Abstract
Most laboratory mouse strains including C57BL/6J do not produce detectable levels of pineal melatonin owing to deficits in enzymatic activity of arylalkylamine N-acetyltransferase (AANAT) and N-acetylserotonin O-methyl transferase (ASMT), two enzymes necessary for melatonin biosynthesis. Here we report that alleles segregating at these two loci in C3H/HeJ mice, an inbred strain producing melatonin, suppress the circadian period-lengthening effect of the Clock mutation. Through a functional mapping approach, we localize mouse Asmt to chromosome X and show that it, and the Aanat locus on chromosome 11, are significantly associated with pineal melatonin levels. Treatment of suprachiasmatic nucleus (SCN) explant cultures from Period2Luciferase (Per2Luc) Clock/+ reporter mice with melatonin, or the melatonin agonist, ramelteon, phenocopies the genetic suppression of the Clock mutant phenotype observed in living animals. These results demonstrate that melatonin suppresses the Clock/+ mutant phenotype and interacts with Clock to affect the mammalian circadian system.
Keywords: arylalkylamine N-acetyltransferase, N-acetylserotonin O-methyl transferase, Clock gene, suprachiasmatic nucleus
Circadian (from the Latin, meaning about a day) rhythms of biochemistry and physiology are innate to virtually all life forms (1, 2). Cell autonomous circadian pacemakers drive these rhythms and synchronize them each day to environmental cues. In mammals, the suprachiasmatic nucleus (SCN) of the hypothalamus contains the central pacemaker that coordinates the local circadian clocks present in tissues throughout the body (3, 4). The molecular clock mechanism within individual cells is composed of transcriptional/translational feedback loops and posttranslational processes (1, 5). The bHLH-PAS transcription factors CLOCK and BMAL1 form the primary loop as they activate transcription of the Period (Per1 and Per2) and Cryptochrome (Cry1 and Cry2) genes (1, 6). The PER and CRY proteins subsequently feed back to abrogate their own transcription by directly inhibiting the CLOCK:BMAL1 complex. A second feedback loop stabilizes the primary loop and is composed of REV-ERBα and RORa, two retinoic acid-related orphan receptors, which repress and activate, respectively, transcription of Bmal1. Whereas approximately a dozen genes involved in this timekeeping system have been identified in mammals, it is clear that other, as-yet unknown genes regulate key properties of circadian rhythms (7, 8).
The SCN imparts daily entraining signals to circadian clocks in cells of peripheral tissues via neural connections and humoral factors (9, 10). One well-characterized circadian rhythm synchronized by the SCN is the synthesis and release at night of the lipophilic pineal hormone melatonin (11). Light information reaches the mammalian pineal gland through a multisynaptic pathway that begins with intrinsically photosensitive melanopsin-containing retinal ganglion cells, which project to the SCN via the retinohypothalamic tract, and ultimately terminates at sympathetic afferents of the pineal gland (12, 13). Lesions of the SCN abolish the melatonin circadian rhythm (14). Two high-affinity melatonin receptors, Mtnr1a and Mtnr1b, have been characterized, and both are expressed in the SCN (15–17). This is interesting as it raises the possibility that melatonin may directly act to affect SCN output. Indeed, studies with Mtnr1a knockout mice show that this receptor mediates acute inhibition of SCN electrical activity by melatonin (18). Furthermore, the phase-shifting effects of exogenous melatonin administration have been associated with the Mtnr1b receptor (19).
Most of the commonly used laboratory mouse strains are deficient in pineal melatonin synthesis, likely as a result of compromised activity of one or both of the major enzymes in the melatonin biosynthesis pathway, AANAT and ASMT (also referred to as hydroxyindole O-methyltransferase, HIOMT) (20–23). While mapping the Clock mutation in mice, which lengthens the circadian period of the locomotor activity rhythm (24), we observed that the Clock/+ behavioral phenotype was significantly suppressed by the C3H/HeJ genetic background. A useful approach in studying the function of a gene is the identification of other loci that modify its mutant phenotype using quantitative trait locus (QTL) mapping (25). For example, the effectiveness of this approach has been demonstrated previously by the identification of Mom-1 (Modifier of Min) in which the expression of Min (multiple intestinal neoplasia) was dramatically affected by genetic background (26). Here we used the QTL modifier approach to map loci that suppress the free-running circadian period of locomotor activity in [(C3H/HeJ × C57BL/6JClock/+)F1 × C57BL/6J]N2 Clock/+ mutant mice, and identify a genetic interaction between the melatonin biosynthetic pathway and the circadian clock mechanism in mammals.
Results
Effect of Strain Background on Circadian Behavior of Clock/+ Mice.
