Abstract
A recently identified variant within the fat mass and obesity-associated (FTO) gene is carried by 46% of Western Europeans and is associated with an ~1.2 kg higher weight, on average, in adults and an ~1 cm greater waist circumference. With >1 billion overweight and 300 million obese persons worldwide, it is crucial to understand the implications of carrying this very common allele for the health of our aging population. FTO is highly expressed in the brain and elevated body mass index (BMI) is associated with brain atrophy, but it is unknown how the obesity-associated risk allele affects human brain structure. We therefore generated 3D maps of regional brain volume differences in 206 healthy elderly subjects scanned with MRI and genotyped as part of the Alzheimer's Disease Neuroimaging Initiative. We found a pattern of systematic brain volume deficits in carriers of the obesity-associated risk allele versus noncarriers. Relative to structure volumes in the mean template, FTO risk allele carriers versus noncarriers had an average brain volume difference of ~8% in the frontal lobes and 12% in the occipital lobes—these regions also showed significant volume deficits in subjects with higher BMI. These brain differences were not attributable to differences in cholesterol levels, hypertension, or the volume of white matter hyperintensities; which were not detectably higher in FTO risk allele carriers versus noncarriers. These brain maps reveal that a commonly carried susceptibility allele for obesity is associated with structural brain atrophy, with implications for the health of the elderly.
Keywords: Alzheimer's Disease Neuroimaging Initiative, body mass index, brain structure, tensor-based morphometry, obesity
Obesity is a major public health concern; it is associated with increased risk for type 2 diabetes, hypertension, stroke, and coronary heart disease (1). Currently, there are >1 billion overweight and 300 million obese persons worldwide (2). The elderly population is particularly vulnerable—40% of men and 45% of women over age 70 suffer from either obesity or type 2 diabetes (3). In addition to increasing one's risk for poor cardiovascular health, obesity is a risk factor for cognitive decline and dementia, including Alzheimer's disease (AD) (4).
Obesity—and the most common and simplest clinical measure of adiposity, body mass index (BMI)—are highly genetically influenced, with genetic factors explaining 50–90% of the variance in BMI, although environmental influences are also important (5). Recently, the genetic bases of obesity and BMI were demonstrated to be in part due to variants within the fat mass and obesity-associated (FTO) gene in three large independent studies (6–8). These studies were somewhat surprising as they discovered a risk allele for obesity that is highly prevalent in the general population. A commonly inherited single-base-pair change at single-nucleotide polymorphism (SNP) rs9939609 is associated with an ~1.2-kg higher weight, on average, in adults, and an ~1-cm greater waist circumference. Carriers of two copies of the risk-conferring variant (18% of Europeans) had a 1.67-fold increased risk of obesity and weighed on average 3 kg more than those not carrying the risk gene (7). This risk polymorphism (7) has population frequencies of 46% in Western and Central Europeans, 51% in Yorubans (West African natives), and 16% in Chinese individuals (9); the effect of the allele on obesity in those with non-European genetic backgrounds is not yet clear (10). Interestingly, the effect of FTO on BMI may be mediated through impaired responsiveness to satiety (11).
FTO has highest expression in the brain in humans, specifically in the cerebral cortex (7). Whether expression changes in the hypothalamus after food deprivation is controversial (12–14). Although the function of the FTO gene is not completely understood, homozygous knockouts from a large region including this gene are not viable, show craniofacial abnormalities, and have abnormal left–right asymmetry and abnormally programmed cell death (15, 16). Duplication of a chromosomal region including FTO in humans leads to obesity and mental retardation (17), and a loss-of-function mutation produces microcephaly and structural brain malformations (18). FTO encodes the 2-oxoglutarate-dependent nucleic acid demethylase protein that localizes to the nucleus and catalyzes demethylation (14).
Obesity is associated with detectable structural differences in the brain in cognitively healthy elderly subjects (19) and in younger adults (20–22). In 94 healthy elderly subjects, BMI, fasting plasma insulin (FPI), and type 2 diabetes were strongly linked with frontal, temporal, and subcortical atrophy (19). Obese subjects (BMI ≥ 30 kg/m2) showed greatest brain tissue deficits, particularly in the frontal lobes, anterior cingulate gyrus, hippocampus, and thalamus when compared to subjects with normal BMI (18.5–25 kg/m2). Another study using voxel-based morphometry showed gray matter deficits in the frontal lobes and postcentral gyri but enlarged volumes in the orbitofrontal white matter (21). Higher BMI was also associated with neuronal and myelin abnormalities in the frontal lobes, using proton magnetic resonance spectroscopy (20). Studies of aging monkeys confirm the effect of diet on brain structure. A calorically restricted diet, without malnutrition, reduced brain atrophy in aging monkeys (23).
FTO is associated with consistent differences in BMI and is highly expressed in the brain, so here we sought to characterize the effect of carrying the FTO risk allele on human brain structure and determine whether these brain differences were consistent with those seen in people with higher BMI. We applied tensor-based morphometry (TBM), a relatively novel method (24–26), to generate 3D maps of whole brain volumetric differences in 206 cognitively healthy elderly subjects scanned with MRI and genotyped as part of the Alzheimer's Disease Neuroimaging Initiative (ADNI). We hypothesized that we would find brain regions with structural deficits in subjects who carry the obesity-associated risk allele, in regions where lower brain tissue volumes were strongly correlated with higher BMI. This study investigates a previously uncharacterized effect of an obesity-associated risk allele on brain structure.
Results
Identification of the Risk Allele.
Two SNPs, rs1421085 and rs17817449, have been previously identified as significantly and reliably associated with obesity (6). The risk alleles for obesity are the C allele for rs1421085 and the G allele for rs17817449. Neither of these SNPs was directly genotyped in our sample; however, we were able to exploit the linkage disequilibrium (LD) between SNPs to find a proxy, or tagging SNP, for these risk alleles. For more details and the LD plot, please see Fig. S1.
Association Between BMI and FTO Genotype.
