Abstract
Hepatocellular carcinoma (HCC) is a highly aggressive cancer with no currently available effective treatment. Understanding of the molecular mechanism of HCC development and progression is imperative for developing novel, effective, and targeted therapies for this lethal disease. In this article, we document that the cellular transcription factor Late SV40 Factor (LSF) plays an important role in HCC pathogenesis. LSF protein was significantly overexpressed in human HCC cells compared to normal hepatocytes. In 109 HCC patients, LSF protein was overexpressed in >90% cases, compared to normal liver, and LSF expression level showed significant correlation with the stages and grades of the disease. Forced overexpression of LSF in less aggressive HCC cells resulted in highly aggressive, angiogenic, and multiorgan metastatic tumors in nude mice. Conversely, inhibition of LSF significantly abrogated growth and metastasis of highly aggressive HCC cells in nude mice. Microarray studies revealed that as a transcription factor, LSF modulated specific genes regulating invasion, angiogenesis, chemoresistance, and senescence. The expression of osteopontin (OPN), a gene regulating every step in tumor progression and metastasis, was robustly up-regulated by LSF. It was documented that LSF transcriptionally up-regulates OPN, and loss-of-function studies demonstrated that OPN plays an important role in mediating the oncogenic functions of LSF. Together, these data establish a regulatory role of LSF in cancer, particularly HCC pathogenesis, and validate LSF as a viable target for therapeutic intervention.
Keywords: metastasis, osteopontin, transcription regulation
Hepatocellular carcinoma (HCC) is one of the five most common cancers worldwide (1). The incidence of HCC is increasing despite a decrease in overall incidence of all cancers (2, 3). In the United States, the estimated new cases of HCC for 2008 were 21,370, of which 18,410 were expected to die (2). The mortality rate of HCC parallels that of its incidence because HCC is a tumor with rapid growth and early vascular invasion that is resistant to conventional chemotherapy, and no systemic therapy is available for the advanced disease (4). As such, understanding the molecular mechanism of HCC development and progression is imperative to establish novel, effective, and targeted therapies for this highly aggressive cancer. Our recent studies reveal that astrocyte elevated gene-1 (AEG-1) is overexpressed in >90% of human HCC patients, compared to normal liver, and AEG-1 plays a key role in regulating development and progression of HCC (5). The transcription factor Late SV40 Factor (LSF) was identified as a downstream gene of AEG-1, and we demonstrated that LSF mediates, in part, AEG-1-induced resistance to 5-fluorouracil (5-FU) in HCC cells (5, 6).
LSF, also known as LBP-1c and TFCP2, regulates diverse cellular and viral promoters (7, 8). A major cellular target of LSF is the thymidylate synthase (TS) gene, which encodes the rate-limiting enzyme in the production of dTTP, required for DNA synthesis (9). Inhibition of LSF abrogates TS induction and induces apoptosis. Thus, LSF plays an important role in DNA synthesis and cell survival. In the liver, LSF is activated by inflammatory cytokines and regulates the expression of acute phase proteins (10, 11). As yet, no studies have linked LSF to the process of tumorigenesis. However, several findings suggest a potential role of LSF in this process. LSF facilitates entry into G1/S phase of the cell cycle, promotes DNA synthesis, and functions as an antiapoptotic factor (9). Overexpression of LSF might augment all of these effects, thus promoting transformation and cancer cell survival. Additionally, because most HCCs are generated in the background of HBV or HCV infection, the activation of LSF by inflammatory cytokines that are secreted upon viral infection suggests that LSF might also play a role in the pathogenesis of inflammatory aspects of HCC. We therefore performed a detailed experimental analysis to elucidate the role of LSF in hepatocarcinogenesis.
Results and Discussion
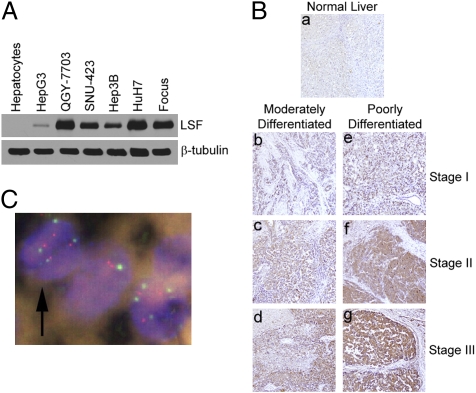
Whereas in normal hepatocytes LSF protein expression was virtually undetected, its expression was robustly up-regulated in human HCC cells, except HepG3 cells, which do not form tumors in nude mice (Fig. 1A) (5). These findings were extended by tissue microarrays containing 86 primary HCC, 23 metastatic HCC, and 9 normal adjacent liver samples that were immunostained using anti-LSF antibody. Little to no LSF immunostaining was detected in the 9 normal liver samples, whereas significant LSF staining was observed in HCC samples (Fig. 1B). LSF expression was detected predominantly in the nucleus (Fig. S1). Among the 109 HCC samples, only 9 scored negative for LSF, and the remaining 100 (91.7%) showed variable levels of LSF that could be significantly correlated with the stages of the disease based on the BCLC staging system (Table 1) (12). Expression of LSF gradually increased with the stages from I to IV (Fig. 1B and Fig. S1), as well as with the grades of differentiation from well differentiated to poorly differentiated (e.g., compare Fig. 1Bb and e, c and f, and d and g).