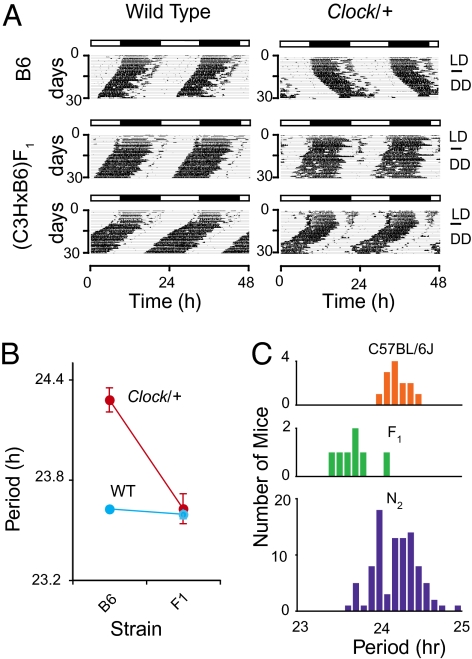
The Clock mutation was originally induced in the coisogenic C57BL/6J background by N-ethyl-N-nitrosourea (ENU) mutagenesis and is caused by a splicing mutation that deletes exon 19 (24, 27). This mutation lengthens the circadian free-running period by ~1 h in heterozygotes and by ~4 h in homozygous Clock mutant mice followed by arrhythmicity in constant darkness (DD). We observed that the circadian behavioral phenotype in Clock/+ animals is often suppressed by crossing to the C3H/HeJ background (Fig. 1A). The free-running period of Clock/+ on a coisogenic C57BL/6J background is 24.3 ± 0.13 h (mean ± SD), whereas that from (C3H/HeJ × C57BL/6J)F1 hybrids is 23.6 ± 0.22 h, a reduction in mutant period of almost 1 h. This period difference was not observed in wild-type littermates from the C57BL/6J (23.6 ± 0.23 h) and (C3H/HeJ × C57BL/6J)F1 (23.6 ± 0.11 h) crosses (Fig. 1B). This suggests that the C3H/HeJ strain carries genetic modifiers, which dominantly suppress the Clock mutant phenotype, but which have no discernible effect on period in wild-type littermates.
Fig. 1.
Effect of genetic background on Clock/+ phenotype. (A) Representative activity records for suppression of Clock mutant circadian behavior in C3H/HeJ background. Locomotor activity records from wild-type (Left) and Clock/+ mice (Right). The genetic backgrounds represented are C57BL/6J (B6) coisogenic animals (Upper, two records) and (C3H/HeJ × C57BL/6J)F1 hybrids (Lower, four records). The period suppression is observed only in Clock/+ mice. (B) Effect of genetic background on free-running period of locomotor activity rhythm. Both strain background and Clock genotype significantly affect circadian period. A two-way ANOVA was highly significant for the Clock genotype [F(1,32) = 56.24, P = 3.00 × 10−8], strain background [F(1,32) = 30.05, P = 6.65 × 10−6] and interaction [F(1,32) = 23.86, P = 3.49 × 10−5]. Each data point represents data from 7 to 25 mice. Error bars represent the mean ± SD. B6: C57BL/6J and F1: (C3H/HeJ × C57BL/6J)F1. (C) Distribution of circadian period for Clock/+ mice of different genetic backgrounds. C57BL/6J (24.3 ± 0.13 h, n = 13; Top), (C3H/HeJ × C57BL/6J)F1 (23.6 ± 0.22 h, n = 7; Middle), and [(C3H/HeJ × C57BL/6J)F1 × C57BL/6J]N2 (24.2 ± 0.27 h, n = 96; Bottom). C3H/HeJ animals carry dominant suppressors of the Clock mutation that segregate in the [(C3H/HeJ × C57BL/6J)F1 × C57BL/6J]N2 genetic background.
We examined the locomotor activity rhythms of [(C3H/HeJ × C57BL/6J)F1 × C57BL/6J]N2 Clock/+ mice to test whether these modifiers segregate in an N2 backcross population (Fig. 1C). Phenotypic variation among [(C3H/HeJ × C57BL/6J)F1 × C57BL/6J]N2 progeny was greater than among the parental animals, C57BL/6J and [(C3H/HeJ × C57BL/6J)F1. Some mice expressed phenotypes similar to Clock/+ from the C57BL/6J coisogenic background whereas others expressed phenotypes similar to Clock/+ from the (C3H/HeJ × C57BL/6J)F1 background. The larger phenotypic variation in the N2 population suggests that Clock suppressors segregate in this population.
Mapping of Clock Suppressors.
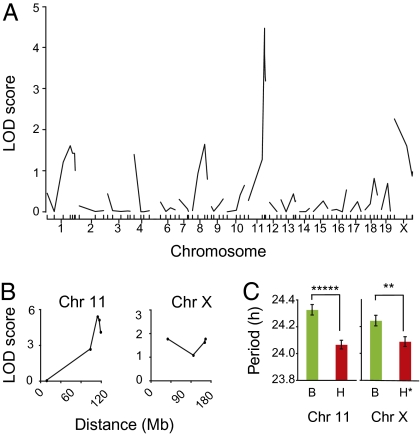
To map modifier loci, we initially genotyped Clock/+ mice from a [(C3H/HeJ × C57BL/6J)F1 × C57BL/6J]N2 panel with simple sequence length polymorphism (SSLP) markers distributed over the genome (Table S1). QTL analysis detected a significant association of period phenotype on distal chromosome 11 (LOD = 4.5) and a weak association on chromosome X (LOD = 2.3) (Fig. 2A). To confirm and strengthen these results, chromosomes 11 and X were analyzed in additional Clock/+ mice (total n = 96) from the [(C3H/HeJ × C57BL/6J)F1 × C57BL/6J]N2 cross (Fig. 2B). The effect of each locus is shown (Fig. 2C).
