Abstract
Hsp104 is a ring-forming AAA+ machine that recognizes both aggregated proteins and prion-fibrils as substrates and, together with the Hsp70 system, remodels substrates in an ATP-dependent manner. Whereas the ability to disaggregate proteins is dependent on the Hsp104 M-domain, the location of the M-domain is controversial and its exact function remains unknown. Here we present cryoEM structures of two Hsp104 variants in both crosslinked and noncrosslinked form, in addition to the structure of a functional Hsp104 chimera harboring T4 lysozyme within the M-domain helix L2. Unexpectedly, we found that our Hsp104 chimera has gained function and can solubilize heat-aggregated β-galactosidase (β-gal) in the absence of the Hsp70 system. Our fitted structures confirm that the subunit arrangement of Hsp104 is similar to other AAA+ machines, and place the M-domains on the Hsp104 exterior, where they can potentially interact with large, aggregated proteins.
Keywords: ATPase, AAA+, chaperone, disaggregasse, ClpB
AAA+ (ATPase associated with diverse cellular activities) ATPases represent an essential group of structurally related but functionally diverse molecular machines that utilize the energy from ATP binding and hydrolysis to facilitate the unfolding or unwinding of proteins and nucleic acids, as well as the remodeling of macromolecular assemblies (1, 2). Members of the AAA+ superfamily usually form hexameric ring structures and share an ATPase domain that harbors canonical Walker A and Walker B motifs, in addition to other conserved, cis- and trans-acting structural elements (2), such as the pore loops required for substrate translocation (3–6), and the so-called Arg-finger residue that facilitates nucleotide hydrolysis by stabilizing the leaving γ-phosphate group (7, 8). It has been shown that the Arg-finger protrudes into the nucleotide binding site of the adjacent protomer (7, 9–12) and, therefore, is a key structural feature that defines the arrangement and orientation of subunits within the hexamer ring (2).
Hsp104 is an AAA+ ATPase that, together with the Hsp70 system (Hsp70/Hsp40), rescues stress-damaged proteins from a previously aggregated state (13, 14). The ability to solubilize amorphous aggregates is shared among orthologous ClpB/Hsp104 proteins and is dependent on the M-domain (15, 16), a hallmark of this chaperone family. Other Clp/Hsp100 proteins that lack the M-domain unfold proteins but, on their own, do not normally solubilize aggregated proteins (17, 18). Whereas the exact function of the M-domain is unclear, it has been suggested that the M-domain mediates interdomain communication (19) and couples the activity of the Hsp70 system to the Hsp104 motor activity (20). However, how this might be accomplished remains poorly understood.
The first atomic insight into a full-length member of the ClpB/Hsp104 family came from the x-ray structure of Thermus thermophilus ClpB (15, 21). The crystal structure consisted of three independent copies of full-length ClpB that formed a helical assembly inside the crystalline lattice (15, 21). Each ClpB monomer possesses two canonical AAA+ ATPase domains that have the same fold as those present in other AAA+ machines (2). The ClpB/Hsp104-specific M-domain forms a long and mobile coiled-coil (15) that extends to the outside of the helical assembly (21). An M-domain location on the ClpB exterior is also consistent with the hexameric ring structure of ClpB, determined independently by single-particle cryoEM (15, 22) and by computational modeling with evolutionary restraints (23), and is corroborated by sulfhydryl crosslinking (15, 20) and fluorescence spectroscopy studies (24).
More recently, the cryoEM structures of yeast Hsp104 were reported (25, 26). Whereas the ATP- and ADP-bound structures were similar (26), the ATPγS-bound structure revealed a largely expanded Hsp104 hexamer (25, 26) that was significantly wider than ClpB (15, 22), ClpA (27), and other AAA+ machines (28, 29). More surprisingly, in the reported Hsp104 cryoEM fits, the proposed subunit arrangement as well as the locations of the Arg-finger residues and conserved substrate binding pore loops (4, 30) differed not only from their known positions in other AAA+ machines (2, 7) but also among the various Hsp104 structures presented (25, 26). For instance, it was proposed that the Hsp104 D1- and D2-large domains move as independent rigid bodies during the ATPase cycle (26), thereby rotating the conserved pore loops from the inside to the outside of the Hsp104 hexamer (26), which is incompatible with their role in substrate translocation through the Hsp104 hexamer (4, 30). Perhaps as a result of those fits, the M-domains were placed either on the inside of Hsp104 or intercalated between neighboring subunits depending on the nucleotide state (25, 26). It is evident that the proposed M-domain location on the Hsp104 interior would likely be inaccessible to the Hsp70 system (20, 31), whereas a location in between subunits would presumably disrupt Hsp104 function by interfering with the Arg-finger.
Here we present the cryoEM structures of two Hsp104 mutant variants in both glutaraldehyde crosslinked and noncrosslinked form, in addition to the cryoEM structure of an Hsp104 chimera that harbors T4 lysozyme within M-domain helix L2. The M-domain densities are partially visible in the Hsp104 chimera and not visible in other cryoEM structures, indicating conformational flexibility. Our fitted structures are consistent with published biochemical and functional data (4–6, 15, 20, 22, 30–33), and place the M-domains on the outside of the Hsp104 hexamer, where they can potentially interact with large, aggregated proteins and components of the Hsp70 system. Surprisingly, we found that our Hsp104 chimera is not only fully functional biochemically but also has gained the capacity to solubilize heat-aggregated β-gal on its own. Taken together, our results demonstrate that Hsp104 and ClpB have a similar 3D structure, consistent with their common ability to solubilize previously aggregated proteins.