Fig. 1.
LSF is overexpressed in HCC. (A) LSF expression was detected by Western blot in the indicated cells. β-tubulin was used as loading control. (B) Analysis of LSF expression in tissue microarray by immunohistochemistry. (C) FISH was performed on human HCC samples for LSF and D12Z3 (probe targeting pericentromeric region of chromosome 12). Red, LSF; green, D12Z3; arrow, cell displaying four signals for each of the probes, indicating four copies of these regions of chromosome 12.
Table 1.
Immunoperoxidase staining of normal liver and different stages of HCC by tissue microarray using anti-LSF antibody: Intensity of LSF staining
| 0 | + | ++ | +++ | Total cases | |
| Normal liver | 4 | 5 | 9 | ||
| Stage I HCC | 4 | 15 | 3 | 1 | 23 |
| Stage II HCC | 1 | 14 | 8 | 2 | 25 |
| Stage III HCC | 3 | 15 | 15 | 5 | 38 |
| Stage IV HCC | 1 | 4 | 8 | 10 | 23 |
To assess the strength of association between LSF expression and stages of HCC, an ordinal logistic regression was conducted with the stage of HCC as the ordinal response and LSF expression as the independent variable in the proportional odds model. The hypothesis of association is highly significant: P < 0.001 using Pearson’s χ2 test with Yates’s continuity correction. A total of 109 HCC cases were analyzed.
Amplification of chromosome band 12q13, the location of the LSF gene, has been reported in some cases of HCC (13, 14). To examine the possibility that copy number gain might be the underlying mechanism of LSF protein overexpression in human HCC patients, dual-color fluorescence in situ hybridization (FISH) was performed on human HCC tissue microarrays containing 9 normal liver samples and 50 HCC samples. Bacterial artificial chromosome (BAC)-derived test probe targeting LSF (red color) was used along with a control probe that is specific for the pericentromeric region of chromosome 12 (D12Z3; green color). The control probe (D12Z3) provided information regarding the number of chromosomes 12 present in the cell. Copy number gains of LSF (amplification or low level gain) were not encountered in any of the HCC samples. However, 34 of 50 HCC samples (68%) exhibited an increased number of signals for both the LSF and D12Z3 probes, suggesting the presence of extra copies of a large region of chromosome 12 or polyploidy. Fig. 1C shows a representative cell (arrow) in which four red and four greet dots are observed. The red dots represent signals from LSF probe, and the green dots represent signals from D12Z3 probe. The presence of four signals from both control (D12Z3) and target (LSF) probes indicates that there are four copies of chromosome 12 indicating polysomy. Thus, chromosome 12 polysomy might be one mechanism of LSF protein overexpression in human HCC in addition to its regulation by AEG-1.
Compared to other HCC cell lines, HepG3 cells express a significantly lower level of LSF. To examine the effect of LSF overexpression in HepG3 cells, we established stable cell lines expressing LSF. Several of these clones were analyzed for LSF overexpression, among which LSF-1 and LSF-17 clones showed LSF expression that is comparable to a naturally LSF-overexpressing cell line, such as QGY-7703 (Fig. 2A). The nuclear expression of LSF was confirmed in LSF-17 clone by immunofluorescence (Fig. S2). The luciferase activity of LSF WT-luc, a luciferase reporter construct containing four LSF-binding sites, was significantly higher in LSF-1 and LSF-17 clones compared to control neomycin-resistant clones Control-8 and Control-13 (Fig. 2B). Both LSF-1 and LSF-17 clones showed higher proliferative activity (Fig. 2C), colony forming ability (Fig. 2D), anchorage-independent growth in soft agar (Fig. 2E), and Matrigel invasion abilities (Fig. 2F) compared to Control-8 and Control-13 clones. Interestingly, LSF overexpression resulted in chromosomal instability in HepG3 cells as evidenced by a significantly increased frequency of micronuclei in the LSF-1 clone (P < 0.05) (Fig. S3).
Fig. 2.
LSF overexpression increases proliferation, anchorage-independent growth, and invasion of HepG3 cells. (A) Control-8 (Cont-8) and Control-13 (Cont-13) clones are neomycin-resistant clones, and LSF-1 and LSF-17 clones are LSF-overexpressing clones of HepG3 cells. Western blot analysis was performed to detect LSF and β-tubulin expression in these cells. (B) LSF WT-Luc, luciferase reporter plasmid preceded by four tandem LSF-binding sites; LSF-MT-Luc, luciferase reporter plasmid preceded by mutated LSF-binding sites. The indicated cells were transfected with either empty pGL3-basic vector or LSF WT-Luc or LSF MT-Luc along with renilla luciferase expression vector. Luciferase assay was performed 2 days later, and firefly luciferase activity was normalized by renilla luciferase activity. (C) Cell viability of the indicated cells at the indicated time points was measured by standard MTT assay. (D) Colony formation assay for the indicated cells. Colony number per 250 cells is shown. (E) Soft agar assay for the indicated cells. For D and E, the colonies were scored 2 weeks after plating. (F) Matrigel invasion assay using the indicated clones. (Inset) Invading cells. For B–F, the data represents mean ± SEM.