Fig. 2.
Genetic mapping of Clock suppressors. (A) Results of QTL analysis (single marker regression) with 55 [(C3H/HeJ × C57BL/6J)F1 × C57BL/6J]N2 animals. A significant association was detected with markers at the distal end of chromosome 11 (LOD = 4.7). A weak association was detected on chromosome X (LOD = 2.3). A significance threshold of LOD = 3.20 for P = 0.05 was determined by analysis of 1,000 permutation tests for this data set. (B) Examination of 41 additional Clock/+ N2 progeny confirms association of Clock suppression and loci on chromosomes 11 and X. Data shown represent 96 total N2 Clock/+ progeny. Peak associations occur with SSLP markers D11Mit12 on chromosome 11 (~113 Mb) and DXMit223 on chromosome X (~163 Mb). (C) Allelic effect of chromosome 11 (Left) and chromosome X (Right) on free-running period in 96 backcross Clock/+ mice. Data shown are mean ± SEM. A one-way ANOVA was significant for genotype at D11Mit12 on chromosome 11 [~113 Mb; ANOVA F(1,95) = 26.24, P = 1.67 × 10−6] and DXMit223 on chromosome X [~163 Mb; ANOVA F(1,95) = 8.33, P = 0.004). For chromosome 11, B stands for B6/B6 and H stands for B6/C3H genotypes. For chromosome X, B stands for either B6/B6 (female) or B6 (male) and H stands for either B6/C3H (female) or C3H (male).
Aanat and Asmt Are Candidate Suppressors of Clock.
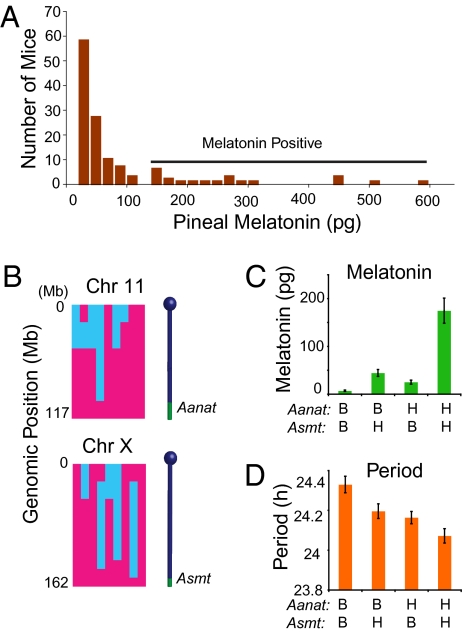
We determined that a potential candidate gene for the QTL on the distal end of chromosome 11 is Aanat. This enzyme is deficient in C57BL/6J mice, but functional in the C3H/HeJ inbred strain (21, 28). We reasoned that if Aanat polymorphisms are responsible for the chromosome 11 QTL, pineal melatonin itself may modify the circadian free-running period of Clock/+ mice. Furthermore, the Asmt locus should also be associated with the circadian free-running period of Clock/+ mice. We therefore functionally mapped the Asmt locus in mice using pineal melatonin as a phenotype in 61 [(C3H/HeJ × C57BL/6J)F1 × C57BL/6J]N2 progeny and 66 [(BALB/cJ × C3H/HeJ)F1 × BALB/cJ]N2 progeny—all wild-type animals. The distribution of pineal melatonin content of the 127 mice examined is skewed toward the low range (Fig. 3A). This suggests that the pineal melatonin content in this population is determined by multiple loci. Among the mice with melatonin content >100 pg/pineal, we genotyped 9 mice (5 from the C57BL/6J × C3H/HeJ cross and 4 from the BALB/cJ × C3H/HeJ cross) with a mouse SNP linkage panel (29). We found that high melatonin content was tightly linked to two loci—one on the distal end of chromosome 11 (Aanat) and another at the distal end of chromosome X (Fig. 3B). We then genotyped the remainder of the population for the two loci and found in both crosses that the pineal melatonin content phenotype was highly associated with the genotype of these two loci. Mice carrying C3H alleles at both loci had higher pineal melatonin content than those lacking C3H alleles at either locus. This result demonstrates that Asmt maps to the distal end of chromosome X.
Fig. 3.
