Abstract
A fundamental goal of systems biology is to identify genetic elements that contribute to complex phenotypes and to understand how they interact in networks predictive of system response to genetic variation. Few studies in plants have developed such networks, and none have examined their conservation among functionally specialized organs. Here we used genetical genomics in an interspecific hybrid population of the model hardwood plant Populus to uncover transcriptional networks in xylem, leaves, and roots. Pleiotropic eQTL hotspots were detected and used to construct coexpression networks a posteriori, for which regulators were predicted based on cis-acting expression regulation. Networks were shown to be enriched for groups of genes that function in biologically coherent processes and for cis-acting promoter motifs with known roles in regulating common groups of genes. When contrasted among xylem, leaves, and roots, transcriptional networks were frequently conserved in composition, but almost invariably regulated by different loci. Similarly, the genetic architecture of gene expression regulation is highly diversified among plant organs, with less than one-third of genes with eQTL detected in two organs being regulated by the same locus. However, colocalization in eQTL position increases to 50% when they are detected in all three organs, suggesting conservation in the genetic regulation is a function of ubiquitous expression. Genes conserved in their genetic regulation among all organs are primarily cis regulated (~92%), whereas genes with eQTL in only one organ are largely trans regulated. Trans-acting regulation may therefore be the primary driver of differentiation in function between plant organs.
Keywords: eQTL, gene network, gene regulation, systems biology, Populus
The pioneering work of King and Wilson (1) described extensive similarity between protein sequences of chimpanzees and humans, providing early evidence that differential gene expression regulation is a critical mechanism producing phenotypic diversity in eukaryotes. The genetic regulation of gene expression has since been uncovered by measuring transcript abundance in segregating populations and identifying gene expression quantitative trait loci (eQTL) that arise as a consequence of genetic variation between parental alleles (2). eQTL mapping studies in humans (3), mice (4), yeast (5, 6), and several higher plants (7–11) showed that genes are frequently regulated by combinations of cis-acting loci of generally large effect and numerous trans-acting elements with smaller contributions to the phenotype. eQTL mapping, in conjunction with traditional trait QTL analysis, has also identified polymorphisms responsible for phenotypic variation in several species (4, 12, 13), reinforcing the role of transcriptional regulation in evolution (14).
Although much has been learned about the genetic regulation of transcription in individual tissues and organs, comparatively little is known about the diversification of cis- and trans-acting factors that control gene expression within an individual. Multicellular organisms have specialized cell types, tissues, and organs in which differential regulation of transcription likely plays a critical role in specification of development, form, and function. The genetic diversity of organism-wide transcriptome regulation has only recently emerged in animals and humans (15–18), and is still unknown in plants where eQTL studies have focused either on whole organisms (9, 19) or single organs (10, 11, 20). Independent of the taxa, natural selection is likely to play a significant role in the evolution of the genetic regulation of transcription in an organism. In primates, mutations that diversify the transcriptional regulation of genes expressed in multiple organs are more likely to be eliminated by negative selection (21). Similarly, orthologs expressed in multiple compartments in Arabidopsis and Populus are more likely to be expressed at similar levels, compared with genes that are specific to a single tissue or organ in both species (22). An extension of these observations would suggest that for genes in which eQTL are detected in multiple organs, the genetic regulation may be more conserved compared with those that are organ specific, which could evolve more rapidly by establishing alternative, trans-acting genetic mechanisms of regulation.
The genetic architecture of individual genes’ expression constitutes only the first level in the hierarchy of transcriptional regulation contributing to the development of an organism. Genome-wide gene expression and eQTL mapping data can also be leveraged to reconstruct transcriptional networks contributing to developmental pathways (2). Analysis of a priori-defined pathways has shown extensive genetic control of underlying gene expression networks in Arabidopsis (23), a finding intricately explored for glucosinolate biosynthesis (24, 25) and flowering time (7). Transcriptional networks can also be identified a posteriori from eQTL data, and their biological roles inferred by identifying overrepresented metabolic and regulatory functions among network members (26). Additional genomic information, including metabolomic, transcription factor binding site (TFBS), and protein–protein interaction data have been incorporated into a few transcriptional network studies (25, 27, 28), increasing the power to identify key network participants (28). The conservation of these networks in an organism has not yet been quantified in any species. Hypothetically, transcriptional networks implicated in essential cell functions may share composition and regulation in different plant parts, whereas those implicated in functions that are specific to certain cell type, tissues, or organs are likely to be unique.
In this study we surveyed the variation in the genetic architecture of gene expression in three plant organs (differentiating xylem, expanding leaves, and mature roots) of Populus trichocarpa and P. deltoides, to assess the role of cis and trans regulation in organ differentiation. Our analysis shows that diversification of the genetic control of transcription is dependent on the extent by which eQTL are detected in single or multiple organs. We also generated cotranscriptional networks and showed that though network membership is sometimes conserved among organs, their genetic regulation is almost invariably organ-specific.
Results
Trans-eQTL Distribution Is Biased Among Linkage Groups.