As complementation to the LSF-overexpressing clones, we established stable clones of QGY-7703 cells expressing a dominant negative LSF (LSFdn, a double amino acid substitution mutant of LSF initially named 234QL/236KE that is unable to bind DNA) (9). An increased level of LSF expression over the control clones indicated expression of LSFdn. LSFdn-8 and LSFdn-15 clones expressed significantly higher levels of LSFdn compared to neomycin-resistant control clones Control-1 and Control-7 (Fig. 3A). The authenticity of these clones was confirmed by lack of activity of LSF WT-luc in LSFdn-8 and LSFdn-15 clones compared to Control-1 and Control-7 clones (Fig. 3B). Compared to Control-1 and Control-7 clones, LSFdn-8 and LSFdn-15 clones had slower proliferation rate (Fig. 3C), less colony formation (Fig. 3D), anchorage-independent growth in soft agar (Fig. 3E), and Matrigel invasion abilities (Fig. 3F).
Fig. 3.
Dominant negative LSF (LSFdn) inhibits proliferation, anchorage-independent growth, and invasion by QGY-7703 cells. (A) Control-1 (Cont-1) and Control-7 (Cont-7) clones are neomycin-resistant clones, and LSFdn-8 (dn-8) and LSFdn-15 (dn-15) clones are dominant negative LSF-overexpressing clones of QGY-7703 cells. Western blot analysis was performed to detect LSF and β-tubulin expression in these cells. (B) The indicated cells were transfected with either empty pGL3-basic vector or LSF WT-Luc or LSF MT-Luc along with renilla luciferase expression vector. Luciferase assay was performed 2 days later, and firefly luciferase activity was normalized by renilla luciferase activity. (C) Cell viability of the indicate cells at the indicated time points was measured by standard MTT assay. (D) Colony formation assay for the indicated cells. Colony number per 250 cells is shown. (E) Soft agar assay for the indicated cells. For (D) and (E), the colonies were scored 2 weeks after plating. (F) Matrigel invasion assay using the indicated clones. (Inset) Invading cells. For B–F, the data represents mean ± SEM.
The tumor-promoting properties of LSF were confirmed by nude mice xenograft assays. Whereas control HepG3 clone Control-8 did not form any tumors, LSF-1 and LSF-17 clones reproducibly generated large and aggressive tumors when implanted s.c. in the flanks of athymic nude mice (Fig. 4 A and B). As a corollary, LSFdn-8 and LSFdn-15 clones of QGY-7703 cells formed significantly smaller s.c. tumors in nude mice compared to the control QGY-7703 clones, Control-1 and Control-7 (Fig. 4 C and D). Analysis of LSF-1 and LSF-17 tumor sections revealed high LSF expression, high proliferation index (analyzed by Ki-67 expression), and increased angiogenesis (determined by CD31 expression indicative of microvessel formation) (Fig. 4E). As expected, LSFdn-15 tumors showed high LSFdn expression, low proliferation index (Ki-67 expression), and decreased angiogenesis (CD31 expression) compared to Control-7 tumors (Fig. 4F). These findings indicate that LSF positively regulates growth and invasion of HCC cells.
Fig. 4.
Overexpression of LSF increases and inhibition of LSF decreases tumorigenesis of human HCC cells in nude mice. Control-8, LSF-1, and LSF-17 clones of HepG3 cells were s.c. implanted in athymic nude mice. Tumor volume (A) and tumor weight (B) were measured 3 weeks after implantation. Control-1, Control-7, LSFdn-8, and LSFdn-15 clones of QGY-7703 cells were s.c. implanted in athymic nude mice. Tumor volume (C) and tumor weight (D) were measured 3 weeks after implantation. Immunofluorescence analysis of LSF, Ki-67, and CD31 in tumor sections of LSF-1 and LSF-17 clones of HepG3 cells (E) and Control-7 and LSFdn-15 clones of QGY-7703 cells (F).