Effect of melatonin on the circadian phenotype of Clock/+ mice. (A) Histogram of pineal melatonin content in N2 hybrid mice. Data are from 61 [(C3H/HeJ × C57BL/6J)F1 × C57BL/6J]N2 and 66 [(C3H/HeJ × BALB/cJ)F1 × BALB/cJ]N2 hybrid mice. A horizontal bar indicates those animals scored as melatonin positive. (B) Loci on the distal ends of chromosomes 11 and X are tightly linked to melatonin content. A total of 9 mice (4 from a C3H/HeJ × BALB/cJ cross and 5 from a C3H/HeJ × C57BL/6J cross) were genotyped with a custom SNP panel described previously (29). Two candidate suppressor loci were identified on these chromosomes: Aanat on 11 and Asmt on X. A two-way ANOVA was significant in the C57BL/6J × C3H/HeJ cross for Aanat genotype [F(1, 57) = 11.65, P = 0.001), Asmt genotype [F(1,57) = 9.37, P = 0.0033] and their interaction [F(1,57) = 5.07, P = 0.028]. In the BALB/cJ × C3H/HeJ cross, a two-way ANOVA was significant for Aanat genotype [F(1,61) = 7.16, P = 0.0095] and Asmt genotype [F(1,61) = 15.85, P = 0.00018]. Their interaction was not significant [F(1,61) = 1.88, P = 0.17]. Blue indicates BALB/cJ or C57BL/6J genome and pink indicates C3H/HeJ genome. (C) Effect of strain background on pineal melatonin content in Clock/+ [(C3H/HeJ × C57BL/6J)F1 × C57BL/6J]N2 mice. A total of 105 progeny were genotyped for the Aanat and Asmt loci. Mice carrying C3H alleles at both the Aanat and Asmt loci had significantly higher pineal melatonin content than animals with one or more B6 alleles. A two-way ANOVA was significant for Aanat genotype [F(1,101) = 21.7, P = 9.6 × 10−6], Asmt genotype [F(1,101) = 34.4, P = 6.0 × 10−8], and their interaction [F(1,101) = 12.4, P = 6.4 × 10−4]. For the Aanat locus, B stands for B6/B6 and H stands for B6/C3H genotypes. For the Asmt locus, B stands for either B6/B6 (female) or B6 (male). H stands for either B6/C3H (female) or C3H (male). (D) Effect of genotype of Aanat and Asmt on circadian free-running period in 157 [(C3H/HeJ × C57BL/6J)F1 × C57BL/6J]N2 mice. Mice carrying C3H alleles at Aanat and Asmt expressed shorter free-running periods than those carrying B6 alleles at either locus. A two-way ANOVA was significant for Aanat genotype [F(1,153) = 15.35, P = 1.3 × 10−4] and Asmt genotype [F(1,153) = 9.25, P = 2.7 × 10−3]. The interaction between Aanat and Asmt was not significant [F(1,153) = 0.31, P = 0.57]. Abbreviations of genotype are the same as in Fig. 3C.
To determine whether pineal melatonin content is also determined by Aanat and Asmt in the Clock/+ background, we tested an association of melatonin with the Aanat and Asmt loci in 105 [(C57BL/6J × C3H)F1 × C57BL/6J]N2 Clock/+ animals (Fig. 3C). Mice carrying C3H alleles at both loci produced higher levels of melatonin compared to those lacking C3H alleles at either locus. Mice carrying C3H alleles only at one of the two loci produced detectable levels of melatonin, but the amount was far lower than animals with C3H alleles at both loci. With respect to circadian period, Clock/+ mice carrying C3H alleles at Aanat and Asmt expressed a significantly shorter period than ones lacking C3H alleles at either locus (Fig. 3D). These observations suggest that melatonin is the causative agent that suppresses the Clock phenotype.
Complementation in SCN Explant Cultures.
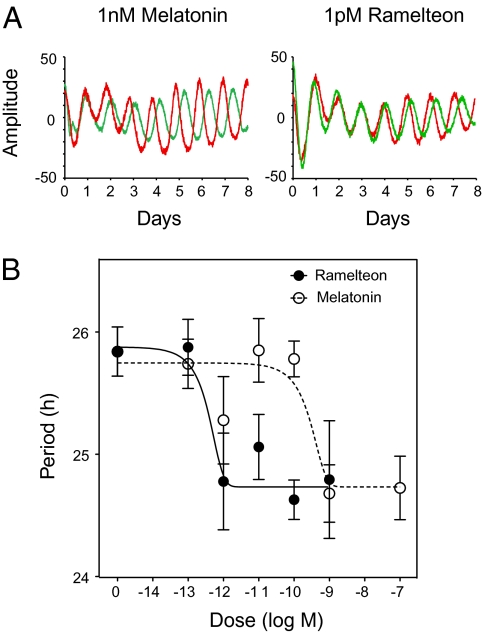
To test for a direct effect of melatonin on the SCN, we treated SCN explant cultures from Per2Luc Clock/+ mice with melatonin or the melatonin receptor-specific agonist, ramelteon (30). In Clock/+ SCN, the circadian period of bioluminescence was shortened by the presence of either melatonin or ramelteon (Fig. 4A), just as the presence of functional AANAT and ASMT in vivo shortens the period of the behavioral rhythm in Clock/+ mice (Fig. 3D). Melatonin treatment shortened circadian period in Clock/+ SCN with an EC50 of 106 pM whereas ramelteon had an EC50 of 0.217 pM (Fig. 4B). These doses are well within the range for specific melatonin receptor activation (31). By contrast, neither melatonin nor ramelteon affected the circadian period of SCN explants from wild-type mice. In addition, there were no effects of either drug on the amplitude of rhythms in Clock/+ or wild-type mice. In conclusion, treatment with either melatonin or ramelteon phenocopies the genetic suppression of Clock/+ mutants by functional Aanat and Asmt strongly implicating the melatonin pathway in this interaction.