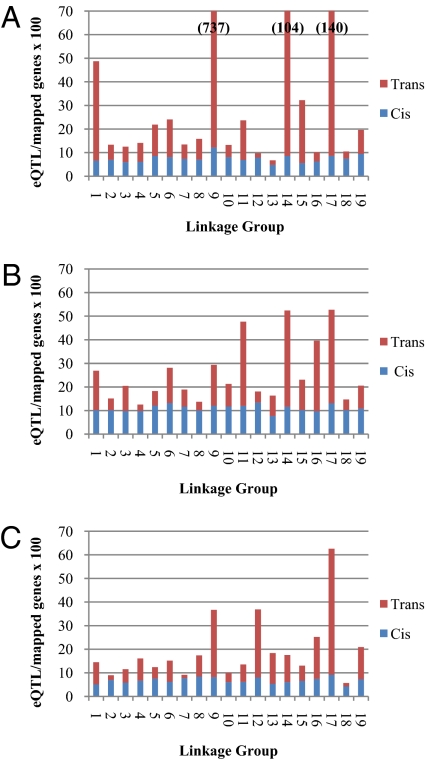
From a set of 192 progeny of family 52–124 (29), we collected 180 xylem, 183 leaf, and 163 root samples and assayed expression using a custom whole-genome microarray (30). QTL analysis of normalized signal intensities identified 36,071 significant eQTL in xylem, 13,403 eQTL in leaf, and 9,137 eQTL in roots representing 30,313, 12,392, and 8,534 genes/ESTs, respectively. eQTL were classified as cis- or trans-acting, based on the overlap of the eQTL peak with the marker interval to which the respective gene model was located in the genome (Table S1). Cis-acting eQTL were detected at a relatively constant rate (~8–10% of genes) independent of the linkage group or organ, when normalized to account for the varying number of genes per chromosome (Fig. 1). In contrast, trans-acting eQTL frequency varied widely between different linkage groups and organs. For instance, in xylem, the number of eQTL ranged from only 76 trans-eQTL on LG XII to 20,935 on LG IX.
Fig. 1.
Global distribution of eQTL across linkage groups for (A) xylem, (B) leaf, and (C) root tissues expressed as the fraction of mapped gene models. In cases where values on the eQTL/mapped genes axis exceed 70, values are provided in brackets. See Table S1 footnotes for description of cis- and trans-eQTL categorization procedure.
Genetic Regulation of Genes with eQTL in Multiple Organs Is Conserved and Occurs Primarily in cis.
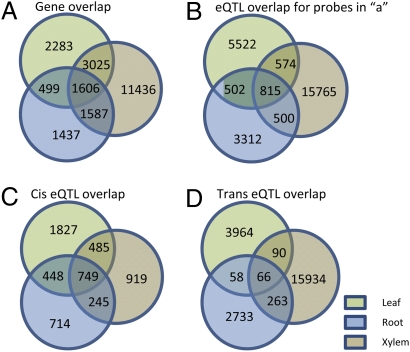
Given the large degree of differentiation among vegetative organs in poplar, yet the seemingly small degree of transcriptional diversity separating them with respect to the contingent of expressed genes (22, 31), we postulated that differential genetic regulation of transcription among organs would be prevalent. Therefore, we determined the degree of overlap between genes with eQTL in multiple organs and compared the location of their eQTL peaks and mode of regulation (i.e., cis or trans; Fig. 2). For 15,156 (69%) genes, eQTL were identified in a single organ, whereas for 6,717 (31%) they were detected in two or more (Fig. 2A). For genes where we identified eQTL in multiple organs, sharing of genetic architecture was highly dependent on whether regulation occurred in cis or trans. For example, of the 4,631 gene models in which eQTL were detected in both leaf and xylem (Fig. 2A), less than one-third (1,389) were regulated by the same genomic interval in the two organs (Fig. 2B). However, the vast majority of these (89%) were cis regulated (Fig. 2 B and C). The proportion of cis regulation increased even more for genes where eQTL were detected in xylem, leaf, and root (92%; Fig. 2 B and C). However, among the 2,391genes that presented overlap in the eQTL position in two or more organs, only 20% are trans regulated (Fig. 2 B and D). The proportion of cis and trans regulation is essentially inverted among genes in which eQTL were detected in either xylem, leaf, or root, where 77% are controlled in trans, suggesting that trans regulation is a more common mechanism governing organ-specific gene expression and possibly developmental differentiation.
Fig. 2.
Overlap between genes and eQTL detected among the three tissues considered. (A) Overlap of genes producing eQTL in each of the three tissues, including genes with physical position undefined in the genome. (B) Cross-tissue conservation of the genomic regulatory region (eQTL location) detected for genes in A. (C) Cross-tissue conservation of genomic regulatory position for genes with cis-eQTL. (D) Cross-tissue conservation of genomic regulatory position for genes with trans-eQTL. Genes in unknown position (ambiguous) in the genome are excluded.
eQTL Hotspots Are Largely Organ-Specific.