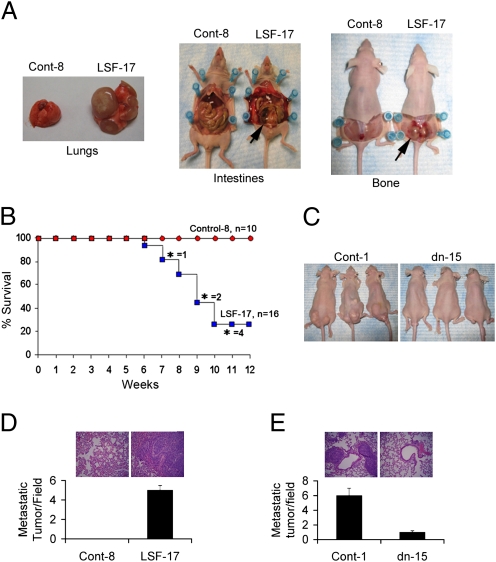
In in vitro assays, the most significant effect of LSF overexpression or inhibition was observed in the Matrigel invasion assay (Figs. 2 and 3). Because invasion is the first step in metastasis, we evaluated the metastasizing capabilities of the established clones by the tail vein metastasis assay. Intravenous injection of LSF-1 and LSF-17 clones (figures shown only for LSF-17 clone) resulted in multiorgan macrometastasis, whereas no metastasis was observed for Control-8 clone of HepG3 cells (Fig. 5A). Metastasis was observed in the lungs (Fig. 5A Left), intestinal regions (Fig. 5A Center, arrow), and liver, and in the lower back region involving the vertebral column (Fig. 5A Right, arrow). The LSF-17-injected animals lost significant body weight (compare the size of the animals in Fig. 5A Center and Right), became cachexic, and started losing ~20% body weight (indication for euthanasia and considered as dead) 6 weeks after injection (Fig. 5B). As demonstrated by Kaplan–Meier survival curves, 80% of the animals injected with LSF-17 clone died by 12 weeks after injection, whereas none of the animals injected with the Control-8 clone of HepG3 cells died (Fig. 5B). Staining of the lungs showed preservation of normal alveolar architecture in Control-8-injected animals, whereas in LSF-17-injected animals the lungs were filled with a solid mass of infiltrating tumor cells adjacent to the blood vessels indicating that the tumor cells had extravasated, lodged into the lungs, and established colonies (Fig. 5D).
Fig. 5.
Overexpression of LSF increases and inhibition of LSF decreases metastasis of human HCC cells in nude mice. (A) Control-8 and LSF-17 clones of HepG3 cells were injected i.v. through the tail vein in athymic nude mice. The internal organs were analyzed 4–6 weeks after injection. (B) Kaplan–Meier survival curve of animals injected with either Control-8 or LSF-17 clones of HepG3 cells. *, mice losing ~20% body weight and euthanized (considered as dead). (C) Control-1 and LSFdn-15 clones of QGY-7703 cells were injected i.v. through the tail vein in athymic nude mice. Metastatic tumors were visible externally in mice injected with the Control-1 clone but not with the LSFdn-15 clone. (D) Graphical representation of metastatic lung nodules in the animals injected with Control-8 and LSF-17 clones of HepG3 cells. (Inset) H&E sections of lungs. (E) Graphical representation of metastatic lung nodules in the animals injected with Control-1 and LSFdn-15 clones of QGY-7703 cells. (Inset) H&E sections of lungs. For (D) and (E), the data represent mean ± SEM.
For QGY-7703 cells, the Control-1 and Control-7 clones gave rise to multiorgan metastatic tumors, whereas LSFdn-8 and LSFdn-15 clones did not show any external signs of metastasis (Fig. 5C). Staining of the lungs identified multiple solid nodules in Control-1-injected animals, whereas normal architecture was preserved in LSFdn-15-injected animals, with only a few isolated metastatic nodules (Fig. 5E).
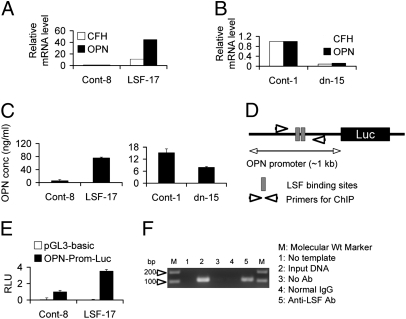
To identify the downstream genes mediating the effects of LSF in HCC cells, gene expression profiles were compared between Control-8 and LSF-17 clones of HepG3 cells by Affymetrix microarray (GEO accession no. GSE19815). With a 2.0-fold cut-off, expression levels of 125 genes were up-regulated and those of 148 genes were down-regulated upon overexpression of LSF. Twenty-one of these genes are directly involved in the process of tumorigenesis (Fig. S4 and Table S1). The most robust induction was observed for SPP1, which encodes osteopontin (OPN), known to be important for regulating every step in metastasis (15). The microarray data were confirmed by quantitative RT-PCR for several genes, showing an ~40-fold increase in OPN mRNA expression in the LSF-17 clone, as compared to the Control-8 clone (Fig. 6A). As a corollary, OPN mRNA expression was markedly down-regulated in the LSFdn-15 clone of QGY-7703 cells compared to the Control-1 clone (Fig. 6B). Another LSF-downstream gene, complement factor H (CFH), also showed a similar trend (Fig. 6 A and B). These findings were confirmed at the protein level by ELISA (Fig. 6C). The robust induction of OPN in LSF-overexpressing clones prompted us to hypothesize that LSF might regulate OPN expression at the transcriptional level. We scanned an ~1-kb region of the OPN promoter and identified two tandem LSF binding sites in this region (Fig. 6D). Consistent with that prediction, OPN promoter-luciferase reporter construct demonstrated significantly higher activity in the LSF-17 clone compared to the Control-8 clone (Fig. 6E) (16). Finally, a chromatin immunoprecipitation (ChIP) assay confirmed LSF binding to the OPN promoter (Fig. 6F).