Fig. 4.
Effect of melatonin and ramelteon on the period of PER2::LUC bioluminescence rhythms from Clock/+ SCN explants. (A) Representative records of bioluminescence rhythms from SCN explants of Clock/+ mice treated with melatonin (Left) or ramelteon (Right). In each panel, red lines represent treatment groups (10−9 M melatonin or 10−12 M ramelteon) and green lines represent vehicle controls. (B) Period analysis of bioluminescence rhythms of cultured Per2Luc SCN explants. For each culture, a single dose of melatonin or ramelteon was administered at the indicated concentration at the beginning of the experiment. Each data point represents the mean ± SEM of 3–14 samples. Nonlinear curve fitting was performed using GraphPad Prism 5.
Discussion
It has been reported that when melatonin or its receptor agonists are present in circumstances under which the master circadian pacemaker is compromised, such as in aging or in simulated jet-lag paradigms (32–34), the effects of melatonin become more apparent. That an effect of melatonin on circadian free-running period is detectable only in Clock/+ mice (genetically compromised circadian system), but not in wild-type animals, affirms this idea. Compensatory mechanisms may obscure the function of melatonin receptor signaling in the SCN in mammals. Thus, it will be of interest to test whether the circadian behavior of other clock mutants (e.g., Cry or Per mutants) are affected by melatonin.
The possibility that melatonin suppresses the effects of the Clock mutation in a strain-specific manner has been suggested by others (35). When the Clock mutation is carried on the melatonin-producing CBA inbred strain, fewer homozygous mutant mice become arrhythmic in DD compared to homozygous mutants of a melatonin-deficient inbred strain. Consistent with our results, Kennaway et al. observed no obvious differences in circadian behavior in wild-type mice from melatonin-producing and melatonin-deficient inbred strains. Although it is possible that the CBA strain segregates genetic loci in addition to Aanat and Asmt, which affect the Clock mutant phenotype, the Kennaway et al. report supports our findings.
To date, two melatonin receptors, Mtrn1a and Mtrn1b, have been identified in mammals (16, 17), and both are expressed in the SCN. Melatonin has two distinct effects on the SCN: acute inhibition and phase shifting of SCN electrical activity (18, 19). The effect of melatonin on phase shifting of the circadian rhythm in the SCN is mediated through protein kinase C activation (36). Although some studies report that melatonin can affect clock gene expression in the SCN (37–39), the signal transduction pathway from melatonin to the core clock remains to be defined. Interestingly, protein kinase C-mediated phosphorylation of CLOCK has been implicated in the resetting of circadian rhythms (40), and RACK1 and protein kinase Cα have been shown to interact with BMAL1 in a circadian manner (41). Thus, it is conceivable that melatonin-induced activation of protein kinase C could ultimately affect CLOCK and BMAL1 to suppress the Clock mutation. Recent work has shown that phosphorylation of CLOCKΔ19 mutant protein is reduced relative to wildtype CLOCK, leading to its stabilization (42). Thus, melatonin could differentially affect CLOCKΔ19 phosphorylation to rescue this defect, which could explain at least in part the selective suppression of the Clock mutant phenotype.
Our results clearly demonstrate that the effect of melatonin on circadian free-running period is genetically determined in mice. Several studies have reported that polymorphisms in clock genes can affect human sleep phenotypes (2). Human genetic variation, however, has not been considered before treatment with melatonin is initiated. Thus, treatment with melatonin or related pharmaceutical compounds such as ramelteon for insomnia (32) or agomelatonine for depression (43) may be more efficacious in some cases by assessing clock gene polymorphisms in patients. Finally, the Clock mutation has also been shown to be associated with metabolic syndrome in mice (44). It is therefore interesting that the melatonin receptor 1B (MTNR1B) gene has been strongly associated with fasting serum glucose levels in humans (45–47). Perhaps, the suppressive effects of melatonin reported here may also have relevance to other phenotypes such as metabolism.
Materials and Methods
Animals.
All animal care and experimental treatments were approved and performed in strict accordance with Northwestern University and University of Texas Southwestern Medical Center guidelines for animal care and use. Animals were raised in a 12-h light/12-h dark cycle (LD 12:12) from birth. After weaning, they were group housed (1–5 mice/cage).
Wheel-Running Behavior Assay.