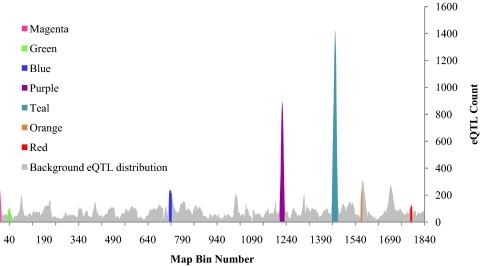
Next we identified eQTL hotspots, or genomic regions regulating accumulation or turnover of large numbers of transcripts. Though the underlying genetic basis of eQTL hotspots has been a topic of ongoing discussion (32), they were shown in some cases to encompass key regulators of gene networks (26, 33). Based on permutation thresholds (Table S1), we detected 67 unique bins corresponding to statistically significant eQTL hotspots in xylem, 97 in leaf, and 88 in root (Fig. 1 and Table S2). Though a large number of unique bins were found to be enriched for eQTL relative to chance, many of these bins were adjacent to one another (Fig. 3) and thus likely correspond to a single hotspot— a result of the limited QTL mapping resolution (34). As expected, eQTL hotspots resulted primarily from the higher accumulation of trans-acting eQTL (Table S1). After normalizing for number of genes per map bin (10), 238/255 of the original bins remained significantly enriched for eQTL.
Fig. 3.
Genome-wide linkage scan of expression traits and demarcation of eQTL hotspots containing coexpressed gene networks in leaf tissue. Similar results were obtained for root and xylem tissue, detailed in Table 1.
We contrasted the localization of eQTL hotspots among vegetative organs and detected weak conservation; only nine hotspot bins were shared between leaf and xylem, 11 between xylem and root, and nine between root and leaf (Table S3). Only two hotspot bins were shared among xylem, leaf, and root, indicating that hotspots are generally organ-specific. In the few cases that hotspots were colocalized between organs, overlap in their gene composition was uncommon (Table S3), further supporting the hypothesis that they are primarily involved in specific functional and developmental processes that differentiate the parental species of the mapping population.
Construction of Organ-Specific, Hotspot-Based Coexpression Networks That Segregate in P. deltoides and P. trichocarpa.
eQTL hotspots frequently correspond to cotranscribed gene sets that are enriched for common functional groups or known biochemical and regulatory pathways (26, 33) and can serve as a foundation to build transcriptional networks that are connected by common genetic regulation (28). In the next step of our analysis we developed such transcriptional networks based on the expression correlation among genes in hotspots (Table 1). Among the 97 leaf eQTL hotspot bins detected, we constructed 51 gene coexpression networks within 38 bins. The leaf coexpression networks encompassed 1,678 distinct genes and ranged in size from 11 to 945 genes (median: 36 genes). Many of the 38 network-producing bins neighbored one another in the genetic map, and the resulting networks were highly redundant (34) (Table S4). Nonetheless, at least nine independent leaf coexpression networks were detected within seven bona fide unique loci (Fig. 3 and Table 2). Similar results were obtained for xylem and root (Table 1 and Table S5).
Table 1.
Summary of tissue-specific eQTL hotspot-based coexpression network construction in family 52–124
| eQTL hotspot bins detected | Bin coexpression networks | Bins with networks | Max network size | Median network size | Total genes in networks | Minimum independent coexpression networks | Minimum independent genomic regions | |
| Leaf | 97 | 51 | 38 | 945 | 36 | 1,678 | 9 | 7 |
| Root | 88 | 75 | 55 | 217 | 33 | 1,188 | 16 | 11 |
| Xylem | 67 | 97 | 62 | 5,787 | 99 | 9,369 | 28 | 16 |
Table 2.
Tissue-specific eQTL hotspots detected in leaf tissue among family 52–124 and characteristics of their associated coexpression networks
| Hotspot locus color | Hotspot eQTL count | Networks constructed | Network size(s) | Most significantly enriched GO category | GO category type | GO enrichment nominal P value | Most significantly enriched cis element | PLACE enrichment nominal P value | LG | Genomic physical interval, Mb |
| Magenta | 275 | 1 | 31 | Tubulin complex | CC | 1.615E-05 | AGATC | 1.236E-04 | 1 | 0.22–1.52 |
| Green | 114 | 1 | 18 | n/a | n/a | n/a | n/a | n/a | 1 | 6.87–? |
| Blue | 239 | 1 | 51 | Chloroplast | CC | 1.370E-40 | AATAAT | 6.129E-05 | 6 | 2.27–3.63 |
| Purple | 892 | 2 | 428 | Chloroplast | CC | 5.703E-05 | AGCGGG | 1.979E-07 | 11 | 4.87–12.24 |
| 36 | Protein aminoacid phosphorylation | MF | 4.458E-08 | TGCAAAG | 1.923E-05 | 11 | 4.87–12.24 | |||
| Teal | 1,421 | 2 | 945 | DNA Recombination | MF | 1.161E-18 | CCACGTCATC | 5.394E-06 | 14 | 4.71–7.1 |
| 27 | n/a | n/a | n/a | CGCGGCAT | 4.426E-05 | 14 | 4.71–7.1 | |||
| Orange | 316 | 1 | 21 | Chloroplast envelope | CC | 2.297E-04 | AATAGAAAA | 2.328E-05 | 16 | 0.04–0.67 |
| Red | 126 | 1 | 11 | Manganese ion binding | MF | 1.043E-17 | n/a | n/a | 19 | 0–2.31 |
Biological annotation of organ-specific gene coexpression networks.