Results
CryoEM Structure of Hsp104.
We examined two Hsp104 mutants, namely full-length Hsp104E285A/E687A (Trap) and an N-terminal domain (NTD)-truncated Hsp104 variant (Hsp104ΔN) that was also used in the previous study (25). Unlike wild-type Hsp104, the Trap variant binds but does not hydrolyze ATP (34) and, therefore, allows visualization of the ATP-bound structure without complications that might arise from ATP hydrolysis, which leads to a mixed nucleotide state. In contrast, Hsp104ΔN is a functional ATPase. The structure of the Hsp104ΔN variant was examined in the presence of ATPγS, a slowly hydrolyzable ATP analog (35, 36), and was compared to the structure of the full-length enzyme (Fig. 1).
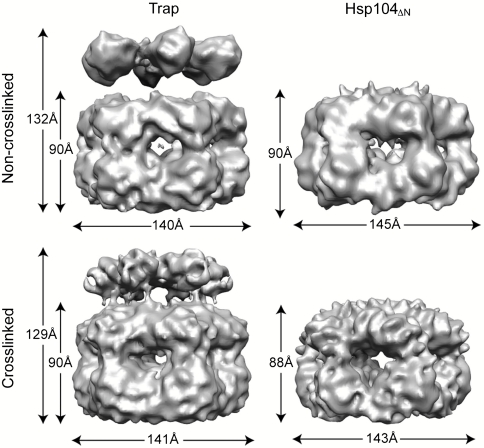
Fig. 1.
CryoEM reconstruction. Isosurface representations of the noncrosslinked (Upper) and glutaraldehyde crosslinked (Lower) Trap-ATP (Left) and Hsp104ΔN-ATPγS hexamer (Right).
Both Hsp104 hexamers were stable and could be examined without glutaraldehyde crosslinking (Fig. S1). Our 3D reconstructions showed that the Trap-ATP and Hsp104ΔN-ATPγS hexamers were similar in width and overall structure, but differed in height due to the lack of the NTDs in the Hsp104ΔN construct (Fig. 1). Notably, we found that the overall dimensions of our Hsp104 reconstructions were comparable to ClpB (15, 22), ClpA (27), and other AAA+ machines whose structures have been determined by cryoEM (28, 29). Full-length Hsp104 formed a three-tiered ring structure composed of two larger rings that make up the main body with a smaller, third ring on top (Fig. 1). The smaller ring was not observed in the Hsp104ΔN reconstruction and, therefore, could be attributed to the NTDs. This arrangement of rings gave rise to an axial channel with a width of 19 Å at the narrowest point. Perhaps most intriguing was the lack of additional mass density on the outside of the Hsp104 hexamer, previously seen in our cryoEM structure of ClpB (15, 22), which might account for the M-domains. It was proposed that the observed differences between the Hsp104 and ClpB cryoEM reconstructions may have arisen from using glutaraldehyde crosslinking to stabilize the ClpB hexamer (26). However, pairwise comparison between glutaraldehyde crosslinked and noncrosslinked Trap and Hsp104ΔN hexamers showed that they were similar in overall shape and dimension (Fig. 1).
To generate an atomic model of the Hsp104 hexamer, we fitted the crystal structure of an intact ClpB monomer [Protein Data Bank (PDB) ID code 1QVR_B] (15) into our 3D reconstruction (Fig. 2 A and C). Remarkably, our cryoEM map closely matched the ClpB domain arrangement seen in the x-ray structure with the exception of the NTD (Fig. 2B). The Hsp104 hexamer was generated subsequently by applying six-fold rotational symmetry.
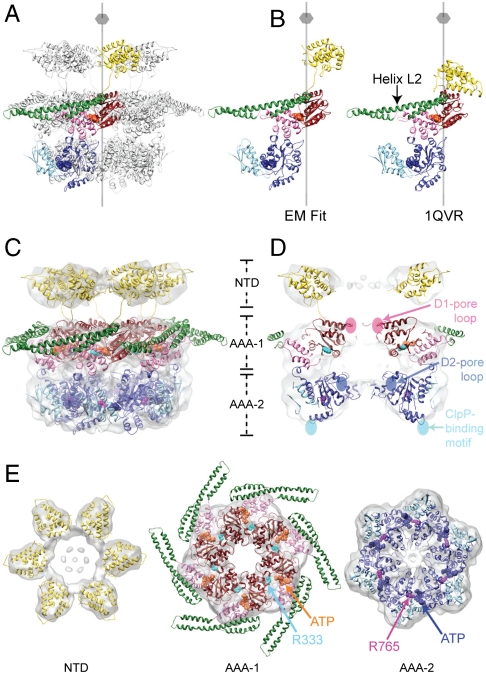
Fig. 2.