At 8–12 weeks of age, mice were transferred to individual cages equipped with running wheels and housed in LD 12:12 conditions. After a minimum of 7 days, animals were transferred to DD conditions for 3 weeks.
Clock Suppressor Mapping Cross.
[(C3H/HeJ × C57BL/6J)F1 × C57BL/6J]N2 Clock/+ mice were bred from (C3H/HeJ × C57BL/6J)F1 Clock/+ × C57BL/6J wild-type mice. Only Clock/+ mice were wheel tested.
ASMT Mapping Cross.
[(C3H/HeJ × C57BL/6J)F1 × C57BL/6J]N2 and [(BALB/cJ × C3H/HeJ)F1 × BALB/cJ]N2 progeny were created for the ASMT mapping cross. They were all wild types. We also created [(C3H/HeJ × C57BL/6J)F1 × C57BL/6J]N2 Clock/+ mice from (C3H/HeJ × C57BL/6J)F1 wild type × C57BL/6J Clock homozygotes. All progeny were Clock/+.
Per2Luc Mice.
The Per2Luc Clock/+ mice used in this study were maintained on the C57BL/6J genetic background at Northwestern University. The genotyping protocol was described previously (48).
Clock Genotyping.
Genomic DNA was prepared by phenol-chloroform extraction from tail biopsies.
Genotyping primers used were: forward 5′-TACCAGCTGCTAATGTCCAGTG-3′; reverse 5′-TACATTGGGCTAGCCTTCCTAAG-3′. PCR conditions were 95 °C for 2 min followed by 32 cycles of 95 °C for 15 s, 55 °C for 30 s, and 72 °C for 15 s. Following amplification, PCR products were digested with 2 units of HincII for 2 h at 37 °C. After restriction digestion, PCR products were resolved on 4% agarose gels. The Clock mutant allele was identified as an ~70-bp product, whereas wild-type animals were identified by the presence of bands at ~50 and ~20 bp.
Free-Running Period Calculation.
The free running period was calculated using a χ2 periodogram as described previously (7).
Genomewide QTL Analysis.
SSLP markers used in this study are listed in Table S1. The genotyping protocol has been described previously (7).
QTL Analysis.
QTL analysis was performed as described R/qtl (http://www.rqtl.org/).
High-Resolution Mapping of Asmt Locus.
Tail DNA was extracted using Gentra Puregene Mouse Tail kit. Genotyping was performed at Harvard University Partners Center for Genetics and Genomics with their custom SNP panel (29).
Pineal Collection and Melatonin Assay.
Mice were raised in LD 12:12 conditions. Mice were killed, decapitated, and enucleated under infrared light at ~ZT22 (~10 h after lights off). Pineal glands were removed under a dissecting microscope and then frozen on dry ice. The pineal samples were maintained at –80 °C until use. Individual pineal glands were dispersed by sonication in 200 μL of PBS containing 0.1% gelatin. The melatonin levels in 20-μL aliquots of each sample were determined by RIA as previously described (49).
Aanat and Asmt Genotyping.
We used a 3′ end primer mismatch method (50) whereby we introduced an artificial mismatch at the third position from the end of the 3′ end of the primer to obtain more significant allele detection than a mismatch at the final 3′ nucleotide alone (51). All reactions were prepared with the ABI SYBR Green PCR kit and carried out real-time PCR on an ABI 7700 with the following conditions: 95 °C for 10 min followed by 40 cycles of 95 °C for 10 s, and 60 °C for 30 s. The primers were as follows:
Aanat.
Aanat-nonC3H: 5′-CCAGCATGACCCAGTCTCAT-3′, Aanat-C3H: 5′-CCAGCATGACCCAGTCTCAC-3′, Aanat-R: 5′-CCCGAGCTGAGAGCTTTTTA-3′.
Asmt.
rs13484098-nonC3H: 5′-CAACAAGGATTTCACCACTAA-3′, rs13484098-C3H: 5′-CAACAAGGATTTCACCACTAC-3′, rs13484098-R: 5′-TGGGCCCATAATAAGCAAAG-3′.
SCN Explant Culture.
SCN slice culture was performed as described (48). The bioluminescence data over time was recorded using a LumiCycle apparatus (Actimetrics). All bioluminescence analyses were performed with the LumiCycle Analysis program as described previously (48).
Supplementary Material
Acknowledgments
This work is supported by Takeda Research Grant 07-030R (to K.S.), National Institutes of Health (NIH) Grant R01 MH61461 (to C.B.G.), NIH Grant U01 MH61915 (to J.S.T.), and Silvio O. Conte Center NIH Grant P50 MH074924 (to C.B.G. and J.S.T.). J.S.T. is an investigator and V.K. was an associate in the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004368107/-/DCSupplemental.
