direct methylation of adenines in regions of DIM-5 association, we adapted DamID for use in N. crassa. Indeed, the DIM-5-Dam fusion efficiently methylated adenines at heterochromatin. We observed limited Dam methylation at euchromatin control regions in the wild-type strain. This is presumably because of nonspecific Dam methylation at “open chromatin,” which has been observed for Dam fusion proteins in other systems (31). This finding demonstrates that DIM-5 preferentially interacts with its genomic substrates and suggests that the DamID method may be particularly useful for investigating weak or transient interactions.
Importantly, we found that DIM-5-Dam recruitment to heterochromatin was abolished in the Δdim-7 mutant, indicating that DIM-7 is required to direct DIM-5 to heterochromatin regions. It is interesting that the background Dam methylation at euchromatin sites, like the more extensive methylation at heterochromatin sites, also required DIM-7. It is tempting to speculate that DIM-7 binds A:T-rich DNA directly to recruit DIM-5 to RIP'd DNA. Alignment of DIM-7 homologs revealed a conserved C2H2 zinc finger-like domain; however, this domain was dispensable for both DNA methylation and interaction with DIM-5. In addition, gel-shift assays using nuclear extracts revealed that a previously identified A:T-rich DNA binding activity was present in Δdim-7 extracts, indicating that DIM-7 is not responsible for this activity (19). It is possible that DIM-7 interacts with another chromatin protein to recruit DIM-5 to heterochromatin. Although we did not observe DIM-7 enrichment at heterochromatin regions by ChIP assays, it seems likely that DIM-7 is localized specifically to heterochromatin because DIM-5 localization is dependent on DIM-7. Thus, the inability to detect DIM-7 localization at heterochromatin by ChIP assays may reflect an indirect or transient DIM-7-chromatin interaction.
Homologs of DIM-7 appear to be restricted to fungi, and even among this group, DIM-7 is not well conserved. Rapid coevolution of centromere sequences and centromere binding proteins has been noted in eukaryotes (35), and telomere binding proteins of Drosophila also show unusually rapid divergence (36). DIM-7 may be undergoing rapid evolution concomitant with rapid sequence evolution of heterochromatic sequences. Notably, the RIP machinery exhibits different di-nucleotide substrate preferences in different fungi, which may shape the sequence content within repetitive DNA in a species-specific manner (4). More generally, it is possible that comparable proteins help recruit histone methyltransferases to incipient heterochromatin in higher eukaryotes.
Materials and Methods
Neurospora Growth and Molecular Analyses.
All strains used in this study are listed in Table S2. N. crassa strains were maintained, grown, and crossed using previously described procedures (37). DNA isolation (38), Southern blotting (18), protein isolation, coimmunoprecipitation (7), and construction of FLAG-tagged strains (27) were performed as described. All primers used in this study are listed in Table S3. A detailed description of plasmid construction is available in the SI Materials and Methods. Immunoprecipitation, ChIP, and Western blot analyses were performed using the following antibodies: α-FLAG (F3165, Sigma), α-HA (mouse; University of Oregon monoclonal facility or rat; clone 3F10, Roche), and α-DIM-5 (provided by X. Cheng, Emory University). ChIP-chip microarray experiments and analyses were performed as described previously (2).
Genetic Analyses.
To select for Dim− mutants, ≈40,000 condia/plate of strain N2977 were spread on 150 × 15-mm Petri plates containing 75 mL of nonselective medium [Vogel's (-NH4NO3), 0.5% proline, 1.5% Agar, 50 ng/mL inositol, 1 mg/mL alanine, 1 mg/mL histidine, 2%Sorbose, 0.05% glucose, 0.05% fructose] (39). Plates were exposed to UV light in a Stratalinker UV cross-linker (Stratagene) for 15 s, which yielded a survival rate of 25%, and were covered in foil to prevent photolyase activation. Conidia were allowed to germinate for 18 h at 32 °C to allow “dilution” of preexisting DNA methylation and covered with 25 mL top agar containing basta (as described above, except containing 1% Agar and 400 μg/mL basta) (39). Basta-resistant colonies were isolated, transferred to slants containing 400 μg/mL basta, and tested for growth on 200 μg/mL hygromycin. Strains that were resistant to both drugs were analyzed further. For complementation tests, each new Dim− strain was coinoculated with other new Dim− strains or previously identified Dim− strains in pairwise fashion to generate forced heterokaryons and each heterokaryon was tested for restoration of DNA methylation.
Supplementary Material
Acknowledgments
We thank the Neurospora Genome Project and the Fungal Genetic Stock Center for providing materials. Libby Asai, Kristin Herring, and Elizabeth Erikson provided technical assistance. Z.A.L. and A.L.S. identified and characterized DIM-7; K.K.A. constructed/characterized the reporter strain. Mass spectrometry of DIM-5-associated proteins was carried out at the Friedrich Miescher Institute (Basel, Switzerland; supported by the Novartis Research Foundation) by Ragna Sack while EUS was hosted by Susan Gasser in 2007. This work was supported by Grant GM025690-22 (to E.U.S.) from the National Institutes of Health. Z.A.L. was previously supported by a Fellowship from the American Cancer Society (PF-04-122-01-GMC) and is currently supported by a Special Fellowship from the Leukemia and Lymphoma Society (3295-09).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000328107/-/DCSupplemental.