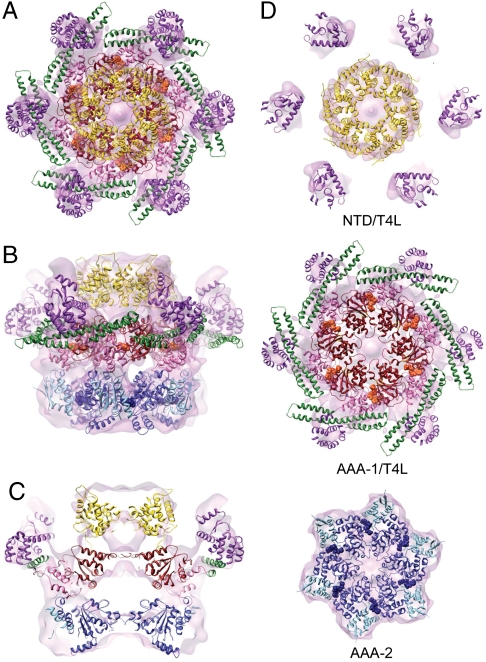
Atomic structure fitting. (A) The Hsp104 hexamer is depicted as ribbon diagram with the NTD (18 kDa, Gold), the D1-large domain (19 kDa, Red), the D1-small domain (10 kDa, Pink), the M-domain (15 kDa, Green), the D2-large domain (23 kDa, Blue), and the D2-small domain (15 kDa, Sky Blue). The six-fold symmetry axis is indicated. (B) Structural comparison between the Hsp104 and ClpB domain arrangement seen in the EM fit and in the crystal (15). (C) CryoEM reconstruction of the Trap-ATP hexamer shown as a semitransparent surface with the structure of the full-length Hsp104 hexamer docked in. The predicted Arg-finger residues, Arg333 (AAA-1, Cyan) and Arg765 (AAA-2, Magenta), and ATP molecules bound to the AAA-1 (Orange) and AAA-2 domains (Blue) are shown as CPK models. (D) Vertical central section depicting the location of the D1- and D2-pore loops and the ClpP-binding motifs. The D2-pore loops are indicated by dotted lines. (E) Top down views of the fitted NTD, AAA-1, and AAA-2 rings. Arg-finger residues and bound ATP molecules are depicted as described in Fig. 2C.
In our fitted structure of the Trap hexamer, the six copies of the NTDs are fully accounted for and, like the AAA-1 and AAA-2 domains, snuggle tightly into the cryoEM map (Fig. 2 C, D, and E). The D2-small domain reaches across the subunit interface and buttresses the D2-large domain of the neighboring subunit (Fig. 2E), thereby stabilizing the Hsp104 hexamer. Fig. 2 D and E show that the D1- and D2-large domains are positioned close to the center with the critical pore-facing loops lining the axial channel, consistent with their proposed role in substrate translocation (4, 30). This orients the D1- and D2-small domains toward the rim of the hexamer and places the M-domain on the outside of Hsp104, as it is in ClpB (15, 22), and keeps the M-domains in continuous sequence-register with the corresponding D1-small domains. Other structural elements, such as the ClpP-binding motifs present in the engineered Hsp104HAP variant (32) (hereafter referred to as the ClpP-binding motifs) and the predicted Arg-finger residues (Arg333 and Arg765) (7, 15, 16, 37), are very close to their expected positions as determined within the resolution limit of our cryoEM reconstruction (Fig. 2 D and E). Notably, Arg333 and Arg765 are located at the interprotomer interface as they are in other AAA+ machines (7, 9–12), and contribute to the ATP-binding pockets of the adjacent protomer.
Probing the Structure of the Hsp104 Hexamer.
To validate our Hsp104 hexamer structure, we probed the surface accessibility of the M-domain and several other key structural elements. To do this, we engineered four distinct Trap variants that harbor an eight-residue Streptavidin binding site (StrepII-tag) in the M-domain (TrapM), the ClpP-binding motif (TrapClpP) (5, 32), the second D1-pore loop (TrapNBD1) (6), or within a loop between strands d4 and d5 in the D2-large domain (TrapNBD2) (15) (Fig. 3A). Care was taken not to disrupt subunit contacts within the Hsp104 hexamer. In our structure, the M-domains and ClpP-binding motifs are fully exposed, whereas the second D1-pore loops and the d4/d5 β-strand regions are located in the interior of the hexamer and, therefore, inaccessible to an antibody probe (Fig. 3B).
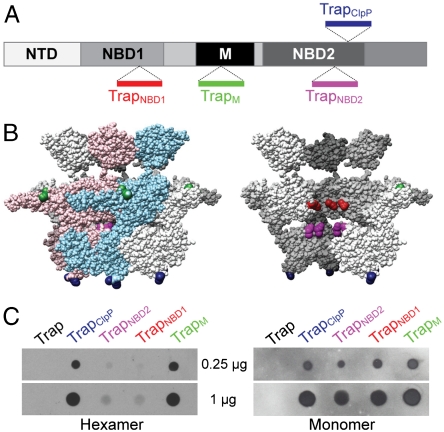
Fig. 3.
Probing the surface accessibility of Hsp104. (A) Schematic diagram summarizing the locations of the inserted StrepII-tag (Trp–Ser–His–Pro–Gln–Phe–Glu–Lys) between Asn292/Gly293 (TrapNBD1, Red), Asn467/Glu468 (TrapM, Green), Gly711/Gln712 (TrapNBD2, Magenta), and Gly740/Ser741 (TrapClpP, Blue). (B) Side-views of the Trap-ATP hexamer showing either all six subunits (Left) or with the two colored subunits removed (Right). The location of the StrepII-tags is indicated using the same color scheme as in Fig. 3A. (C) Dot blots of purified StrepII-tagged Trap hexamers (Left) and monomers (Right) probed with a monoclonal antibody against the StrepII-tag.