After controlling for age and sex, we found an association between BMI and carrying at least one copy of the risk allele at the FTO tagging SNP (n = 206; β = 1.480; P = 0.0185) in our population of cognitively healthy elderly subjects. This finding was in the direction expected from prior studies (risk allele genotype giving higher BMI). This result gives yet another replication, in an independent sample of subjects, that the FTO gene variants are indeed associated with BMI. Additionally, we found that education was not correlated with either BMI or the FTO genotype (see SI Text for more details).
Obesity-Associated Risk Alleles and Brain Structure.
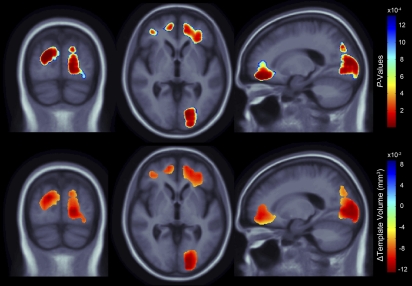
Structural brain differences in gray and white matter were statistically related to carrying at least one risk allele of the FTO gene (Fig. 1; critical uncorrected P = 1.31 × 10−3). We used a standard false discovery rate (FDR) correction (27) for multiple statistical comparisons across voxels of the brain, at the conventionally accepted level of q = 0.05. Significance maps are shown (Fig. 1 Upper) and the corresponding regression coefficient (unstandardized beta) maps are shown below them, representing the average volume difference of brain tissue deficit or excess relative to the template for those subjects carrying at least one copy of the obesity-associated risk allele, versus noncarriers. The regression coefficients can equivalently be interpreted as the average percentage of volume difference relative to the template between carriers and noncarriers upon a simple conversion (SI Text), and both representations are discussed.
Fig. 1.
(Upper) 3D maps show areas where regional brain tissue volumes were significantly associated with carrying the risk allele at the obesity-associated tagging SNP, rs3751812, in healthy elderly subjects (n = 206). (Lower) In the significant areas, the regression coefficients (unstandardized beta values) are shown at each voxel. These values represent the estimated degree of tissue excess or deficit at each voxel (in cubic millimeters relative to the template) that is associated with the presence of the risk allele, after statistically controlling for effects of age and sex on brain structure. Images are in radiological convention (left side of the brain shown on the right) and are displayed over the MDT.
BMI and Brain Structure.
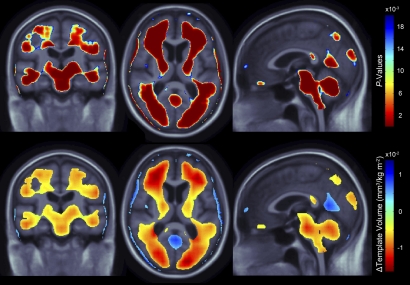
As found in previous studies (19), subjects with higher BMI had significantly lower regional brain volumes (FDR q = 0.05, critical uncorrected P = 0.0202) in many areas (Fig. 2). With every 1-unit increase in BMI, there was an associated 1–1.5% average brain tissue reduction (after statistically controlling for age and sex) in several broadly distributed frontal, temporal, parietal, and occipital lobe regions. Atrophy was also detected in the brainstem and cerebellar regions.
Fig. 2.
(Upper) 3D maps show areas where regional brain volumes were significantly associated with BMI in healthy elderly subjects (n = 206). (Lower) In the significant areas, the regression coefficients (unstandardized beta values or “slopes”) are shown at each voxel. These represent the estimated degree of tissue excess or deficit at each voxel (in cubic millimeters relative to the template) for each unit gain in body mass index, after statistically controlling for effects of age and sex, on brain structure. Images are displayed in radiological convention (left side of the brain shown on the right) and are displayed over the MDT.
White Matter Burden and Brain Structure.
To understand whether the FTO effect on brain structure was attributable to microvascular damage in the white matter, a measure of whole brain white matter burden (WMB) was regressed against brain structure after controlling for both age and sex (Fig. S2). See SI Text for details about how WMB was assessed. Subjects with higher WMB had significantly lower regional brain volumes in the frontal lobe and the precuneus brain regions (FDR q = 0.05 level, critical uncorrected P = 0.0016). With every unit increase in WMB, there was an estimated 10% reduction in brain tissue (Fig. S2, red colors) in a localized region in the frontal lobe and the precuneus brain region. At the edge of the ventricles, a 15–20% increase in ventricular volume is also observed; this effect includes some nonventricular voxels at the interface between the ventricles and brain tissue, due to partial volume effects that arise from the limited spatial resolution of the deformation fields. Ventricular expansion is due to widespread atrophy of the surrounding brain tissue (24, 28).
Conjunction Analysis.
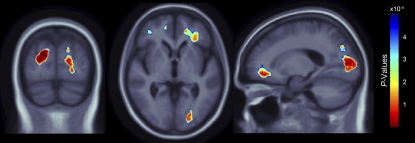
A conjunction analysis (29) was used to separately test the hypotheses that (i) the FTO risk allele and BMI affect brain volume at the same locations and (ii) FTO and WMB affect brain volume at the same locations. The null hypothesis was rejected in case i, so we can be reasonably confident that there is an effect of both the FTO risk allele and BMI on the voxels colored in Fig. 3 (FDR q = 0.05, critical uncorrected P = 5.007 × 10−4). Even so, the null hypothesis was not rejected in case ii, using an FDR correction for multiple comparisons. As we did not detect a significant volume difference from both the FTO risk allele and WMB at the same voxels, it is fair to speculate that the FTO effect on the brain is not completely attributable to microvascular white matter damage.
Fig. 3.
3D maps show regions of significant brain volume differences from both presence of the FTO risk allele and higher BMI using a conjunction analysis displayed over a study-specific template. Images are displayed in radiological convention (left side of the brain shown on the right). Images are corrected for multiple comparisons on the basis of FDR (FDR q-level = 0.05, critical uncorrected P = 5.007 × 10−4). The same slices as in Fig. 1 are shown.