References
- 1.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 2.Lewis ZA, et al. Relics of repeat-induced point mutation direct heterochromatin formation in Neurospora crassa. Genome Res. 2009;19:427–437. doi: 10.1101/gr.086231.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selker EU, et al. The methylated component of the Neurospora crassa genome. Nature. 2003;422:893–897. doi: 10.1038/nature01564. [DOI] [PubMed] [Google Scholar]
- 4.Galagan JE, Selker EU. RIP: the evolutionary cost of genome defense. Trends Genet. 2004;20:417–423. doi: 10.1016/j.tig.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 6.Freitag M, Hickey PC, Khlafallah TK, Read ND, Selker EU. HP1 is essential for DNA methylation in neurospora. Mol Cell. 2004;13:427–434. doi: 10.1016/s1097-2765(04)00024-3. [DOI] [PubMed] [Google Scholar]
- 7.Honda S, Selker EU. Direct interaction between DNA methyltransferase DIM-2 and HP1 is required for DNA methylation in Neurospora crassa. Mol Cell Biol. 2008;28:6044–6055. doi: 10.1128/MCB.00823-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brower-Toland B, et al. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pal-Bhadra M, et al. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 10.Ting AH, Schuebel KE, Herman JG, Baylin SB. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat Genet. 2005;37:906–910. doi: 10.1038/ng1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- 12.Okada T, et al. CENP-B controls centromere formation depending on the chromatin context. Cell. 2007;131:1287–1300. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 13.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sripathy SP, Stevens J, Schultz DC. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol Cell Biol. 2006;26:8623–8638. doi: 10.1128/MCB.00487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 16.Freitag M, et al. DNA methylation is independent of RNA interference in Neurospora. Science. 2004;304:1939. doi: 10.1126/science.1099709. [DOI] [PubMed] [Google Scholar]
- 17.Adhvaryu KK, Selker EU. Protein phosphatase PP1 is required for normal DNA methylation in Neurospora. Genes Dev. 2008;22:3391–3396. doi: 10.1101/gad.1738008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miao VP, Freitag M, Selker EU. Short TpA-rich segments of the zeta-eta region induce DNA methylation in Neurospora crassa. J Mol Biol. 2000;300:249–273. doi: 10.1006/jmbi.2000.3864. [DOI] [PubMed] [Google Scholar]
- 19.Tamaru H, Selker EU. Synthesis of signals for de novo DNA methylation in Neurospora crassa. Mol Cell Biol. 2003;23:2379–2394. doi: 10.1128/MCB.23.7.2379-2394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rountree MR, Selker EU. DNA methylation inhibits elongation but not initiation of transcription in Neurospora crassa. Genes Dev. 1997;11:2383–2395. doi: 10.1101/gad.11.18.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irelan JT, Selker EU. Cytosine methylation associated with repeat-induced point mutation causes epigenetic gene silencing in Neurospora crassa. Genetics. 1997;146:509–523. doi: 10.1093/genetics/146.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fulci V, Macino G. Quelling: post-transcriptional gene silencing guided by small RNAs in Neurospora crassa. Curr Opin Microbiol. 2007;10:199–203. doi: 10.1016/j.mib.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 23.Orbach MJ, Porro EB, Yanofsky C. Cloning and characterization of the gene for beta-tubulin from a benomyl-resistant mutant of Neurospora crassa and its use as a dominant selectable marker. Mol Cell Biol. 1986;6:2452–2461. doi: 10.1128/mcb.6.7.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis ZA, et al. High-density detection of restriction-site-associated DNA markers for rapid mapping of mutated loci in Neurospora. Genetics. 2007;177:1163–1171. doi: 10.1534/genetics.107.078147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colot HV, et al. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA. 2006;103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiu PK, Raju NB, Zickler D, Metzenberg RL. Meiotic silencing by unpaired DNA. Cell. 2001;107:905–916. doi: 10.1016/s0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- 27.Honda S, Selker EU. Tools for fungal proteomics: multifunctional neurospora vectors for gene replacement, protein expression and protein purification. Genetics. 2009;182:11–23. doi: 10.1534/genetics.108.098707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36(Database issue):D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horn PJ, Bastie JN, Peterson CL. A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev. 2005;19:1705–1714. doi: 10.1101/gad.1328005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogel MJ, Peric-Hupkes D, van Steensel B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat Protoc. 2007;2:1467–1478. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- 32.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y, et al. Ubiquitin ligase components Cullin4 and DDB1 are essential for DNA methylation in Neurospora crassa. J Biol Chem. 2010;285:4355–4365. doi: 10.1074/jbc.M109.034710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 35.Talbert PB, Bryson TD, Henikoff S. Adaptive evolution of centromere proteins in plants and animals. J Biol. 2004;3:18. doi: 10.1186/jbiol11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao G, et al. HipHop interacts with HOAP and HP1 to protect Drosophila telomeres in a sequence-independent manner. EMBO J. 2010;29:819–829. doi: 10.1038/emboj.2009.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis RH. Neurospora: Contributions of a Model Organism. Oxford: Oxford University Press; 2000. [Google Scholar]
- 38.Pomraning KR, Smith KM, Freitag M. Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods. 2009;47:142–150. doi: 10.1016/j.ymeth.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 39.Pall ML. The use of Ignite (Basta;glufosinate;phosphinothricin) to select transformants of bar-containing plasmids in Neurospora crassa. Fungal Genet Newsl. 1993;40:58. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.