It was shown previously that the Hsp104 hexamer is stabilized in the presence of adenine nucleotides (38), but destabilized at high ionic strength (39). Consistent with this notion, we found that all of our engineered StrepII-tagged Trap variants formed hexamers in the presence of ATP as determined by size-exclusion chromatography (Fig. S2), demonstrating that the insertion of the StrepII-tag did not impair oligomerization. To assess the surface exposure of StrepII-tag containing structural elements, we prepared dot blots of hexameric and monomeric Trap variants, and probed the blot with a commercial monoclonal antibody that specifically recognizes the StrepII-tag. Strikingly, the antibody showed strong binding only to the TrapM and TrapClpP hexamers, whereas binding to hexameric TrapNBD1 and TrapNBD2 was insignificant (Fig. 3C, Left). Conversely, all monomeric Trap variants were bound equally by the antibody probe in a control experiment (Fig. 3C, Right).
CryoEM Structure of an Hsp104T4L Chimera.
Whereas our fitted cryoEM structures, combined with dot blot analysis, place the M-domain outside of Hsp104, we still lacked physical evidence. We reasoned that a small, globular protein, when placed within helix L2 of the M-domain, might function as a fiducial marker that would enable us to locate the M-domain. At the same time, if the six copies of the M-domain are located on the inside of Hsp104 or are intercalated between neighboring subunits, the additional mass would likely prevent oligomerization as a result of steric clash.
T4 lysozyme (T4L) is an ideal candidate for such a marker, because it is a small, stably folded protein of known 3D structure (40). To locate the M-domain, we engineered an Hsp104-T4L chimera by inserting the T4L gene between residues Asn467 and Glu468 in the middle of the M-domain helix L2 in both wild-type (Hsp104T4L) and Trap mutant Hsp104 (TrapT4L). The chimera was designed so that the T4L moieties would be surface exposed on the outside of the Hsp104 hexamer, as would be expected from our fitted structure.
We then examined the structure of TrapT4L in the ATP state by cryoEM (Fig. 4). Unlike the Trap variant, however, the TrapT4L hexamer could only be imaged after crosslinking the preassembled hexamer. As noted above, we already showed that this procedure did not markedly alter the Hsp104 hexamer structure (Fig. 1). Fig. 4A depicts a representative area of a digital micrograph of TrapT4L embedded in vitreous ice. Our data confirmed that TrapT4L assembled into hexamers, suggesting that the presence of T4L did not prevent oligomerization. As in previous samples, TrapT4L-ATP hexamers were well distributed, showed no signs of aggregation and displayed a wide range of orientations (Fig. S3). Our final cryoEM reconstruction of the TrapT4L hexamer was determined to a resolution of 11 Å at a Fourier shell correlation (FSC) of 0.5 (Fig. S4).
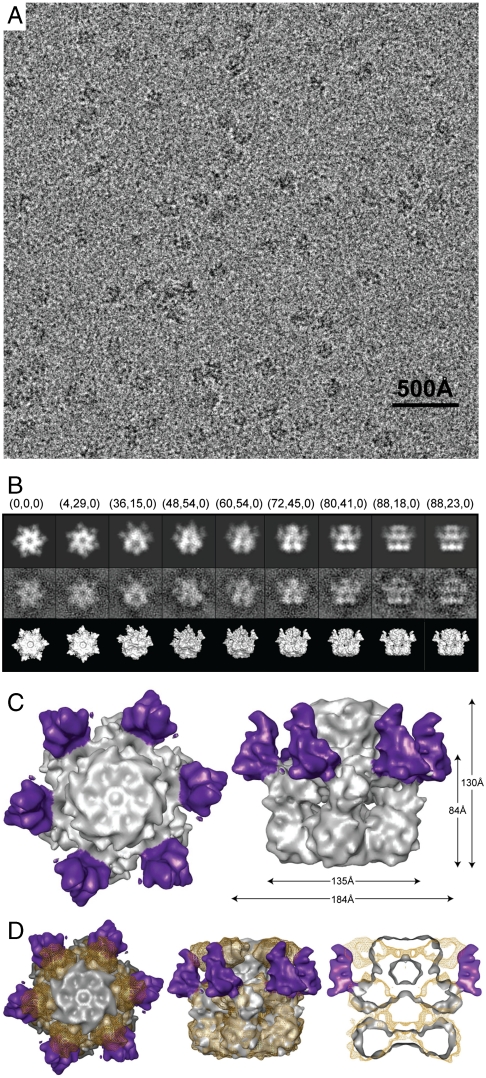
Fig. 4.
CryoEM analysis of the TrapT4L-ATP hexamer. (A) Digital micrograph of the TrapT4L hexamer embedded in vitreous ice. (B) Selected projection views (Upper), corresponding class averages (Middle), and 3D reconstructions in the same view (Lower). Corresponding Euler angles (alt, az, phi) in EMAN convention are given. (C) Top down (Left) and side view of the TrapT4L hexamer reconstruction (Right), with the six T4L moieties colored purple. (D) Top down view (Left), side view (Middle), and vertical central section (Right) of the superimposed cryoEM maps of the Trap (Golden Mesh) and TrapT4L hexamer (Gray Solid).
Like the Trap hexamer, TrapT4L has a three-tiered ring structure (Fig. 4C, Right). However, the top ring of the TrapT4L hexamer is smaller and more compact. Most importantly, in our TrapT4L reconstruction, we observed six additional mass densities on the outside of the hexamer surrounding the AAA-1 ring (Fig. 4 C and D). These additional mass densities were not observed with the Trap hexamer and, therefore, can be attributed to the folded T4L moiety that was engineered into the M-domain of each of the six subunits.