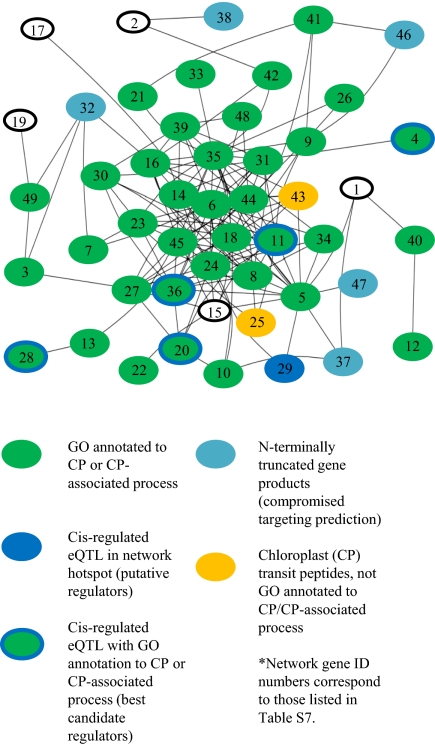
To elucidate biological relevance of coexpression networks, we tested for enrichment of GO categories represented within each network, based on Fisher's exact test (Penrichment), Bonferroni corrected for multiple testing. For the 51 coexpression networks identified in leaf, we detected 42 with at least one significant GO category enrichment (median Penrichment = 7.35 × 10−7). Analogous results were obtained for xylem (75/98 networks, Penrichment(median)= 1.24 × 10−5) and root (63/75 networks, Penrichment(median) = 9.58 × 10−6). Among the most significant enrichments, the leaf “blue” hotspot locus on linkage group (LG) VI (Fig. 3, bin 734) revealed a network with an overrepresentation of genes associated with chloroplast biogenesis and function (Penrichment = 1.37 × 10−40). Of the 49 GO annotated genes within the coexpressed network, 35 (68.6%) were GO annotated as being localized to the chloroplast (Fig. 4 and Table S6), a >5.5× enrichment over the number of chloroplast-localized genes expected by chance (4504/36,688 ≈ 12.27% or 6.25 chloroplast localized genes). Chloroplast stroma (Penrichment = 3.16 × 10−22), thylakoid lumen (Penrichment = 1.97 × 10−6), and chloroplast envelope (Penrichment = 5.68 × 10−10) cellular components were also significantly enriched in this network (Table S6), reinforcing the inference that this LG VI locus plays an important role in chloroplast biogenesis and/or function. Using this strategy on all 180 networks with at least one GO categorical enrichment, we identified 183 enriched GO categories, representing 1,212 combinations of significant organ-specific networks and enriched GO categories (Table S6).
Fig. 4.
Leaf coexpression network constructed from the “blue” eQTL hotspot and enriched for chloroplast-related Gene Ontogeny categories. Hub colors indicate annotation nature of network members, and edges indicate Pearson correlation between coexpressed genes of |r| > 0.80.
eQTL-based prediction of putative network regulators.
Developing transcriptional networks represents an initial step toward understanding the relationships between genes in a biological system. However, one must identify a network key regulator(s) to modify downstream network functions. Discovering coexpression networks on the basis of eQTL hotspots facilitates the identification of regulators, because differential transcript accumulation is predicted to occur due to a genetic variant underlying the eQTL hotspot position. Therefore, cis-regulated genes belonging to a network defined by an eQTL hotspot represent initial candidate regulators. Though this strategy is incomplete in that it will not identify network regulators differentially controlled outside the realm of transcription, it has previously offered direct evidence to define putative network regulators for downstream investigation (15, 28). Using this strategy, we identified putative regulators for 43 of the 62 leaf coexpression networks, 38 of the 75 root networks, and 50 of the 98 xylem networks (Table S6). Frequently, more than one putative regulator was identified for each network. For the LG VI leaf coexpression network previously shown to be associated with chloroplast function, we identified six network members with eQTL in cis to the hotspot locus. Among these six genes, five produced chloroplast-localized protein products, three of which are known structural components of the chloroplast. These three genes represent the best predicted candidates for network regulation (Fig. 4 and Table S7). Of particular interest is FtsZ2, a gene with well-described roles in chloroplast structure, biogenesis, and division (35, 36), which may play a key regulatory role in this cotranscriptional network.
Enrichment of transcription factor binding sites in coexpression networks.
In addition to the information obtained from GO annotation enrichment, the functional roles of transcriptional networks may be inferred from conserved TFBS in coregulated genes (28). We used the publicly available plant cis-acting regulatory element (37) database (PLACE) to identify significant enrichment of regulatory motifs upstream of the start codon of genes in each coexpression network. We detected enrichment for 27 motifs in 35 of the 62 leaf gene coexpression networks, 32 motifs in 21 of the 75 root networks, and 36 motifs in 29 of the 98 xylem networks. Networks were significantly enriched for as few as one to as many as 26 cis elements (median of three motifs enriched per network). A total of 419 combinations of 85 organ-specific networks and 72 enriched motifs were detected among the dataset (Table S8).