References
- 1.Lowrey PL, Takahashi JS. Mammalian circadian biology: Elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stratmann M, Schibler U. Properties, entrainment, and physiological functions of mammalian peripheral oscillators. J Biol Rhythms. 2006;21:494–506. doi: 10.1177/0748730406293889. [DOI] [PubMed] [Google Scholar]
- 4.Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: Cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 6.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 7.Shimomura K, et al. Genome-wide epistatic interaction analysis reveals complex genetic determinants of circadian behavior in mice. Genome Res. 2001;11:959–980. doi: 10.1101/gr.171601. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi JS. Finding new clock components: Past and future. J Biol Rhythms. 2004;19:339–347. doi: 10.1177/0748730404269151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buijs RM, Kalsbeek A. Hypothalamic integration of central and peripheral clocks. Nat Rev Neurosci. 2001;2:521–526. doi: 10.1038/35081582. [DOI] [PubMed] [Google Scholar]
- 10.Kalsbeek A, Buijs RM. Output pathways of the mammalian suprachiasmatic nucleus: Coding circadian time by transmitter selection and specific targeting. Cell Tissue Res. 2002;309:109–118. doi: 10.1007/s00441-002-0577-0. [DOI] [PubMed] [Google Scholar]
- 11.Stehle JH, von Gall C, Korf HW. Melatonin: A clock-output, a clock-input. J Neuroendocrinol. 2003;15:383–389. doi: 10.1046/j.1365-2826.2003.01001.x. [DOI] [PubMed] [Google Scholar]
- 12.Moore RY. Neural control of the pineal gland. Behav Brain Res. 1996;73:125–130. doi: 10.1016/0166-4328(96)00083-6. [DOI] [PubMed] [Google Scholar]
- 13.Hankins MW, Peirson SN, Foster RG. Melanopsin: An exciting photopigment. Trends Neurosci. 2008;31:27–36. doi: 10.1016/j.tins.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Klein DC, Moore RY. Pineal N-acetyltransferase and hydroxyindole-O-methyltransferase: Control by the retinohypothalamic tract and the suprachiasmatic nucleus. Brain Res. 1979;174:245–262. doi: 10.1016/0006-8993(79)90848-5. [DOI] [PubMed] [Google Scholar]
- 15.Weaver DR, Rivkees SA, Reppert SM. Localization and characterization of melatonin receptors in rodent brain by in vitro autoradiography. J Neurosci. 1989;9:2581–2590. doi: 10.1523/JNEUROSCI.09-07-02581.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reppert SM, Weaver DR, Ebisawa T. Cloning and characterization of a mammalian melatonin receptor that mediates reproductive and circadian responses. Neuron. 1994;13:1177–1185. doi: 10.1016/0896-6273(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 17.Reppert SM, et al. Molecular characterization of a second melatonin receptor expressed in human retina and brain: The Mel1b melatonin receptor. Proc Natl Acad Sci USA. 1995;92:8734–8738. doi: 10.1073/pnas.92.19.8734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu C, et al. Molecular dissection of two distinct actions of melatonin on the suprachiasmatic circadian clock. Neuron. 1997;19:91–102. doi: 10.1016/s0896-6273(00)80350-5. [DOI] [PubMed] [Google Scholar]
- 19.Jin X, et al. Targeted disruption of the mouse Mel(1b) melatonin receptor. Mol Cell Biol. 2003;23:1054–1060. doi: 10.1128/MCB.23.3.1054-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goto M, Oshima I, Tomita T, Ebihara S. Melatonin content of the pineal gland in different mouse strains. J Pineal Res. 1989;7:195–204. doi: 10.1111/j.1600-079x.1989.tb00667.x. [DOI] [PubMed] [Google Scholar]
- 21.Roseboom PH, et al. Natural melatonin ‘knockdown’ in C57BL/6J mice: Rare mechanism truncates serotonin N-acetyltransferase. Brain Res Mol Brain Res. 1998;63:189–197. doi: 10.1016/s0169-328x(98)00273-3. [DOI] [PubMed] [Google Scholar]
- 22.Vivien-Roels B, et al. Daily variations in pineal melatonin concentrations in inbred and outbred mice. J Biol Rhythms. 1998;13:403–409. doi: 10.1177/074873098129000228. [DOI] [PubMed] [Google Scholar]
- 23.Ebihara S, Marks T, Hudson DJ, Menaker M. Genetic control of melatonin synthesis in the pineal gland of the mouse. Science. 1986;231:491–493. doi: 10.1126/science.3941912. [DOI] [PubMed] [Google Scholar]
- 24.Vitaterna MH, et al. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nadeau JH. Modifier genes in mice and humans. Nat Rev Genet. 2001;2:165–174. doi: 10.1038/35056009. [DOI] [PubMed] [Google Scholar]
- 26.Dietrich WF, et al. Genetic identification of Mom-1, a major modifier locus affecting Min-induced intestinal neoplasia in the mouse. Cell. 1993;75:631–639. doi: 10.1016/0092-8674(93)90484-8. [DOI] [PubMed] [Google Scholar]