To generate an atomic model for TrapT4L, we docked the structure of the full-length Trap hexamer into the mass density of the TrapT4L reconstruction (Fig. 5). Our Trap hexamer structure fit very well and required only minor adjustments, indicating that the presence of T4L did not significantly alter the structure of the main hexamer body. Markedly, the additional mass density observed on the outside of the Hsp104 hexamer closely matched the predicted location of the T4L moiety when modeled into the ClpB monomer crystal structure (PDB ID code 1QVR_B) (15), further supporting the validity of our fitted Hsp104 structure. However, in contrast to the Trap hexamer, the NTDs are more closely packed (Fig. 5) but, nevertheless, the observed mass densities are large enough to account for six copies of an intact NTD. Because the NTDs can adopt different orientations even in the same nucleotide state (15), the importance of the observed structural difference is currently unknown.
Fig. 5.
Atomic structure fitting of the TrapT4L hexamer. (A–D) The cryoEM reconstruction is shown as a semitransparent surface, with the structure of the full-length TrapT4L hexamer docked in. T4L (18 kDa) is colored magenta and bound ATP molecules orange (AAA-1) and blue (AAA-2), respectively. (A) Top down, (B) side, and (C) vertical central section of the fitted TrapT4L hexamer. (D) Top down views of the fitted NTD/T4L, AAA-1/T4L, and AAA-2 rings.
The Hsp104T4L Chimera is a Functional Protein Disaggregase.
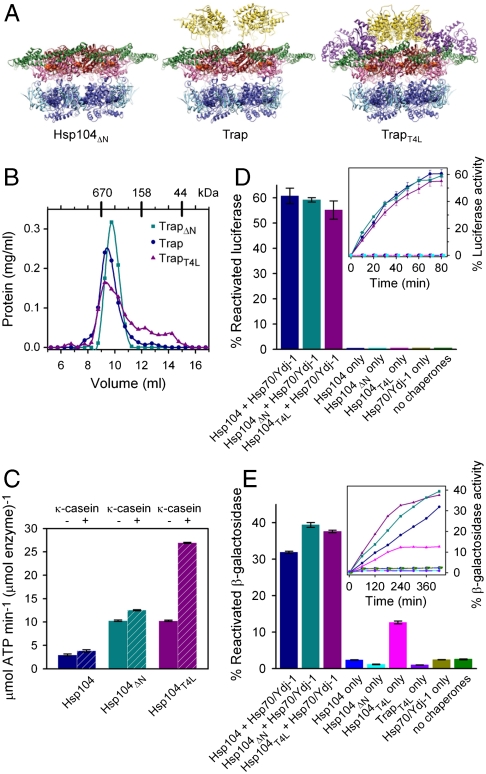
To show that our TrapT4L hexamer structure does not represent an off-pathway state, we asked whether our chimera is functional biochemically. First, we analyzed whether TrapT4L forms a hexamer in the absence of glutaraldehyde crosslinking by examining the oligomeric state of TrapT4L in the presence of ATP by size-exclusion chromatography. As shown in Fig. 6B, TrapT4L forms a hexamer, although a shoulder and several smaller peaks were also observed, indicating that the chimera is somewhat less stable than Trap.
Fig. 6.
Biochemical analysis of Hsp104, Hsp104ΔN, and Hsp104ΔT4L. (A) Fitted structures of the Hsp104ΔN-ATPγS, Trap-ATP, and TrapT4L-ATP hexamers. (B) Hexamer assembly of TrapΔN (Teal), Trap (Blue), and TrapT4L (Magenta) analyzed by size-exclusion chromatography. (C) ATPase activities of Hsp104 (Blue), Hsp104ΔN (Teal), and Hsp104T4L (Magenta) measured in the absence (Solid Bars) or presence (Hashed Bars) of 0.2 mg/mL κ-casein. Standard errors from three independent measurements are shown. (D) Reactivation of chemically denatured firefly luciferase by Hsp104 (Blue), Hsp104ΔN (Teal), or Hsp104T4L (Magenta) either alone or together with Hsp70/Ydj-1. Amount of reactivated luciferase is shown after 80 min as percentage of a native control (Bars) or as time course analysis (Inset). Standard errors of three independent assays are shown. (E) Solubilization of heat-aggregated β-gal. The color scheme is the same as in (D), with TrapT4L alone (Violet) added as another control. Amount of reactivated β-gal is shown after 420 min as percentage of a native control (Bars) or as time course analysis (Inset). Standard errors of three independent assays are shown.
Next, we asked whether our engineered chimera has ATPase activity. We found that Hsp104T4L has 3.5-fold higher ATPase activity than wild-type Hsp104, which can be further stimulated 2.6-fold by κ-casein (Fig. 6C). The reason for the higher ATPase activities is currently unknown. To establish whether our chimera is a functional protein disaggregase, we asked whether Hsp104T4L cooperates with the Hsp70 chaperone system to solubilize aggregated model substrates. To test this, we used an in vitro refolding assay with either chemically denatured firefly luciferase or heat-aggregated β-gal as substrate. Consistent with previous studies (14), we found that Hsp104T4L cooperates with the Hsp70 chaperone system to solubilize either substrate (Fig. 6 D and E). Unexpectedly, Hsp104T4L, but not wild-type Hsp104 or TrapT4L, could also solubilize heat-aggregated β-gal in the absence of the Hsp70 system (Fig. 6E), indicating that our chimera has gained function.