Genetic Regulation of Gene Expression Networks Is Organ Specific, but Gene Membership Is Shared Among Networks in Xylem, Leaf, and Root.
The analysis of eQTL and eQTL hotspot sharing between organs suggests that regulation of individual genes and networks by a shared locus is uncommon. However, even if networks are regulated by distinct loci in separate plant compartments, they could still share members. To address this question, we tested for significant membership overlap among networks detected in xylem, leaf, and root using a χ2 analysis, accounting for the number of genes in each network and shared between them. From all 20,931 cross-organ pairwise comparisons (97 leaf networks × 67 xylem networks = 6,499; 97 leaf networks × 88 root networks = 8,536; 67 xylem networks × 88 root networks = 5,896) we initially selected 1,012 comparisons where at least five genes were shared between networks. Of these, a total of 974 significant instances of shared membership were observed after correcting for multiple testing using a Bonferroni threshold of 0.01/n, where n is the number of pairwise comparisons. Network members detected in one organ were also frequently detected as multiple subgroups, genetically controlled by distinct loci in other organs. For instance, a network with 428 genes on LG XI in leaf (interval 1226, network 1) shared 156 gene members with three networks in xylem, including 91 in a network on LG I (interval 252, network 1; P ≈ 0), 53 on LG XV (interval 1557, network 1; P ≈ 0), and 25 on LG XIV (interval 1386, network 1; P < 1.832 × 10−9). Thirteen leaf network genes appeared in both the LG XV and LG I xylem networks. In total, we identified 414 network pairs that were statistically enriched for shared genes among leaf and xylem, 304 between root and leaf, and 256 between root and xylem (Table S9). These findings suggest that distinct trans-acting factors might control expression of coordinate groups of genes in different plant compartments, and also indicate that combinations of biological subnetworks could be differentially combined to drive cell type, tissue, and organ diversification.
Discussion
Unraveling the orchestrated action of genes, and modeling their interactions in a biological system, is among the most significant challenges of genomics, and the ultimate goal of biology. Here we used an established quantitative genetics framework to (i) characterize the genetic architecture of gene expression in xylem, roots and leaves, (ii) identify networks that describe gene interactions, and (iii) infer their biological function and mechanism of regulation. In our study, the detection of eQTL reflects allelic differences in the genetic regulation of gene expression between parental individuals from two species, P. deltoides and P. trichocarpa. These effects appear to have evolved differently depending on whether eQTL were detected in single or multiple organs. Where eQTL for individual genes were detected in two or more vegetative organs, their regulation occurred far more frequently through common cis rather than trans regulatory loci (29% compared with 7%), supporting observations previously made in mice and humans (38–40). When considering only genes with eQTL detected in xylem, leaf, and root (1,606; Fig. 2A), more than half are regulated by the same locus (815/1,606; Fig. 2B), and the majority of these (749/815, Fig. 2C) are controlled in cis in all three organs. Therefore, genetic divergence in gene expression regulation appears to be constrained to local (cis) versus distant (trans) variants for genes transcribed in multiple organs and differentially regulated between P. deltoides and P. trichocarpa. In contrast, genes for which eQTL were detected in a single organ were largely regulated in trans [Fig. 2A; leaf (1,462/2,283), root (1,155/1,437), xylem (10,772/11,436)]. In a human association genetics study, cell-type-specific eQTL were shown to localize at greater distances from transcription binding sites, leading to the proposition that enhancers are the primary drivers of cell-specific expression differentiation (16). Though that study did not attempt to detect trans regulation, it agrees with our observation that genetic polymorphisms in regulators located at greater distances from the coding region play a more significant role in cell or organ-specific differentiation, compared with those that are im-mediately adjacent to it.
To further understand the role of trans-eQTL in the developmental diversity of plant organs, we built transcriptional networks on the basis of eQTL hotspots (4, 28, 41, 42), and detected significant enrichment for genes in GO functional categories and transcriptional regulatory elements. eQTL hotspots were largely specific to xylem, leaf, or root. In the few cases where hotspots colocalized in multiple organs, gene membership overlap was very limited. However, as previously detected in Drosophila (43) and murine (41) genetic systems, there was significant overlap in gene membership between transcriptional networks detected in different organs, implicating gene groups that may be subject to multiple points of regulation governing variation between vegetative organs. It has been previously suggested that the high frequency of tissue specificity among trans-eQTLs results from tissue-specific gene regulatory networks (40). We showed that organ-specific modulation of transcriptional networks may represent a key role in differentiation plant development and individuals in higher plants. This greatly increases the potential for regulatory complexity to drive phenotypic diversification. A seemingly small number of genes, each regulated in a tightly controlled cell-, tissue-, or organ-specific manner by complex assortments of multiple cis-acting elements and trans-acting regulatory factors, exponentially increase the number of combinations that can act in concert to generate phenotypic variation.