Whereas the conjunction analysis shows that a subset of the FTO-associated brain regions is consistent with a subset of the BMI-associated brain regions, an additional regression analysis was conducted to determine if there were statistically significant brain regions related to carrying at least one risk allele of the FTO gene, after adjustment for age, sex, and BMI. There were no significant brain regions associated with the risk allele, after FDR correction at 5%; however, unthresholded maps of the regression coefficients (Fig. S4 Lower) are consistent with the equivalent thresholded maps (Fig. S4 Upper) also displayed in Fig. 1. Additionally, BMI was correlated with brain volume in the different genotype groups (Fig. S5). Subject demographics (Table S1) and unstandardized beta maps are presented for each genotype group (Fig. S5, first and second rows). In the nonrisk group, BMI was not statistically associated with brain volume after FDR correction at 5% (Fig. S5, fourth row).
Discussion
Here we showed a substantial effect on brain structure of a commonly carried variant within the FTO gene, which is associated with obesity and carried by 46% of Western Europeans. In a large sample of healthy elderly subjects scanned with brain MRI, we identified structural brain atrophy in carriers of the FTO risk allele. Those carrying at least one copy of the risk allele showed brain tissue deficits in the frontal and the occipital lobes, areas that are also associated with volume reductions in subjects with higher BMI. Carriers of the obesity-associated risk allele showed an average 8% deficit in brain tissue versus noncarriers in the bilateral frontal lobe and an average 12% deficit in the bilateral occipital lobe, where the percentage units are expressed in terms of the average volumes seen in the general population (of carriers and noncarriers). Subjects with higher BMI also showed brain volume deficits in frontal, temporal, parietal, and occipital lobe regions and in the brainstem and cerebellum—areas identified in past obesity studies, including our own in an independent, nonoverlapping sample (19). Greater WMB was also associated with an ~10% brain volume deficit in the frontal lobes and precuneus, where the percentage units are expressed in terms of the average volumes seen in the general population (of carriers and noncarriers). Even so, WMB did not explain the effect of the FTO risk allele on brain atrophy. Intriguingly, WMB was not statistically higher in FTO risk allele carriers than in noncarriers, and the effects of WMB and the FTO risk allele on brain atrophy were in different regions.
We investigated the genetic contribution to brain structure from a tagging SNP to verified obesity-associated risk alleles and found deficits in largely the same brain regions where higher BMI is associated with lower tissue volumes. Volume deficits within the frontal lobes, which reached 8% here in FTO risk allele carriers (Fig. 1), have been associated with decreased executive functioning (30). Further, individuals with obesity have generally poorer executive function than their healthy counterparts (31–33).
Temporal lobe atrophy was detected in subjects with higher BMI but not in subjects who carry the obesity-associated risk allele (Figs. 1–3). Other studies have found that midlife BMI is associated with temporal lobe atrophy 24 years later, suggesting that BMI is correlated with future differences in brain structure (34). Temporal lobe structures, including the hippocampus and entorhinal cortex, are among the earliest affected by neurofibrillary tangle pathology in Alzheimer's disease. FTO effects may either not target the temporal lobe or not be detectable in our sample due to the limited sample size. The obesity-associated risk allele may have different effects on brain structure in elderly versus younger groups; further studies are necessary to clarify this.
The occipital lobe volume deficits in subjects with greater BMI and those who carry one or more copies of the obesity-associated risk allele are clearly evident (Figs. 1–3). BMI effects on occipital lobe tissue were found previously (19, 22, 34). Studies of types 1 and 2 diabetes have also shown that diabetes is associated with greater brain atrophy. One study reported that patients with diabetic retinopathy, a complication of type 1 diabetes, showed reduced gray matter density in the inferior frontal gyrus and right occipital lobe when compared patients without retinopathy (35). Patients with type 2 diabetes also had 3% less gray matter and ~17% increase in CSF in the parieto-occipital region (36).
For the subjects examined in this study (Table 1), neither hypertension nor cholesterol levels were statistically different on the basis of presence of the FTO risk allele; therefore, we do not have evidence to suggest FTO risk alleles increase the odds for some of the symptoms associated with metabolic syndrome; this result counters the notion that the FTO risk allele might be merely a surrogate indicator of having symptoms often associated with metabolic syndrome. An important finding is that, between the two genetic groups of FTO carriers and noncarriers in our sample, there were no intergroup statistical differences in cholesterol, serum glucose levels, diabetes, or MRI-based measures of white matter burden. The FTO effects are not therefore merely reflecting some more fundamental difference in these parameters, at least when measured at the time of the scan. Even so, it cannot be ruled out that FTO may be associated with a different natural history of cholesterol or glucose levels in the past, even though such a relationship was not detectable at the time of the scan. As these factors are also related to mortality, given the old age of the participants studied here, we may have sampled a somewhat restricted range of natural histories for these risk factors. In conclusion, the FTO effects cannot be dismissed as reflecting the more conventionally studied risk factors assessed at the time of the scan, but FTO could be associated with these risk factors at other times throughout life or in other populations.
Table 1.