Discussion
Hsp104 is Not an Atypical AAA+ ATPase.
Hsp104 and ClpB are orthologous proteins that share the ability to rescue proteins from a previously aggregated state. Our Hsp104 structures are comparable with the overall dimensions of ClpB (15, 22), ClpA (27), and other more distantly related AAA+ ATPases (28, 29), but differ substantially from the previously published Hsp104 models (25, 26). Notably, in our fitted Hsp104 structure, the predicted Arg-finger residues are located at the interprotomer interface, as they are in other AAA+ machines; the ClpP-binding motifs (5, 32) are exposed at the bottom surface of the AAA-2 ring as required for stable association of the Hsp104HAP variant with the oligomeric ClpP protease (31, 32) (Fig. 2 D and E), and the D1/2-pore loops line the central channel, which is compatible with their role in substrate threading through the Hsp104 hexamer (4, 30–32). Our fit places the M-domains on the outside of the Hsp104 hexamer, as they are in ClpB (15, 22), and is supported by dot blot analysis using purified components (Fig. 3C). Finally, placing the M-domain on the outside of the Hsp104 hexamer is also consistent with the ClpB x-ray structure and keeps the M-domain in sequence register with the remainder of the D1-small domain.
The M-domain is Located on the Outside of Hsp104.
In our Hsp104 cryoEM reconstructions, no mass density was observed that could account for the M-domains. This is surprising because the latter were at least partially resolved in cryoEM reconstructions of the T. thermophilus ClpB hexamer (15, 22). While the exact reason for this discrepancy is currently unknown, it has previously been suggested that the ClpB M-domains may undergo a transition from a folded to an unfolded state (20), and perhaps more so in Hsp104.
To obtain physical evidence to support an M-domain location on the outside of Hsp104, we engineered a biochemically functional Hsp104 chimera (Fig. 6 D and E) that harbors T4L within the M-domain helix L2. The rationale behind this approach was to introduce a stably folded marker that would allow us to pinpoint the location of the M-domain. Most remarkably, our cryoEM reconstruction of the TrapT4L chimera revealed six additional mass densities surrounding the Hsp104 hexamer (Fig. 4C), which were not observed in the cryoEM reconstruction of Trap. The location of these mass densities matched the predicted location of the T4L moieties when modeled into the ClpB monomer x-ray structure (PDB ID code 1QVR_B) (15). While part of the M-domain also becomes visible (Fig. 5), no other significant structural differences were observed in the main hexamer body (Fig. 6A).
Hsp104T4L has Gained the Ability to Solubilize Heat-aggregated β-gal.
Like wild-type Hsp104, our Hsp104-T4L chimera cooperates with the Hsp70 chaperone system to solubilize and reactivate amorphous aggregates of known Hsp104 model substrates (Fig. 6 D and E). Remarkably, we found that Hsp104T4L alone could also disaggregate heat-aggregated β-gal (Fig. 6E), which resembles more closely the physiological substrate, but not chemically denatured firefly luciferase (Fig. 6D). The latter might suggest possible differences in the physical properties between aggregated model substrates (41) or distinct Hsp70/Hsp40-dependencies for protein reactivation following solubilization by Hsp104.
Interestingly, neither wild-type Hsp104 nor TrapT4L mutant could substitute for Hsp104T4L to solubilize heat-aggregated β-gal on its own (Fig. 6E), indicating that the observed gain of function is ATP-dependent and specific to the Hsp104T4L chimera. Because wild-type Hsp104 can solubilize heat-aggregated β-gal in the presence of the Hsp70 system, it is conceivable that the T4L moiety may mimic an Hsp70, Hsp40, and/or substrate interaction that unleashes the protein remodeling activity of Hsp104.
Conclusion
Our combined structural and biochemical findings demonstrate that yeast Hsp104 is not an atypical AAA+ machine but has the same subunit arrangement as bacterial ClpB, consistent with their similar biological function. In our structure the M-domains are placed on the outside of the Hsp104 hexamer, as they are in ClpB, where they can potentially interact with large protein aggregates. How this might be accomplished is subject to further investigations.
Materials and Methods
The cloning, mutagenesis, protein expression, and purification of Saccharomyces cerevisiae Hsp104 wild-type, chimera, and mutant variants, as well as procedures for the dot blot and biochemical assays are described in SI Text.
CryoEM.
CryoEM samples were prepared as described in SI Text. Frozen hydrated grids were imaged at -180 °C using a JEOL JEM-2010F electron microscope operated at 200 keV. Images were recorded on a 4K × 4K CCD camera in low dose mode (14–16 e-/Å2) at an effective magnification of 82,800, resulting in a pixel size of 1.81 Å at the specimen level with defocus ranging from 1.0 to 5.5 μm.
Image Processing and 3D Reconstruction.
All image processing, analysis, and 3D reconstructions were done using EMAN (42–44). Particles were selected from digital micrographs using boxer. The contrast transfer function (CTF) correction parameters were first determined automatically using fitctf, and then manually adjusted using ctfit. An initial model for the Trap hexamer was generated from reference-free class averages using startcsym. A multimodel refinement step was included at the beginning (SI Text) essentially as previously described (45). Further refinement was performed with a subset of particles using single-model refinement cycles with 6-fold symmetry. In each cycle, a new model was constructed from the class averages of the CTF amplitude and phase corrected particle images with Wiener filtration. A soft Gaussian mask was used throughout the refinement and resolution analysis, except for early cycles of refinement. In each cycle, the mask was calculated automatically from the mass densities with threshold about half of the isosurface rendering plus an additional 20 Å layer. The resolution of our final reconstructions was determined at a threshold of 0.5 from the FSC between reconstructions from even and odd halves of the datasets (Table S1 and Fig. S4).