A key goal of systems biology is to generate testable predictions of system-wide behavior in response to genetic variation, and the consequences to growth and development. The construction of organ-specific transcriptional networks based on coexpressed genes in eQTL hotspots in this Populus hybrid population represents an initial step toward this goal. Previous studies in yeast have shown that transcriptional networks developed based on eQTL data are more predictive of genetic and pharmacological perturbation than those produced based on gene expression information alone (28). Accordingly, the networks produced in this study are similarly predictive of system behavior in response to interspecific genetic variants. Efforts are now underway to validate the role of specific candidate regulators and networks in the phenotypic diversity seen among individuals in this population.
Materials and Methods
Plant Materials and Growth Conditions.
A pseudobackcross progeny (family 52–124) of 396 individuals from a cross of P. trichocarpa × P. deltoides (genotype 52–225) and P. deltoides (genotype D-124) were propagated and grown as described (29). From a common set of 192 randomly selected individuals we collected 180 samples of differentiating xylem, 183 expanding leaves, and 163 whole roots for gene expression analysis. Collected tissues were immediately flash-frozen in liquid nitrogen and stored at −80 °C until lyophilization and RNA extraction. We favored use of a single biological sample (instead of biological replicates) of each individual in the progeny to maximize the size of the population and meiotic events sampled. Because this experiment reflects the analysis of a segregating population, each allele is biologically replicated in approximately half of the individuals of the population.
RNA Isolation and Microarray Analysis.
RNA was extracted from each lyophilized sample by a standard protocol (44), converted to double-stranded cDNA, labeled with cy3, and hybridized to microarrays (30). Hybridizations were carried out using a previously described four-plex NimbleGen microarray platform [Gene Expression Omnibus (GEO) accession no. GPL7234] using probes designed to minimize the effects of sequence polymorphism on the estimates of gene expression (30). The microarray comprised one probe per gene for 55,793 previously described gene models derived from the annotation of the genome sequence of P. trichocarpa clone Nisqually-1 (version 1.1) (31) and a set of nonannotated ESTs. Raw data from hybridizations were background subtracted, log2 transformed, and quantile normalized separately on a tissue-by-tissue basis as previously described (30). Raw and normalized gene expression data are publically available (GEO accession nos. GSE12623, GSE20117, and GSE20118).
eQTL Analysis.
Each quantile-normalized gene expression value was analyzed using composite interval mapping (45, 46) implemented in QTL Cartographer (47) (walk speed: 2 cM) using a previously established, high-quality genetic map of family 52–124 (29, 30). The genetic map is based primarily upon microsatellite (SSR) markers; however, a number of previously identified microarray-based polymorphic features (30) were also included to expand map coverage in regions of low SSR density (29). The map covers at least 85% of the assembled P. trichocarpa genome and exhibits no major regions of segregation distortion (29).
Significance of eQTL logarithm of odds (LOD) values was estimated for xylem, leaf, and root using a global permutation threshold (9), reported in the footnotes of Table S1. eQTL were declared on the basis of a strategy wherein eQTL composed of unimodal LOD curves are located by the peak position (10). Bimodal peaks were declared as separate eQTL if the trough between them exceeded 2 LOD.
The eQTL were classified as cis or trans regulated based on colocalization of the eQTL LOD peak with the genetic map marker bin containing the gene model in the Nisqually-1 genome sequence. Though the family 52–124 map encompasses >85% of the assembled genome sequence (29, 30), 23,116 of the 55,793 gene models/ESTs assessed by our microarray are located on unassembled genomic scaffolds (17,726) or outside the coverage of the genetic map (5,390). eQTL for these probes were designated “undefined” for the purposes of declaring cis vs. trans regulation.
eQTL Hotspot Detection and Analysis.
To identify significant eQTL hotspots, we permuted the per-bin total eQTL peak counts for xylem, leaf, and root across the 1,840 ~2-cM bins of the genetic map 1,000 times, and determined the 95th percentile of these permutations. Each 2-cM bin with a total absolute eQTL peak count greater than this permutation threshold was declared an eQTL hotspot. To eliminate differential gene density as an explanatory factor for eQTL hotspots (i.e., more genes per genetic distance), we used a χ2 test as previously described (10).
Hotspot-Based Coexpression Network Construction.
We constructed coexpression networks conditioned on the bins declared as eQTL hotspots. For each ~2- cM map bin identified as an eQTL hotspot, we selected all genes whose eQTL LOD values surpassed the organ-specific permutation thresholds for eQTL significance (Table S1 footnotes). We isolated the log2-transformed, normalized expression values for these genes in the respective organ of interest and computed pairwise Pearson correlations between them. Networks were declared when no fewer than 10 genes in a given hotspot bin demonstrated a Pearson correlation of |r| > 0.80. Network edges were constructed and tallied between network members displaying correlations surpassing this threshold.
GO Annotation and Enrichment Testing.