Demographic information for the 206 subjects
| Genotype groups | Nonrisk genotype (G/G) | Risk genotype (T/G or T/T) | Statistical results | |
| Sample size (n) | 78 (44 M/34 F) | 128 (67 M/61 F) | χ21 = 0.323; P = 0.57 | |
| Age (years) | 76.1 ± 4.7 | 76.2 ± 5.2 | F1,204 = 0.030; P = 0.86 | |
| Body mass index (BMI) | 25.6 ± 4.1 | 27.1 ± 4.5 | F1,204 = 5.33; P = 0.022 | |
| Education (years) | 16.3 ± 2.5 | 16.1 ± 2.9 | F1,204 = 0.257; P = 0.61 | |
| Glucose (mg/dL) | 58.0 ± 9.7 (n = 32) | 60.6 ± 9.0 (n = 64) | F1,94 = 1.75; P = 0.19 | |
| Cholesterol (mg/dL) | 195.0 ± 36.5 (n = 77) | 195.6 ± 41.0 (n = 124) | F1,199 = 0.012; P = 0.91 | |
| White matter hyperintensity (cm3) | 2.4 ± 1.8 (n = 69) | 2.9 ± 2.9 (n = 100) | F1,167 = 0.265; P = 0.61 | |
| History of hypertension | (37/78) 47.4% | (54/128) 42.2% | χ21 = 0.541; P = 0.46 | |
| History of stroke | (2/78) 2.6% | (0/128) 0% | χ21 = 3.31; P = 0.069 | |
| History of cardiovascular disease | (53/78) 67.9% | (82/128) 64.1% | χ21 = 0.324; P = 0.57 | |
| Global clinical dementia rating | 0 | 0 | NA | |
| Mini mental state examination | 29.3 ± 0.9 | 29.1 ± 0.9 | *χ21 = 1.14; P = 0.2852 | |
| BMI groups (kg/m2) | BMI < 25 | 25 ≤ BMI < 30 | BMI ≥ 30 | |
| Global clinical dementia rating | 0 | 0 | 0 | NA |
| Mini mental state examination | 29.4 ± 0.8 | 29.0 ± 0.9 | 29.1 ± 1.0 | *χ22 = 7.43; P = 0.024 |
The mean ± SD is shown for each measure; clinical measures are categorized by number of risk alleles at the obesity-associated tagging SNP and body mass index groups. ANOVAs, performed on each variable, show whether the mean clinical measure differed significantly across groups. χ2 tests were performed for categorical variables, where subscripts indicate the number of degrees of freedom. A nonparametric Kruskal–Wallis one-way ANOVA was used to assess measures that were not normally distributed [e.g., mini mental state examination (MMSE) scores]. White matter hyperintensity measures were log10 transformed before statistical tests. F, females; M, males; NA, not analyzed. Boldface type indicates statistical significance (P < 0.05).
Previous studies relating the level of white matter hyperintensity (WMH) to cortical atrophy in healthy elderly subjects show gray matter reduction and ventricular expansion (37, 38) consistent with the WMB maps in Fig. S2. DeCarli et al. also found that WMH volume was associated with increased ventricular volume, reduced brain volume, and reduced cognitive scores in healthy subjects (37). Greater WMH volume was also associated with significantly lower frontal lobe metabolism, higher blood pressure, and lower scores on neuropsychological tests known to rely heavily on the frontal lobes. Although WMH is known to be associated with a characteristic pattern of brain atrophy, this pattern is not consistent with that shown here to be associated with the presence of the FTO risk allele as determined through a conjunction analysis.
The mechanism behind this difference in brain structure from a single-base-pair change in the genome is not currently known, and it is not possible to say where the brain differences lie in the causal chain of factors that influence obesity. A strong hypothesis would be that BMI affects brain structure and that FTO exerts some additive detectable effect over and above whatever the BMI of the person happens to be. A weak hypothesis might be that BMI exerts an effect on brain structure and that FTO tracks this effect because is it associated with BMI. We made some efforts to see if we could detect any independent (additive) effect of FTO, above and beyond what is explainable by BMI, which FTO is known to affect. If FTO effects are not seen after adjustment for BMI (Fig. S4), it is either because the effects are present but too small to be detected, given the high collinearity of BMI and FTO, or because FTO has a causal effect on BMI but no independent statistical effect on brain volume after adjusting for BMI.
BMI had an independent effect on the brain, over and above what is explainable by FTO, because its effect was seen in separate groups of people with the same genotype (Fig. S5; see Table S1 for subject demographics). Perhaps surprisingly, a strong effect of BMI was seen in carriers (with one or more FTO risk alleles), but it was not detected in noncarriers of the FTO allele (Fig. S5, unthresholded maps), even though the sample size for the noncarriers was larger (n = 78) than the homozygous at-risk group (n = 33) where BMI effects were still seen. Although the lack of a detectable effect of BMI in FTO noncarriers does not imply that it would not be present in a large sample, it cannot be ruled out that FTO status may influence the effect of BMI on the brain, which should be investigated when larger samples are available.
Here we modeled the influence of previously identified risk alleles in the FTO gene on the brain, on the basis of a proxy, or tagging SNP, that is 98.8% accurate in predicting the verified risk alleles according to a similar population sample (Fig. S1). Therefore, although we are not measuring the verified risk allele, we have an acceptable proxy to measure its effect on brain structure. The tagging SNP is very highly linked to the previously associated polymorphisms, so little would be gained by using imputation techniques to estimate the allele at the associated locus on the basis of the surrounding genotype data in this study.
Recent studies have shown that physical activity and diet may provide promising outcomes for individuals who carry the obesity-associated risk allele (39). A study of middle-aged individuals showed that a high-fat diet and low leisure-time physical activity might increase susceptibility for higher BMI in those carrying the FTO allele (39). In another study of 704 healthy adults, weight increase attributable to the presence of the risk allele was numerically less, although not significantly less, in subjects who were physically active (40). Another study showed that physical activity attenuated the genetic effect of carrying the obesity-associated risk allele on BMI and waist circumference (41). Even in individuals who are genetically susceptible to obesity, physical activity can play a major role in controlling obesity and waist circumference.
The present finding has important implications for obesity research and for combating the progression of neurodegeneration.
Materials and Methods
Subjects.
Neuroimaging and genetic data were acquired from 818 subjects as part of the ADNI, a large 5-year study launched in 2004 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies, and nonprofit organizations (42, 43). The goal of the ADNI study is to determine biological markers of Alzheimer's disease through neuroimaging, genetics, neuropsychological tests, and other measures to develop new treatments and monitor their effectiveness and lessen the time of clinical trials. Subjects were recruited from 58 sites in the United States. The study was conducted according to the Good Clinical Practice guidelines, the Declaration of Helsinki, and U.S. 21 Code of Federal Regulations Part 50, Protection of Human Subjects, and Part 56, Institutional Review Boards. Written informed consent was obtained from all participants before protocol-specific procedures were performed. All data acquired as part of this study are publicly available (http://www.loni.ucla.edu/ADNI/).