Fig. 6.
OPN expression is transcriptionally induced by LSF. (A and B) Real-time PCR analysis of CFH and OPN mRNA expression in Control-8 and LSF-17 clones of HepG3 cells (A) and Control-1 and LSFdn-15 clones of QGY-7703 cells (B). (C) OPN expression was detected by ELISA in Control-8 and LSF-17 clones of HepG3 cells (Left) and Control-1 and LSFdn-15 clones of QGY-7703 cells (Right). (D) Schematic diagram of OPN promoter-luciferase construct showing the location of LSF binding sites in the promoter and primers designed for ChIP assay. (E) Control-8 and LSF-17 clones of HepG3 cells were transfected with pGL3-basic vector or OPN-Prom-luc along with renilla luciferase expression vector. Luciferase assay was performed 2 days later, and firefly luciferase activity was normalized by renilla luciferase activity. (F) ChIP assay to detect LSF binding to the OPN promoter.
To confirm the role of OPN in mediating LSF effect, we established stable OPN shRNA-expressing clones in the background of LSF-17 clone of HepG3 cells (LSF17-OPNsh). Two independent clones LSF-17-OPNsh-6 and LSF-17-OPNsh-18 showed marked down-regulation of OPN mRNA and protein expression (Fig. 7 A and B, respectively) when compared to the parental LSF-17 clone or LSF-17consh-15 clone that stably expresses control scrambled shRNA. LSF expression remained unchanged in LSF-17, LSF-17consh-15, and LSF-17-OPNsh clones (Fig. S5). Compared to parental LSF-17 and LSF-17consh-15 clone, LSF17-OPNsh-6 and LSF17-OPNsh-18 clones had a significantly slower proliferation rate (Fig.7C), less colony formation (Fig. 7D), anchorage-independent growth in soft agar (Fig. 7E), and Matrigel invasion abilities (Fig. 7F). LSF17-OPNsh-6 and LSF17-OPNsh-18 clones formed significantly smaller s.c. tumors in nude mice compared to the parental LSF-17 and LSF-17consh-15 clone (Fig. 8A). These studies were further corroborated by experimental metastasis assays demonstrating significantly decreased numbers of metastatic nodules in the lungs in mice injected with LSF17-OPNsh-18 clone compared with those injected with LSF-17consh-15 clone (Fig. 8B).
Fig. 7.
Inhibition of OPN abrogates augmentation of proliferation, anchorage-independent growth, and invasion by LSF. LSF-17-OPNsh-6 (OPNsh-6) and LSF-17-OPNsh-18 (OPNsh-18) clones stably express OPN shRNA and were generated in the background of LSF-17 clone of HepG3 cells. LSF-17consh-15 (Consh-15) clone stably expresses control scrambled shRNA and was also generated in LSF-17 background. (A) OPN mRNA expression detected by real-time PCR in the indicated clones. (B) OPN protein expression detected by ELISA in the indicated clones. (C) Cell viability of the indicated cells at the indicated time points were measured by standard MTT assay. (D) Colony formation assay for the indicated cells. (E) Soft agar assay for the indicated cells. (F) Matrigel invasion assay of the indicated clones. The data represents mean ± SEM.
Fig. 8.
Inhibition of OPN abrogates LSF-induced tumorigenesis and metastasis. The indicated clones were s.c. implanted in athymic nude mice. (A) Tumor volume and tumor weight were measured 3 weeks after implantation. (B) Graphical representation of metastatic lung nodules in the animals injected with the LSF-17consh-15 and LSF-17-OPNsh-18 clones via tail vein. The data represent mean ± SEM. (Inset) H&E sections of lungs of animals injected with the indicated clones. (C) Parental HepG3 cells were treated with conditioned media from Control-8 or LSF-17 clone of HepG3 cells and then subjected to Matrigel invasion. (D) Matrigel invasion assay using LSF-17 clone of HepG3 cells in the presence of neutralizing antibodies. Integrin, anti-αvβ3 integrin antibody; CD44, anti-CD44 antibody. The data represent mean ± SEM.
Because OPN is a secreted protein, we checked whether conditioned media from LSF-17 clones might augment the invasive ability of the parental HepG3 cells. Indeed, conditioned media from the LSF-17 clone, but not from the Control-8 clone, significantly increased invasion by HepG3 cells (Fig. 8C). OPN works through αvβ3 integrin and CD44 receptors (15). We blocked these receptors on LSF-17 clone of HepG3 cells with neutralizing antibodies and performed Matrigel invasion assay. Whereas normal IgG did not affect the invasive ability of the LSF-17 cells, anti-αvβ3 integrin or anti-CD44 antibody significantly inhibited invasion and the combination of the two antibodies decreased the invasion, further confirming that OPN working through its canonical receptors plays a key role in regulating LSF function (Fig. 8D). It should be noted that for all of the assays described in this article using isolated clones, similar in vitro phenotypes, although less pronounced because of transfection efficiency, were observed with transient transfection assays without selection, thereby ruling out any clonal bias arising from the selection procedure.