Atomic Structure Fitting.
The Trap hexamer was generated by fitting the atomic structure of a T. thermophilus ClpB monomer (PDB ID code 1QVR_B) (15) (SI Text). The Hsp104ΔN and TrapT4L hexamers were generated from the fitted Trap hexamer and slightly adjusted to reflect differences in diameter and height (Figs. 1A and 4C). The NTDs were either deleted (Hsp104ΔN) or fitted into the more compact mass density on top of the AAA rings (TrapT4L). T4L (PDB ID code 1L63) (40) was modeled into the extra mass density protruding from the main body. Our fit places T4L in sequence register with helix L2. Part of the M-domain is also visible and occupies the lower part of the density. All figures were generated with Chimera (46).
Supplementary Material
Acknowledgments.
We thank E. Fuh and H. Kim for generating constructs, D. Chen and S.J. Ludtke for advice and help with EMAN, and A. Biter for critical reading of this manuscript. We are grateful to W. Chiu for access to the cryoEM facility of the National Center for Macromolecular Imaging. Work in the laboratories of F.T.F.T. and S.L. is supported by National Institutes of Health Grants AI076239 and GM067672, the Department of Defense, the American Cancer Society, the Welch Foundation Grant Q-1530, and the American Heart Association. B.S. was a recipient of a training fellowship (DK071505) from the Pharmacoinformatics Training Program of the Keck Center of the Gulf Coast Consortia.
Footnotes
The authors declare no conflict of interest.
Data deposition: The cryoEM maps reported in this paper have been deposited with the Electron Microscopy Data Bank (EMDB) under accession codes EMD-1631 (Trap), EMD-1629 (Hsp104ΔN), and EMD-1630 (TrapT4L).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003572107/-/DCSupplemental.
References
- 1.Hanson PI, Whiteheart SW. AAA+ proteins: Have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 2.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 3.Wang J, et al. Crystal structure of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism. Structure. 2001;9:177–184. doi: 10.1016/s0969-2126(01)00570-6. [DOI] [PubMed] [Google Scholar]
- 4.Lum R, Tkach JM, Vierling E, Glover JR. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem. 2004;279:29139–29146. doi: 10.1074/jbc.M403777200. [DOI] [PubMed] [Google Scholar]
- 5.Weibezahn J, et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 6.Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL. Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell. 2005;121:1029–1041. doi: 10.1016/j.cell.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 7.Ogura T, Whiteheart SW, Wilkinson AJ. Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J Struct Biol. 2004;146:106–112. doi: 10.1016/j.jsb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Wigley DB. The “glutamate switch” provides a link between ATPase activity and ligand binding in AAA+ proteins. Nat Struct Mol Biol. 2008;15:1223–1227. doi: 10.1038/nsmb.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, et al. Structure of the AAA ATPase p97. Mol Cell. 2000;6:1473–1484. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 10.De LaBarre B, Brünger AT. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat Struct Biol. 2003;10:856–863. doi: 10.1038/nsb972. [DOI] [PubMed] [Google Scholar]
- 11.DeLaBarre B, Brünger AT. Nucleotide dependent motion and mechanism of action of p97/VCP. J Mol Biol. 2005;347:437–452. doi: 10.1016/j.jmb.2005.01.060. [DOI] [PubMed] [Google Scholar]
- 12.Davies JM, Brünger AT, Weis WI. Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure. 2008;16:715–726. doi: 10.1016/j.str.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 13.Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 14.Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 15.Lee S, et al. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- 16.Mogk A, et al. Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J Biol Chem. 2003;278:17615–17624. doi: 10.1074/jbc.M209686200. [DOI] [PubMed] [Google Scholar]
- 17.Dougan DA, Reid BG, Horwich AL, Bukau B. ClpS, a substrate modulator of the ClpAP machine. Mol Cell. 2002;9:673–683. doi: 10.1016/s1097-2765(02)00485-9. [DOI] [PubMed] [Google Scholar]
- 18.Haslberger T, et al. Protein disaggregation by the AAA+ chaperone ClpB involves partial threading of looped polypeptide segments. Nat Struct Mol Biol. 2008;15:641–650. doi: 10.1038/nsmb.1425. [DOI] [PubMed] [Google Scholar]
- 19.Cashikar AG, et al. Defining a pathway of communication from the C-terminal peptide binding domain to the N-terminal ATPase domain in a AAA protein. Mol Cell. 2002;9:751–760. doi: 10.1016/s1097-2765(02)00499-9. [DOI] [PubMed] [Google Scholar]