The sample examined in this study consisted of a subsample of the entire ADNI cohort. Only Caucasian (non-Hispanic) cognitively healthy elderly subjects were included (n = 207). Caucasian subjects were selected to reduce population stratification effects (44). One subject whose height was not recorded was excluded, as BMI could not be calculated. The sample analyzed in this study involved 206 subjects (Table 1).
DNA Isolation, SNP Genotyping Methods, and Genetic Analysis.
For details about DNA isolation and SNP genotyping methods, please refer to SI Text. Genomewide genotype information was collected at 620,901 markers; however, this study deals only with one SNP, rs3751812. The allele on the forward strand is reported. This marker met quality criteria based on previous genomewide association studies (45): genotype call rate >95%, nonsignificant deviation from Hardy–Weinberg equilibrium P > 5.7 × 10−7, minor allele frequency >0.01, and a platform-specific recommended quality control score of >0.15. LD patterns with previously identified mutations in the general population were explored in the HapMap sample (9), using allele frequency information from a European population (CEU; http://www.hapmap.org/; Phase III/Release #2, Feb 09, NCBI b36 assembly).
MRI Acquisition, Image Calibration, and Correction.
All subjects were scanned at multiple ADNI sites according to a standardized protocol developed after a major effort to evaluate 3D T1-weighted sequences for morphometric analyses (46, 47). High-resolution structural brain MRI scans were acquired using 1.5- and 3-Tesla (T) MRI scanners; however, because the majority of subjects were scanned at 1.5 T, and to avoid any potentially confounding effect of scanner field strength on tissue volume quantification (48), we restricted our analysis to 1.5-T MRI scans. See SI Text for scanner vendor information and image correction across sites.
Tensor-Based Morphometry (TBM) and 3D Jacobian Maps.
Images were preprocessed according to standard protocol discussed previously (25). A minimal deformation template (MDT) was created from the MRI scans of 40 cognitively healthy ADNI subjects (See Table S2 for subject demographics) to enable automated image registration and reduce statistical bias (24, 25). All scans were nonlinearly aligned to the study-specific template so that they would all share a common coordinate system. The local expansion factor of the 3D elastic warping transform (26), calculated as the determinant of the Jacobian matrix of the deformation, was plotted for each subject. These 3D expansion factor maps show relative volume differences between each individual and the common template, and reveal areas of structural volume deficits or expansions, relative to the population average. Additionally, log10 transformations of the Jacobian determinant were computed and all regression analyses were run using the transformed data (Fig. S6). Such a transformation can remove skewness in the Jacobian distribution, although the dependent variable is a logged volume rather than a true volume (49).
Regression of Structural Brain Differences with the Obesity-Associated Tagging SNP and with BMI.
At each voxel, the relation of the Jacobian values (which represent brain tissue deficit or excess relative to the standard template) to the presence or absence of the risk allele of the SNP rs3751812 (Fig. 1), BMI (Fig. 2), and WMB (Fig. S2) controlling for age and sex was tested using the general linear model. The significance maps were corrected for multiple comparisons using the FDR method (27). The critical uncorrected P value is reported, which represents the highest P-value threshold for which the surviving voxels are expected to have no more than 5% false positives. The critical P-value threshold is generally larger for greater effect sizes in the map. Further details of the statistical models are found in SI Text. We estimated the minimal number of subjects (n = 96) needed to repeat the finding by successively removing subjects, at random but without bias, from our sample and calculating the critical P value for FDR threshold, until the results were no longer significant. This gives a rough estimate of how many subjects are needed to find the effect in an independent sample (on the premise that in very small samples, the FTO effect would not explain enough of the variance) as in prior studies (50). A cumulative distribution function (CDF) plot displays the results from smaller sample sizes having lower critical P values than those computed from larger sample sizes (Fig. S7).
Conjunction Analysis.
Conjunction analysis was conducted by creating a map composed of the maximum (least associated) P value at each voxel from both conditions of interest (29). An FDR correction on the maximum P value (minimum statistic) map was used to correct for multiple comparisons (27). Voxels surviving multiple comparisons are displayed in Fig. 3.
CDF Plots.
CDF plots, based on the above correlation tests, were used to rank the proportion of significant voxels in association with SNP rs3751812, BMI, and WMB (Fig. S3) (24, 25, 51). See SI Text.
Supplementary Material
Acknowledgments
Data collection and sharing for this project were funded by the Alzheimer's Disease Neuroimaging Initiative (National Institutes of Health Grant U01 AG024904). The Alzheimer's Disease Neuroimaging Initiative is funded by the National Institute on Aging and the National Institute of Biomedical Imaging and Bioengineering and through generous contributions from the following: Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson and Johnson, Eli Lilly and Co., Medpace, Inc., Merck and Co., Inc., Novartis AG, Pfizer Inc, F. Hoffman-La Roche, Schering-Plough, Synarc, Inc., and Wyeth, as well as nonprofit partners the Alzheimer's Association and Alzheimer's Drug Discovery Foundation, with participation from the U.S. Food and Drug Administration. Private sector contributions to the Alzheimer's Disease Neuroimaging Initiative are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. The Alzheimer's Disease Neuroimaging Initiative data are disseminated by the Laboratory for Neuroimaging at the University of California, Los Angeles. This research was also supported by National Institutes of Health Grants P30 AG010129 and K01 AG030514 and the Dana Foundation. Algorithm development for this study was also funded by the National Institute on Aging, the National Institute on Biomedical Imaging and Bioengineering, the National Institute of Child Health and Human Development, the National Library of Medicine, and the National Center for Research Resources (AG016570, EB01651, HD050735, LM05639, and RR019771, to P.M.T.). J.L.S. was also funded by National Institutes of Health/National Institute on Drug Abuse Grant 1-T90-DA022768:02, the Achievement Rewards for College Scientists foundation, and the National Institute of Mental Health (1F31MH087061). A.J.H. was also funded by National Institutes of Health/National Institute on Drug Abuse Grant 1-T90-DA022768:02, the National Science Foundation Graduate Research Fellowship Program, and the Achievement Rewards for College Scientists foundation.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
†Data used in the preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (www.loni.ucla.edu/ADNI). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators is available at http://www.loni.ucla.edu/ADNI/Collaboration/ADNI_Manuscript_Citations.pdf.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910878107/DCSupplemental.