Our present findings reveal a role of LSF in the process of hepatocarcinogenesis. We demonstrate that by augmenting transcription of OPN, LSF promotes aggressive progression of HCC. OPN levels can be used as a sensitive and specific marker in predicting disease progression in diverse cancers, including HCC, and OPN is known to promote every step in metastasis as well as growth of the primary tumor (15, 17). By regulating OPN expression, LSF functions as a key regulator of HCC development and progression. In addition, LSF also activates two important cell survival-regulating pathways, MEK/ERK and NF-κB (Fig. S6), and inhibition of the MEK/ERK pathway significantly abrogates invasion by LSF-17 cells (Fig. S7). Activation of NF-κB by LSF suggests its potential role in regulating the inflammatory aspects of HCC (18). Our present findings thus strongly suggest that LSF might be a viable target, and that small-molecule inhibitors targeting the DNA binding domains of LSF might be an effective HCC therapeutic. Additionally, the correlation of LSF expression with the stages and grades of HCC suggests that LSF might be used as a prognostic marker for this disease. Finally, the observation that LSF is overexpressed in cancer indications other than HCC indicates a potential oncogenic function of LSF in diverse other cancers (Fig. S8).
Materials and Methods
Cell Lines, Culture Condition, Viability, Colony Formation Assays, Anchorage-Independent Growth in Soft Agar, and Matrigel Invasion Assays.
Primary rat hepatocytes were isolated and cultured as described in ref. 19. SNU-423 cells were obtained from ATCC and cultured as instructed. HepG3, QGY-7703, Hep3B, HuH7, Focus, and HEK-293 cells were cultured as described in ref. 5 and 20. Cell viability was determined by standard MTT assays as described in ref. 5. Colony formation, anchorage-independent growth in soft agar, and Matrigel invasion assays were performed exactly as described in ref. 5.
Tissue Microarray.
Human HCC tissue microarrays were obtained from Imgenex. Two tissue microarrays were used: one containing 40 primary HCC, 10 metastatic HCC, and 9 normal adjacent liver samples (IMH-360; Imgenex), the other containing 46 primary HCC and 13 metastatic HCC (IMH-318; Imgenex) for immunohistochemistry. IMH-360 was used for fluorescence in situ hybridization analysis (FISH).
Construction of Stable Cell Lines.
LSF and dominant negative LSF (LSFdn) expression constructs were described in ref. 9. HepG3 clones stably expressing LSF and QGY-7703 clones stably expressing LSFdn were created by transfecting the corresponding expression constructs using Lipofectamine 2000 (Invitrogen) and selection with neomycin. An empty pcDNA3.1(+)-Neo plasmid was used similarly to establish the control clones. The LSF-17 clone of HepG3 cells was transduced with a pool of three to five lentiviral vector plasmids, each encoding target-specific 19- to 25-nt (plus hairpin) shRNAs designed to knock down osteopontin (OPN) gene expression (Santa Cruz Biotechnology). Individual colonies were selected by puromycin. Lentiviral particles expressing scrambled shRNA were used to similarly establish LSF-17Consh clones.
Transient Transfection and Luciferase Assay.
Transfection was carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. For LSF luciferase reporter assays, cells were plated into 24-well plates and the next day transfected with empty vector (pGL3-basic), pGL3B-WT4-E1b (luciferase reporter plasmid containing four tandem LSF-binding site; LSF WT-Luc), or pGL3B-MT4-E1b (luciferase reporter plasmid containing four tandem mutated LSF-binding site; LST MT-Luc) and renilla luciferase expression plasmid for transfection control (9). For the NF-κB luciferase reporter assay, cells were plated into 24-well plates and the next day transfected with 3κB-luc (luciferase reporter plasmid containing three tandem repeats of NF-κB binding site) and renilla luciferase expression plasmid for transfection control (21). Cells were incubated in the absence or presence of TNF-α (10 ng/mL) for 12 h. For the OPN promoter luciferase assay, cells were transfected with OPN-Prom-Luc construct containing ~1 kb of OPN promoter upstream of the luciferase gene (kindly provided by Paul C. Kuo, Duke University) along with a renilla luciferase expression plasmid (16). Luciferase assays were measured using a Dual Luciferase Reporter Assay kit (Promega) according to the manufacturer’s protocol, and firefly luciferase activity was normalized by renilla luciferase activity.
Preparation of Whole-Cell Lysates and Western Blot Analyses.