- 20.Haslberger T, et al. M domains couple the ClpB threading motor with the DnaK chaperone activity. Mol Cell. 2007;25:247–260. doi: 10.1016/j.molcel.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 21.Lee S, Sowa ME, Choi JM, Tsai FTF. The ClpB/Hsp104 molecular chaperone-a protein disaggregating machine. J Struct Biol. 2004;146:99–105. doi: 10.1016/j.jsb.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 22.Lee S, Choi JM, Tsai FTF. Visualizing the ATPase cycle in a protein disaggregating machine: Structural basis for substrate binding by ClpB. Mol Cell. 2007;25:261–271. doi: 10.1016/j.molcel.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diemand AV, Lupas AN. Modeling AAA+ ring complexes from monomeric structures. J Struct Biol. 2006;156:230–243. doi: 10.1016/j.jsb.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 24.Watanabe Y, Takano M, Yoshida M. ATP binding to nucleotide binding domain (NBD)1 of the ClpB chaperone induces motion of the long coiled-coil, stabilizes the hexamer, and activates NBD2. J Biol Chem. 2005;280:24562–24567. doi: 10.1074/jbc.M414623200. [DOI] [PubMed] [Google Scholar]
- 25.Wendler P, et al. Atypical AAA+ subunit packing creates an expanded cavity for disaggregation by the protein-remodeling factor Hsp104. Cell. 2007;131:1366–1377. doi: 10.1016/j.cell.2007.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wendler P, et al. Motor mechanism for protein threading through Hsp104. Mol Cell. 2009;34:81–92. doi: 10.1016/j.molcel.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishikawa T, Maurizi MR, Steven AC. The N-terminal substrate-binding domain of ClpA unfoldase is highly mobile and extends axially from the distal surface of ClpAP protease. J Struct Biol. 2004;146:180–188. doi: 10.1016/j.jsb.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 28.Rouiller I, et al. Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nat Struct Biol. 2002;9:950–957. doi: 10.1038/nsb872. [DOI] [PubMed] [Google Scholar]
- 29.Rockel B, Jakana J, Chiu W, Baumeister W. Electron cryo-microscopy of VAT, the archaeal p97/CDC48 homologue from Thermoplasma acidophilum. J Mol Biol. 2002;317:673–681. doi: 10.1006/jmbi.2002.5448. [DOI] [PubMed] [Google Scholar]
- 30.Lum R, Niggemann M, Glover JR. Peptide and protein binding in the axial channel of Hsp104: Insights into the mechanism of protein unfolding. J Biol Chem. 2008;283:30139–30150. doi: 10.1074/jbc.M804849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tipton KA, Verges KJ, Weissman JS. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol Cell. 2008;32:584–591. doi: 10.1016/j.molcel.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tessarz P, Mogk A, Bukau B. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol Microbiol. 2008;68:87–97. doi: 10.1111/j.1365-2958.2008.06135.x. [DOI] [PubMed] [Google Scholar]
- 33.Doyle SM, Wickner S. Hsp104 and ClpB: Protein disaggregating machines. Trends Biochem Sci. 2009;34:40–48. doi: 10.1016/j.tibs.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 34.Bosl B, Grimminger V, Walter S. Substrate binding to the molecular chaperone Hsp104 and its regulation by nucleotides. J Biol Chem. 2005;280:38170–38176. doi: 10.1074/jbc.M506149200. [DOI] [PubMed] [Google Scholar]
- 35.Doyle SM, et al. Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat Struct Mol Biol. 2007;14:114–122. doi: 10.1038/nsmb1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaupp AMM, Grimminger V, Bösl B, Walter S. Processing of proteins by the molecular chaperone Hsp104. J Mol Biol. 2007;370:674–686. doi: 10.1016/j.jmb.2007.04.070. [DOI] [PubMed] [Google Scholar]
- 37.Bosl B, Grimminger V, Walter S. The molecular chaperone Hsp104—a molecular machine for protein disaggregation. J Struct Biol. 2006;156:139–148. doi: 10.1016/j.jsb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 38.Parsell DA, Kowal AS, Lindquist S. Saccharomyces cerevisiae Hsp104 protein. J Biol Chem. 1994;269:4480–4487. [PubMed] [Google Scholar]
- 39.Hattendorf DA, Lindquist S. Cooperative kinetics of both Hsp104 ATPase domains and interdomain communication revealed by AAA sensor-1 mutants. EMBO J. 2002;21:12–21. doi: 10.1093/emboj/21.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eriksson AE, Baase WA, Matthews BW. Similar hydrophobic replacements of Leu99 and Phe153 within the core of T4 lysozyme have different structural and thermodynamic consequences. J Mol Biol. 1993;229:747–769. doi: 10.1006/jmbi.1993.1077. [DOI] [PubMed] [Google Scholar]
- 41.Lewandowska A, Matuszewska M, Liberek K. Conformational properties of aggregated polypeptides determine ClpB-dependence in the disaggregation process. J Mol Biol. 2007;371:800–811. doi: 10.1016/j.jmb.2007.05.057. [DOI] [PubMed] [Google Scholar]
- 42.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 43.Ludtke SJ, Jakana J, Song JL, Chuang DT, Chiu W. A 11.5 Å single particle reconstruction of GroEL using EMAN. J Mol Biol. 2001;314:253–262. doi: 10.1006/jmbi.2001.5133. [DOI] [PubMed] [Google Scholar]
- 44.Ludtke SJ, Chen DH, Song JL, Chuang DT, Chiu W. Seeing GroEL at 6 Å resolution by single particle electron cryomicroscopy. Structure. 2004;12:1129–1136. doi: 10.1016/j.str.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 45.Chen DH, Song JL, Chuang DT, Chiu W, Ludtke SJ. An expanded conformation of single-ring GroEL–GroES complex encapsulates an 86 kDa substrate. Structure. 2006;14:1711–1722. doi: 10.1016/j.str.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 46.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.