Contributor Information
Michael Weiner, ADNI.
Leon Thal, ADNI.
Ronald Petersen, ADNI.
Clifford R. Jack, Jr., ADNI.
William Jagust, ADNI.
John Trojanowki, ADNI.
Arthur W. Toga, ADNI.
Laurel Beckett, ADNI.
Robert C. Green, ADNI.
Anthony Gamst, ADNI.
William Z. Potter, ADNI.
Tom Montine, ADNI.
Dale Anders, ADNI.
Matthew Bernstein, ADNI.
Joel Felmlee, ADNI.
Nick Fox, ADNI.
Paul Thompson, ADNI.
Norbert Schuff, ADNI.
Gene Alexander, ADNI.
Dan Bandy, ADNI.
Robert A. Koeppe, ADNI.
Norm Foster, ADNI.
Eric M. Reiman, ADNI.
Kewei Chen, ADNI.
John Trojanowki, ADNI.
Les Shaw, ADNI.
Virginia M.-Y. Lee, ADNI.
Magdalena Korecka, ADNI.
Arthur W. Toga, ADNI.
Karen Crawford, ADNI.
Scott Neu, ADNI.
Danielle Harvey, ADNI.
Anthony Gamst, ADNI.
John Kornak, ADNI.
Zaven Kachaturian, ADNI.
Richard Frank, ADNI.
Peter J. Snyder, ADNI.
Susan Molchan, ADNI.
Jeffrey Kaye, ADNI.
Remi Vorobik, ADNI.
Joseph Quinn, ADNI.
Lon Schneider, ADNI.
Sonia Pawluczyk, ADNI.
Bryan Spann, ADNI.
Adam S. Fleisher, ADNI.
Helen Vanderswag, ADNI.
Judith L. Heidebrink, ADNI.
Joanne L. Lord, ADNI.
Kris Johnson, ADNI.
Rachelle S. Doody, ADNI.
Javier Villanueva-Meyer, ADNI.
Munir Chowdhury, ADNI.
Yaakov Stern, ADNI.
Lawrence S. Honig, ADNI.
Karen L. Bell, ADNI.
John C. Morris, ADNI.
Mark A. Mintun, ADNI.
Stacy Schneider, ADNI.
Daniel Marson, ADNI.
Randall Griffith, ADNI.
Beverly Badger, ADNI.
Hillel Grossman, ADNI.
Cheuk Tang, ADNI.
Jessica Stern, ADNI.
Leyla deToledo-Morrell, ADNI.
Raj C. Shah, ADNI.
Julie Bach, ADNI.
Ranjan Duara, ADNI.
Richard Isaacson, ADNI.
Silvia Strauman, ADNI.
Marilyn S. Albert, ADNI.
Julia Pedroso, ADNI.
Jaimie Toroney, ADNI.
Henry Rusinek, ADNI.
Mony J de Leon, ADNI.
Susan M De Santi, ADNI.
P. Murali Doraiswamy, ADNI.
Jeffrey R. Petrella, ADNI.
Marilyn Aiello, ADNI.
Christopher M. Clark, ADNI.
Cassie Pham, ADNI.
Jessica Nunez, ADNI.
Charles D. Smith, ADNI.
Curtis A. Given II, ADNI.
Peter Hardy, ADNI.
Steven T. DeKosky, ADNI.
MaryAnn Oakley, ADNI.
Donna M. Simpson, ADNI.
M. Saleem Ismail, ADNI.
Anton Porsteinsson, ADNI.
Colleen McCallum, ADNI.
Steven C. Cramer, ADNI.
Ruth A. Mulnard, ADNI.
Catherine McAdams-Ortiz, ADNI.
Ramon Diaz-Arrastia, ADNI.
Kristen Martin-Cook, ADNI.
Michael DeVous, ADNI.
Allan I. Levey, ADNI.
James J. Lah, ADNI.
Janet S. Cellar, ADNI.
Jeffrey M. Burns, ADNI.
Heather S. Anderson, ADNI.
Mary M. Laubinger, ADNI.
George Bartzokis, ADNI.
Daniel H.S. Silverman, ADNI.
Po H. Lu, ADNI.
Rita Fletcher, ADNI.
Francine Parfitt, ADNI.
Heather Johnson, ADNI.
Martin Farlow, ADNI.
Scott Herring, ADNI.
Ann M. Hake, ADNI.
Christopher H. van Dyck, ADNI.
Martha G. MacAvoy, ADNI.
Laurel A. Bifano, ADNI.
Howard Chertkow, ADNI.
Howard Bergman, ADNI.
Chris Hosein, ADNI.
Sandra Black, ADNI.
Simon Graham, ADNI.
Curtis Caldwell, ADNI.
Howard Feldman, ADNI.
Michele Assaly, ADNI.
Ging-Yuek R. Hsiung, ADNI.
Andrew Kertesz, ADNI.
John Rogers, ADNI.
Dick Trost, ADNI.
Charles Bernick, ADNI.
Darren Gitelman, ADNI.
Nancy Johnson, ADNI.
Marsel Mesulam, ADNI.
Carl Sadowsky, ADNI.
Teresa Villena, ADNI.
Scott Mesner, ADNI.
Paul S. Aisen, ADNI.
Kathleen B. Johnson, ADNI.
Kelly E. Behan, ADNI.
Reisa A. Sperling, ADNI.
Dorene M. Rentz, ADNI.
Keith A. Johnson, ADNI.
Allyson Rosen, ADNI.
Jared Tinklenberg, ADNI.