Preparation of whole-cell lysates and Western blot analyses were performed as described in ref. 5. The primary antibodies used were anti-LSF (1:2,000, mouse monoclonal; BD Biosciences), anti-pERK (1:2,000, rabbit polyclonal; Cell Signaling), anti-ERK (1:2,000, rabbit polyclonal; Cell Signaling), anti-pAKT (1:1,000, rabbit polyclonal; Cell Signaling), and anti-AKT (1:1,000, rabbit polyclonal; Cell Signaling). Blots were stripped and normalized by reprobing with anti-β-tubulin (1:1,000, mouse monoclonal; Sigma).
Immunostaining.
Immunofluorescence analysis in tumor sections was performed essentially as described in ref. 5. Anti-LSF (1:200, mouse monoclonal; BD Biosciences), anti-Ki-67 (1:200, mouse monoclonal; BD Biosciences), and anti-CD31 (1:200, mouse monoclonal; Dako) antibodies were used. Images were analyzed using an Olympus immunofluorescence microscope. For the tissue microarray (IMH-360 and IMH-318; Imgenex), anti-LSF antibody was used at 1:100 dilution and the signals were developed by avidin-biotin-peroxidase complexes with a DAB substrate solution (Vector Laboratories).
Nude Mice Xenograft Studies.
Subcutaneous xenografts were established in the flanks of athymic nude mice using 1 × 106 human HCC cells and the clones. Tumor volume was measured twice weekly with a caliper and calculated using the formula π/6 × larger diameter × (smaller diameter)2. Mice were followed for 3 weeks. For the metastasis assays, 1 × 106 cells were i.v. injected through the tail vein in nude mice. The lungs, intestines, liver, bone, and other organs were isolated and analyzed after 4 weeks. All experiments were performed with at least five mice in each group and repeated three times.
Total RNA Extraction, Real-Time PCR, and Microarray Assay.
Total RNA was extracted using Qiagen miRNeasy mini kit. Real-time PCR was performed using an ABI 7900 fast real-time PCR system and Taqman gene expression assays for OPN, CFH, and GAPDH according to the manufacturer’s protocol (Applied Biosystems). An Affymetrix oligonucleotide microarray (GeneChip Human Genome U133A 2.0) analysis was performed to compare gene expression between Control-8 and LSF-17 clones of HepG3 cells using standard Affymetrix protocol (22).
Fluorescence in Situ Hybridization and Micronuclei Analysis.
Dual-color fluorescence in situ hybridization (FISH) was performed as previously described on hepatocellular carcinoma tissue microarrays (23). Bacterial artificial chromosome (BAC)-derived test probes targeting LSF (12q13, RP11-142E3; BACPAC Resources Center) were paired with an enumeration probe for the pericentromeric region of chromosome 12 (D12Z3) for dual-target hybridization. For micronuclei analysis, interphase nuclei from the parental HepG3 cells and LSF-1 clones were harvested and slides were prepared according to standard procedures using the criteria of Fenech (24). The frequency of micronuclei present in the cell lines was compared using a χ2 test with a significance level of α = 0.05.
ChIP Assays.
ChIP assays were performed using a commercially available kit from Active Motif according to the manufacturer’s protocol. OPN promoter-specific primers used were sense 5′-ACACGCTTATGCGGGTATGT-3′ and antisense 5′-GAACATTTGGTAGGGGGAAA-3′.
Statistical Analysis.
Data were represented as the mean ± SEM and analyzed for statistical significance using one-way ANOVA followed by Newman–Keuls test as a post hoc test. To assess the strength of association between LSF expression and stages of HCC, an ordinal logistic regression was conducted with the stage of HCC as the ordinal response and LSF expression as the independent variable in the proportional odds model using Pearson’s χ2 test with Yates’s continuity correction.
Supplementary Material
Acknowledgments
We thank Mr. Nicollaq Vozhilla for excellent technical assistance. This work was supported in part by grants from the Goldhirsh Foundation and the Dana Foundation and National Cancer Institute Grant R01 CA138540-01A1 (to D.S.), National Institutes of Health Grant R01 CA134721 (to P.B.F.), the Samuel Waxman Cancer Research Foundation (to P.B.F.), National Institute of Environmental Health Sciences Grant R01 ES12074 (to C.J.-C.), and the Liver Tissue Cell Distribution System (National Institutes of Health Contract N01-DK-7-0004/HHSN267200700004C). Human tissues, patient consents, and clinical data were obtained through collaboration with the Tissue and Data Acquisition and Analysis Core–Massey Cancer Center and the Department of Pathology, Virginia Commonwealth University, with support from the Tissue Acquisition System to Support Cancer Research protocol. D.S. is the Harrison Endowed Scholar in Cancer Research. P.B.F. holds the Thelma Newmeyer Corman Chair in Cancer Research and is a Samuel Waxman Cancer Research Foundation Investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The Affymetrix microarray data have been deposited in the Gene Expression Omnibus (GEO) database (GEO accession no. GSE19815).
This article contains supporting information online at www.pnas.org/cgi/content/full/1000374107/DCSupplemental.