- 27.King DP, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ebihara S, Hudson DJ, Marks T, Menaker M. Pineal indole metabolism in the mouse. Brain Res. 1987;416:136–140. doi: 10.1016/0006-8993(87)91505-8. [DOI] [PubMed] [Google Scholar]
- 29.Moran JL, et al. Utilization of a whole genome SNP panel for efficient genetic mapping in the mouse. Genome Res. 2006;16:436–440. doi: 10.1101/gr.4563306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirai K, et al. Ramelteon (TAK-375) accelerates reentrainment of circadian rhythm after a phase advance of the light-dark cycle in rats. J Biol Rhythms. 2005;20:27–37. doi: 10.1177/0748730404269890. [DOI] [PubMed] [Google Scholar]
- 31.Kato K, et al. Neurochemical properties of ramelteon (TAK-375), a selective MT1/MT2 receptor agonist. Neuropharmacology. 2005;48:301–310. doi: 10.1016/j.neuropharm.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 32.Roth T, et al. Effects of ramelteon on patient-reported sleep latency in older adults with chronic insomnia. Sleep Med. 2006;7:312–318. doi: 10.1016/j.sleep.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 33.Sack RL. Clinical practice. Jet lag. N Engl J Med. 2010;362:440–447. doi: 10.1056/NEJMcp0909838. [DOI] [PubMed] [Google Scholar]
- 34.Arendt J. Managing jet lag: Some of the problems and possible new solutions. Sleep Med Rev. 2009;13:249–256. doi: 10.1016/j.smrv.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 35.Kennaway DJ, Boden MJ, Voultsios A. Reproductive performance in female Clock Delta19 mutant mice. Reprod Fertil Dev. 2004;16:801–810. doi: 10.1071/rd04023. [DOI] [PubMed] [Google Scholar]
- 36.Dubocovich ML, Markowska M. Functional MT1 and MT2 melatonin receptors in mammals. Endocrine. 2005;27:101–110. doi: 10.1385/ENDO:27:2:101. [DOI] [PubMed] [Google Scholar]
- 37.Poirel VJ, et al. Contrary to other non-photic cues, acute melatonin injection does not induce immediate changes of clock gene mRNA expression in the rat suprachiasmatic nuclei. Neuroscience. 2003;120:745–755. doi: 10.1016/s0306-4522(03)00344-0. [DOI] [PubMed] [Google Scholar]
- 38.Agez L, Laurent V, Pévet P, Masson-Pévet M, Gauer F. Melatonin affects nuclear orphan receptors mRNA in the rat suprachiasmatic nuclei. Neuroscience. 2007;144:522–530. doi: 10.1016/j.neuroscience.2006.09.030. [DOI] [PubMed] [Google Scholar]
- 39.Agez L, et al. Endogenous melatonin provides an effective circadian message to both the suprachiasmatic nuclei and the pars tuberalis of the rat. J Pineal Res. 2009;46:95–105. doi: 10.1111/j.1600-079X.2008.00636.x. [DOI] [PubMed] [Google Scholar]
- 40.Shim HS, et al. Rapid activation of CLOCK by Ca2+-dependent protein kinase C mediates resetting of the mammalian circadian clock. EMBO Rep. 2007;8:366–371. doi: 10.1038/sj.embor.7400920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robles MS, Boyault C, Knutti D, Padmanabhan K, Weitz CJ. Identification of RACK1 and protein kinase Calpha as integral components of the mammalian circadian clock. Science. 2010;327:463–466. doi: 10.1126/science.1180067. [DOI] [PubMed] [Google Scholar]
- 42.Yoshitane H, et al. Roles of CLOCK phosphorylation in suppression of E-box-dependent transcription. Mol Cell Biol. 2009;29:3675–3686. doi: 10.1128/MCB.01864-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arendt J, Rajaratnam SM. Melatonin and its agonists: An update. Br J Psychiatry. 2008;193:267–269. doi: 10.1192/bjp.bp.108.050955. [DOI] [PubMed] [Google Scholar]
- 44.Turek FW, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bouatia-Naji N, et al. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet. 2009;41:89–94. doi: 10.1038/ng.277. [DOI] [PubMed] [Google Scholar]
- 46.Lyssenko V, et al. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet. 2009;41:82–88. doi: 10.1038/ng.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prokopenko I, et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009;41:77–81. doi: 10.1038/ng.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoo SH, et al. PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rollag MD, Niswender GD. Radioimmunoassay of serum concentrations of melatonin in sheep exposed to different lighting regimens. Endocrinology. 1976;98:482–489. doi: 10.1210/endo-98-2-482. [DOI] [PubMed] [Google Scholar]
- 50.Germer S, Higuchi R. Homogeneous allele-specific PCR in SNP genotyping. Methods Mol Biol. 2003;212:197–214. doi: 10.1385/1-59259-327-5:197. [DOI] [PubMed] [Google Scholar]
- 51.Zhou G, et al. Quantitative detection of single nucleotide polymorphisms for a pooled sample by a bioluminometric assay coupled with modified primer extension reactions (BAMPER) Nucleic Acids Res. 2001;29:E93. doi: 10.1093/nar/29.19.e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.