Wes Ashford, ADNI.
Marwan Sabbagh, ADNI.
Donald Connor, ADNI.
Sanja Obradov, ADNI.
Ron Killiany, ADNI.
Alex Norbash, ADNI.
Thomas O. Obisesan, ADNI.
Annapurni Jayam-Trouth, ADNI.
Paul Wang, ADNI.
Alexander P. Auchus, ADNI.
Juebin Huang, ADNI.
Robert P. Friedland, ADNI.
Charles DeCarli, ADNI.
Evan Fletcher, ADNI.
Owen Carmichael, ADNI.
Smita Kittur, ADNI.
Seema Mirje, ADNI.
Sterling C. Johnson, ADNI.
Michael Borrie, ADNI.
T-Y Lee, ADNI.
Sanjay Asthana, ADNI.
Cynthia M. Carlsson, ADNI.
Steven G. Potkin, ADNI.
Diane Highum, ADNI.
Adrian Preda, ADNI.
Dana Nguyen, ADNI.
Pierre N. Tariot, ADNI.
Barry A. Hendin, ADNI.
Douglas W. Scharre, ADNI.
Maria Kataki, ADNI.
David Q. Beversdorf, ADNI.
Earl A. Zimmerman, ADNI.
Dzintra Celmins, ADNI.
Alice D. Brown, ADNI.
Sam Gandy, ADNI.
Marjorie E. Marenberg, ADNI.
Barry W. Rovner, ADNI.
Godfrey Pearlson, ADNI.
Karen Blank, ADNI.
Karen Anderson, ADNI.
Andrew J. Saykin, ADNI.
Robert B. Santulli, ADNI.
Nadia Pare, ADNI.
Jeff D. Williamson, ADNI.
Kaycee M. Sink, ADNI.
Huntington Potter, ADNI.
B. Ashok Raj, ADNI.
Amy Giordano, ADNI.
Brian R. Ott, ADNI.
Chuang-Kuo Wu, ADNI.
Ronald Cohen, ADNI.
Kerri L. Wilks, ADNI.
References
- 1.Must A, et al. The disease burden associated with overweight and obesity. JAMA. 1999;282:1523–1529. doi: 10.1001/jama.282.16.1523. [DOI] [PubMed] [Google Scholar]
- 2.WHO . Obesity and Overweight. Geneva: World Health Organization; 2009. [Google Scholar]
- 3.Ceska R. Clinical implications of the metabolic syndrome. Diab Vasc Dis Res. 2007;4(Suppl 3):S2–S4. doi: 10.3132/dvdr.2007.049. [DOI] [PubMed] [Google Scholar]
- 4.Elias MF, Elias PK, Sullivan LM, Wolf PA, D'Agostino RB. Obesity, diabetes and cognitive deficit: The Framingham Heart Study. Neurobiol Aging. 2005;26(Suppl 1):11–16. doi: 10.1016/j.neurobiolaging.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 5.Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav Genet. 1997;27:325–351. doi: 10.1023/a:1025635913927. [DOI] [PubMed] [Google Scholar]
- 6.Dina C, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 7.Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scuteri A, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frazer KA, et al. International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hennig BJ, et al. FTO gene variation and measures of body mass in an African population. BMC Med Genet. 2009;10:21. doi: 10.1186/1471-2350-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wardle J, et al. Obesity associated genetic variation in FTO is associated with diminished satiety. J Clin Endocrinol Metab. 2008;93:3640–3643. doi: 10.1210/jc.2008-0472. [DOI] [PubMed] [Google Scholar]
- 12.Fischer J, et al. Inactivation of the Fto gene protects from obesity. Nature. 2009;458:894–898. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- 13.Fredriksson R, et al. The obesity gene, FTO, is of ancient origin, up-regulated during food deprivation and expressed in neurons of feeding-related nuclei of the brain. Endocrinology. 2008;149:2062–2071. doi: 10.1210/en.2007-1457. [DOI] [PubMed] [Google Scholar]
- 14.Gerken T, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peters T, Ausmeier K, Dildrop R, Rüther U. The mouse Fused toes (Ft) mutation is the result of a 1.6-Mb deletion including the entire Iroquois B gene cluster. Mamm Genome. 2002;13:186–188. doi: 10.1007/s00335-001-2142-7. [DOI] [PubMed] [Google Scholar]
- 16.van der Hoeven F, et al. Programmed cell death is affected in the novel mouse mutant Fused toes (Ft) Development. 1994;120:2601–2607. doi: 10.1242/dev.120.9.2601. [DOI] [PubMed] [Google Scholar]
- 17.Stratakis CA, et al. Anisomastia associated with interstitial duplication of chromosome 16, mental retardation, obesity, dysmorphic facies, and digital anomalies: Molecular mapping of a new syndrome by fluorescent in situ hybridization and microsatellites to 16q13 (D16S419-D16S503) J Clin Endocrinol Metab. 2000;85:3396–3401. doi: 10.1210/jcem.85.9.6776. [DOI] [PubMed] [Google Scholar]
- 18.Boissel S, et al. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am J Hum Genet. 2009;85:106–111. doi: 10.1016/j.ajhg.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raji C, et al. Brain structure and obesity. Hum Brain Mapp. 2010;31:353–364. doi: 10.1002/hbm.20870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gazdzinski S, Kornak J, Weiner MW, Meyerhoff DJ. Body mass index and magnetic resonance markers of brain integrity in adults. Ann Neurol. 2008;63:652–657. doi: 10.1002/ana.21377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pannacciulli N, et al. Brain abnormalities in human obesity: A voxel-based morphometric study. Neuroimage. 2006;31:1419–1425. doi: 10.1016/j.neuroimage.2006.01.047. [DOI] [PubMed] [Google Scholar]
- 22.Taki Y, et al. Relationship between body mass index and gray matter volume in 1,428 healthy individuals. Obesity. 2008;16:119–124. doi: 10.1038/oby.2007.4. [DOI] [PubMed] [Google Scholar]