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 3.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 4.Bruix J, Sherman M Practice Guidelines Committee, American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma. Hepatology. 2005;42:1208–1236. doi: 10.1002/hep.20933. [DOI] [PubMed] [Google Scholar]
- 5.Yoo BK, et al. Astrocyte elevated gene-1 regulates hepatocellular carcinoma development and progression. J Clin Invest. 2009;119:465–477. doi: 10.1172/JCI36460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoo BK, et al. Identification of genes conferring resistance to 5-fluorouracil. Proc Natl Acad Sci USA. 2009;106:12938–12943. doi: 10.1073/pnas.0901451106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veljkovic J, Hansen U. Lineage-specific and ubiquitous biological roles of the mammalian transcription factor LSF. Gene. 2004;343:23–40. doi: 10.1016/j.gene.2004.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen U, Owens L, Saxena UH. Transcription factors LSF and E2Fs: Tandem cyclists driving G0 to S? Cell Cycle. 2009;8:2146–2151. doi: 10.4161/cc.8.14.9089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Powell CM, Rudge TL, Zhu Q, Johnson LF, Hansen U. Inhibition of the mammalian transcription factor LSF induces S-phase-dependent apoptosis by downregulating thymidylate synthase expression. EMBO J. 2000;19:4665–4675. doi: 10.1093/emboj/19.17.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang JH, Liao WS. Synergistic induction of mouse serum amyloid A3 promoter by the inflammatory mediators IL-1 and IL-6. J Interferon Cytokine Res. 1999;19:1403–1411. doi: 10.1089/107999099312867. [DOI] [PubMed] [Google Scholar]
- 11.Bing Z, Huang JH, Liao WS. NFkappa B interacts with serum amyloid A3 enhancer factor to synergistically activate mouse serum amyloid A3 gene transcription. J Biol Chem. 2000;275:31616–31623. doi: 10.1074/jbc.M005378200. [DOI] [PubMed] [Google Scholar]
- 12.Llovet JM, Brú C, Bruix J. Prognosis of hepatocellular carcinoma: The BCLC staging classification. Semin Liver Dis. 1999;19:329–338. doi: 10.1055/s-2007-1007122. [DOI] [PubMed] [Google Scholar]
- 13.Zondervan PE, et al. Molecular cytogenetic evaluation of virus-associated and non-viral hepatocellular carcinoma: Analysis of 26 carcinomas and 12 concurrent dysplasias. J Pathol. 2000;192:207–215. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH690>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 14.Wilkens L, et al. Cytogenetic aberrations in primary and recurrent fibrolamellar hepatocellular carcinoma detected by comparative genomic hybridization. Am J Clin Pathol. 2000;114:867–874. doi: 10.1309/BMTT-JBPD-D13H-1UVD. [DOI] [PubMed] [Google Scholar]
- 15.Bellahcene A, Castronovo V, Ogbureke KU, Fisher LW, Fedarko NS. Small integrin-binding ligand N-linked glycoproteins (SIBLINGs): Multifunctional proteins in cancer. Nat Rev Cancer. 2008;8:212–226. doi: 10.1038/nrc2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takami Y, et al. Sp1 regulates osteopontin expression in SW480 human colon adenocarcinoma cells. Surgery. 2007;142:163–169. doi: 10.1016/j.surg.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan HW, et al. Overexpression of osteopontin is associated with intrahepatic metastasis, early recurrence, and poorer prognosis of surgically resected hepatocellular carcinoma. Cancer. 2003;98:119–127. doi: 10.1002/cncr.11487. [DOI] [PubMed] [Google Scholar]
- 18.Pikarsky E, et al. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431:461–466. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- 19.Bissell DM, Guzelian PS. Phenotypic stability of adult rat hepatocytes in primary monolayer culture. Ann NY Acad Sci. 1980;349:85–98. doi: 10.1111/j.1749-6632.1980.tb29518.x. [DOI] [PubMed] [Google Scholar]
- 20.Su ZZ, Luo ZY, Guo LP, Li JZ, Liu YL. Inhibitory effect of parvovirus H-1 on cultured human tumour cells or transformed cells. Sci Sin B. 1988;31:69–80. [PubMed] [Google Scholar]
- 21.Sarkar D, et al. Molecular basis of nuclear factor-kappaB activation by astrocyte elevated gene-1. Cancer Res. 2008;68:1478–1484. doi: 10.1158/0008-5472.CAN-07-6164. [DOI] [PubMed] [Google Scholar]
- 22.Irizarry RA, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuller CE, Wang H, Zhang W, Fuller GN, Perry A. High-throughput molecular profiling of high-grade astrocytomas: The utility of fluorescence in situ hybridization on tissue microarrays (TMA-FISH) J Neuropathol Exp Neurol. 2002;61:1078–1084. doi: 10.1093/jnen/61.12.1078. [DOI] [PubMed] [Google Scholar]
- 24.Fenech M. Cytokinesis-block micronucleus assay evolves into a “cytome” assay of chromosomal instability, mitotic dysfunction and cell death. Mutat Res. 2006;600:58–66. doi: 10.1016/j.mrfmmm.2006.05.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.