For each coexpression network constructed, we annotated member genes for Gene Ontology (GO) (48) categories by conducting a BLASTX search of the poplar gene model transcripts against The Arabidopsis Information Resource (TAIR) proteins Release v. 8.0. A significant BLAST match was declared at an E-value threshold of <1 × 10−5, with transcripts returning E > 1 × 10−5 annotated as “no hits.” The GO annotation of the closest Arabidopsis putative homolog was assigned to the respective poplar gene. We identified putative orthologs for 45,648 genes on the microarray, of which 36,688 included at least one GO designation in TAIR's database.
Overrepresentation of GO categories was tested within each network by producing 2 × 2 contingency tables for each GO category represented within the network, followed by a right-tailed Fisher's exact test (calculating the probability of observing an equal or higher frequency of the category in the network, relative to the genome frequency of the GO category). To avoid introducing bias due to differential expression of specific categories of genes in a given tissue, contingency tables were developed using the full complement of 36,688 GO-annotated gene models. Because each network was tested for a distinct number of GO enrichments, a Bonferroni correction for multiple testing was applied separately for each network tested, which was computed using the formula Pcorr = 0.05/n for networks comprising n distinct GO categories.
Cis Element Detection and Enrichment Testing.
To annotate the presence and absence of common plant cis-acting elements in the promoters of the gene models from the P. trichocarpa genome, we extracted the promoter sequences upstream of the start codon for the 55,793 genes represented in the microarray. Uninterrupted sequence of 1,500 bp of length could be identified for 49,066 genes, as the position of the remaining 6,727 was less than 1,500 bp from an unresolved sequence region or a whole genome shotgun scaffold or contig end. To avoid bias associated with cis motifs that may be located at preferential distances from the start codon, we did not consider these 6,727 genes in our statistical analysis. We downloaded the plant cis-acting regulatory element sequence database (PLACE) (37) and determined the presence and absence of motifs within all 49,066 gene promoters by using Patmatch (31). Among 469 PLACE database elements, we detected 360 in at least one gene promoter region included in our analysis. For each of these motifs, we tested each coexpression network for enrichment of genes bearing the motif in question using a right-tailed Fisher's exact test. Multiple testing was corrected using a Bonferroni threshold of P < 0.05/360 = 1.389 × 10−4 to judge significance of resulting enrichments.
Supplementary Material
Acknowledgments
The authors thank Karen Koch (University of Florida), Ron Sederoff (North Carolina State University), and two anonymous reviewers for constructive comments to improve the manuscript. This work was supported by the Department of Energy, Office of Science, Office of Biological and Environmental Research Grant DE-FG02-05ER64114 (to M.K.) and the National Science Foundation, Genes and Genomes System Cluster in the Division of Molecular and Cellular Biosciences Grant 0817900 (to M.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (Platform GPL7234 and Gene Expression Series GSE12623, GSE20117, and GSE20118).
This article contains supporting information online at www.pnas.org/cgi/content/full/0914709107/DCSupplemental.
References
- 1.King MC, Wilson AC. Evolution at 2 levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 2.Jansen RC, Nap JP. Genetical genomics: The added value from segregation. Trends Genet. 2001;17:388–391. doi: 10.1016/s0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]
- 3.Morley M, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schadt EE, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 5.Brem RB, Kruglyak L. The landscape of genetic complexity across 5,700 gene expression traits in yeast. Proc Natl Acad Sci USA. 2005;102:1572–1577. doi: 10.1073/pnas.0408709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 7.Keurentjes JJ, et al. Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci. Proc Natl Acad Sci USA. 2007;104:1708–1713. doi: 10.1073/pnas.0610429104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vuylsteke M, van Eeuwijk F, Van Hummelen P, Kuiper M, Zabeau M. Genetic analysis of variation in gene expression in Arabidopsis thaliana. Genetics. 2005;171:1267–1275. doi: 10.1534/genetics.105.041509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.West MA, et al. Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics. 2007;175:1441–1450. doi: 10.1534/genetics.106.064972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potokina E, et al. Gene expression quantitative trait locus analysis of 16,000 barley genes reveals a complex pattern of genome-wide transcriptional regulation. Plant J. 2008;53:90–101. doi: 10.1111/j.1365-313X.2007.03315.x. [DOI] [PubMed] [Google Scholar]
- 11.Kirst M, Basten CJ, Myburg AA, Zeng ZB, Sederoff RR. Genetic architecture of transcript-level variation in differentiating xylem of a eucalyptus hybrid. Genetics. 2005;169:2295–2303. doi: 10.1534/genetics.104.039198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mehrabian M, et al. Integrating genotypic and expression data in a segregating mouse population to identify 5-lipoxygenase as a susceptibility gene for obesity and bone traits. Nat Genet. 2005;37:1224–1233. doi: 10.1038/ng1619. [DOI] [PubMed] [Google Scholar]