- 23.Colman RJ, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hua X, et al. 3D characterization of brain atrophy in Alzheimer's disease and mild cognitive impairment using tensor-based morphometry. Neuroimage. 2008;41:19–34. doi: 10.1016/j.neuroimage.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hua X, et al. Tensor-based morphometry as a neuroimaging biomarker for Alzheimer's disease: an MRI study of 676 AD, MCI, and normal subjects. Neuroimage. 2008;43:458–469. doi: 10.1016/j.neuroimage.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leow A, et al. Inverse consistent mapping in 3D deformable image registration: Its construction and statistical properties. Inf Process Med Imaging. 2005;19:493–503. doi: 10.1007/11505730_41. [DOI] [PubMed] [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 28.Hua X, et al. Detecting brain growth patterns in normal children using tensor-based morphometry. Hum Brain Mapp. 2009;30:209–219. doi: 10.1002/hbm.20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nichols T, Brett M, Andersson J, Wager T, Poline JB. Valid conjunction inference with the minimum statistic. Neuroimage. 2005;25:653–660. doi: 10.1016/j.neuroimage.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 30.Raz N, Gunning-Dixon FM, Head D, Dupuis JH, Acker JD. Neuroanatomical correlates of cognitive aging: Evidence from structural magnetic resonance imaging. Neuropsychology. 1998;12:95–114. doi: 10.1037//0894-4105.12.1.95. [DOI] [PubMed] [Google Scholar]
- 31.Cserjési R, Luminet O, Poncelet AS, Lénárd L. Altered executive function in obesity. Exploration of the role of affective states on cognitive abilities. Appetite. 2009;52:535–539. doi: 10.1016/j.appet.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 32.Elias MF, Elias PK, Sullivan LM, Wolf PA, D'Agostino RB. Lower cognitive function in the presence of obesity and hypertension: The Framingham heart study. Int J Obes Relat Metab Disord. 2003;27:260–268. doi: 10.1038/sj.ijo.802225. [DOI] [PubMed] [Google Scholar]
- 33.Gunstad J, et al. Elevated body mass index is associated with executive dysfunction in otherwise healthy adults. Compr Psychiatry. 2007;48:57–61. doi: 10.1016/j.comppsych.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Gustafson D, Lissner L, Bengtsson C, Björkelund C, Skoog I. A 24-year follow-up of body mass index and cerebral atrophy. Neurology. 2004;63:1876–1881. doi: 10.1212/01.wnl.0000141850.47773.5f. [DOI] [PubMed] [Google Scholar]
- 35.Wessels AM, et al. Voxel-based morphometry demonstrates reduced grey matter density on brain MRI in patients with diabetic retinopathy. Diabetologia. 2006;49:2474–2480. doi: 10.1007/s00125-006-0283-7. [DOI] [PubMed] [Google Scholar]
- 36.Last D, et al. Global and regional effects of type 2 diabetes on brain tissue volumes and cerebral vasoreactivity. Diabetes Care. 2007;30:1193–1199. doi: 10.2337/dc06-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeCarli C, et al. The effect of white matter hyperintensity volume on brain structure, cognitive performance, and cerebral metabolism of glucose in 51 healthy adults. Neurology. 1995;45:2077–2084. doi: 10.1212/wnl.45.11.2077. [DOI] [PubMed] [Google Scholar]
- 38.Wen W, Sachdev PS, Chen X, Anstey K. Gray matter reduction is correlated with white matter hyperintensity volume: A voxel-based morphometric study in a large epidemiological sample. Neuroimage. 2006;29:1031–1039. doi: 10.1016/j.neuroimage.2005.08.057. [DOI] [PubMed] [Google Scholar]
- 39.Sonestedt E, et al. Fat and carbohydrate intake modify the association between genetic variation in the FTO genotype and obesity. Am J Clin Nutr. 2009;90:1418–1425. doi: 10.3945/ajcn.2009.27958. [DOI] [PubMed] [Google Scholar]
- 40.Rampersaud E, et al. Physical activity and the association of common FTO gene variants with body mass index and obesity. Arch Intern Med. 2008;168:1791–1797. doi: 10.1001/archinte.168.16.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vimaleswaran KS, et al. Physical activity attenuates the body mass index-increasing influence of genetic variation in the FTO gene. Am J Clin Nutr. 2009;90:425–428. doi: 10.3945/ajcn.2009.27652. [DOI] [PubMed] [Google Scholar]
- 42.Mueller SG, et al. The Alzheimer's disease neuroimaging initiative. Neuroimaging Clin N Am. 2005;15:869–877. xi–xii. doi: 10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mueller SG, et al. Ways toward an early diagnosis in Alzheimer's disease: The Alzheimer's Disease Neuroimaging Initiative (ADNI) Alzheimers Dement. 2005;1:55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 45.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jack CR, Jr, et al. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leow AD, et al. Longitudinal stability of MRI for mapping brain change using tensor-based morphometry. Neuroimage. 2006;31:627–640. doi: 10.1016/j.neuroimage.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ho A, et al. Comparing 3 T and 1.5 T MRI for tracking AD progression with tensor-based morphometry. Hum Brain Mapp. 2010 doi: 10.1002/hbm.20882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leow AD, et al. Statistical properties of Jacobian maps and the realization of unbiased large-deformation nonlinear image registration. IEEE Trans Med Imaging. 2007;26:822–832. doi: 10.1109/TMI.2007.892646. [DOI] [PubMed] [Google Scholar]
- 50.Morra JH, et al. Automated 3D mapping of hippocampal atrophy and its clinical correlates in 400 subjects with Alzheimer's disease, mild cognitive impairment, and elderly controls. Hum Brain Mapp. 2009;30:2766–2788. doi: 10.1002/hbm.20708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morra JH, et al. Validation of a fully automated 3D hippocampal segmentation method using subjects with Alzheimer's disease mild cognitive impairment, and elderly controls. Neuroimage. 2008;43:59–68. doi: 10.1016/j.neuroimage.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.