- 13.Sonderby IE, et al. A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PLoS One. 2007;2:e1322. doi: 10.1371/journal.pone.0001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 15.Chesler EJ, et al. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet. 2005;37:233–242. doi: 10.1038/ng1518. [DOI] [PubMed] [Google Scholar]
- 16.Dimas AS, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobrin R, et al. Multi-tissue coexpression networks reveal unexpected subnetworks associated with disease. Genome Biol. 2009;10:R55. doi: 10.1186/gb-2009-10-5-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petretto E, et al. Heritability and tissue specificity of expression quantitative trait loci. PLoS Genet. 2006;2:e172. doi: 10.1371/journal.pgen.0020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kliebenstein DJ, et al. Genomic survey of gene expression diversity in Arabidopsis thaliana. Genetics. 2006;172:1179–1189. doi: 10.1534/genetics.105.049353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeCook R, Lall S, Nettleton D, Howell SH. Genetic regulation of gene expression during shoot development in Arabidopsis. Genetics. 2006;172:1155–1164. doi: 10.1534/genetics.105.042275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khaitovich P, Enard W, Lachmann M, Paabo S. Evolution of primate gene expression. Nat Rev Genet. 2006;7:693–702. doi: 10.1038/nrg1940. [DOI] [PubMed] [Google Scholar]
- 22.Quesada T, et al. Comparative analysis of the transcriptomes of Populus trichocarpa and Arabidopsis thaliana suggests extensive evolution of gene expression regulation in angiosperms. New Phytol. 2008;180:408–420. doi: 10.1111/j.1469-8137.2008.02586.x. [DOI] [PubMed] [Google Scholar]
- 23.Kliebenstein DJ, et al. Identification of QTLs controlling gene expression networks defined a priori. BMC Bioinformatics. 2006;7:308. doi: 10.1186/1471-2105-7-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wentzell AM, Boeye I, Zhang Z, Kliebenstein DJ. Genetic networks controlling structural outcome of glucosinolate activation across development. PLoS Genet. 2008;4:e1000234. doi: 10.1371/journal.pgen.1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wentzell AM, et al. Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLoS Genet. 2007;3:1687–1701. doi: 10.1371/journal.pgen.0030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu C, et al. Gene set enrichment in eQTL data identifies novel annotations and pathway regulators. PLoS Genet. 2008;4:e1000070. doi: 10.1371/journal.pgen.1000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu J, et al. System-wide molecular evidence for phenotypic buffering in Arabidopsis. Nat Genet. 2009;41:166–167. doi: 10.1038/ng.308. [DOI] [PubMed] [Google Scholar]
- 28.Zhu J, et al. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat Genet. 2008;40:854–861. doi: 10.1038/ng.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Novaes E, et al. Quantitative genetic analysis of biomass and wood chemistry of Populus under different nitrogen levels. New Phytol. 2009;182:878–890. doi: 10.1111/j.1469-8137.2009.02785.x. [DOI] [PubMed] [Google Scholar]
- 30.Drost DR, et al. A microarray-based genotyping and genetic mapping approach for highly heterozygous outcrossing species enables localization of a large fraction of the unassembled Populus trichocarpa genome sequence. Plant J. 2009;58:1054–1067. doi: 10.1111/j.1365-313X.2009.03828.x. [DOI] [PubMed] [Google Scholar]
- 31.Tuskan GA, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 32.Breitling R, et al. Genetical genomics: Spotlight on QTL hotspots. PLoS Genet. 2008;4:e1000232. doi: 10.1371/journal.pgen.1000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yvert G, et al. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- 34.Mackay TF. The genetic architecture of quantitative traits. Annu Rev Genet. 2001;35:303–339. doi: 10.1146/annurev.genet.35.102401.090633. [DOI] [PubMed] [Google Scholar]
- 35.McAndrew RS, Froehlich JE, Vitha S, Stokes KD, Osteryoung KW. Colocalization of plastid division proteins in the chloroplast stromal compartment establishes a new functional relationship between FtsZ1 and FtsZ2 in higher plants. Plant Physiol. 2001;127:1656–1666. [PMC free article] [PubMed] [Google Scholar]
- 36.Stokes KD, McAndrew RS, Figueroa R, Vitha S, Osteryoung KW. Chloroplast division and morphology are differentially affected by overexpression of FtsZ1 and FtsZ2 genes in Arabidopsis. Plant Physiol. 2000;124:1668–1677. doi: 10.1104/pp.124.4.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 39.Gerrits A, et al. Expression quantitative trait loci are highly sensitive to cellular differentiation state. PLoS Genet. 2009;5:e1000692. doi: 10.1371/journal.pgen.1000692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hubner N, et al. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat Genet. 2005;37:243–253. doi: 10.1038/ng1522. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grieve IC, et al. Genome-wide co-expression analysis in multiple tissues. PLoS One. 2008;3:e4033. doi: 10.1371/journal.pone.0004033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ayroles JF, et al. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet. 2009;41:299–307. doi: 10.1038/ng.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993;11:117–121. [Google Scholar]
- 45.Zeng ZB. Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc Natl Acad Sci USA. 1993;90:10972–10976. doi: 10.1073/pnas.90.23.10972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeng ZB. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang S, Basten CJ, Zeng ZB. Windows QTL Cartographer 2.5 . North Carolina State University, Raleigh, NC: Department of Statistics; 2007. Available at http://statgen.ncsu.edu/qtlcart/WQTLCart.htm. [Google Scholar]
- 48.Ashburner M, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.