Abstract
Quantitative trait loci analysis of natural Arabidopsis thaliana accessions is increasingly exploited for gene isolation. However, to date this has mostly revealed deleterious mutations. Among them, a loss-of-function allele identified the root growth regulator BREVIS RADIX (BRX). Here we present evidence that BRX and the paralogous BRX-LIKE (BRXL) genes are under selective constraint in monocotyledons as well as dicotyledons. Unexpectedly, however, whereas none of the Arabidopsis orthologs except AtBRXL1 could complement brx null mutants when expressed constitutively, nearly all monocotyledon BRXLs tested could. Thus, BRXL proteins seem to be more diversified in dicotyledons than in monocotyledons. This functional diversification was correlated with accelerated rates of sequence divergence in the N-terminal regions. Population genetic analyses of 30 haplotypes are suggestive of an adaptive role of AtBRX and AtBRXL1. In two accessions, Lc-0 and Lov-5, seven amino acids are deleted in the variable region between the highly conserved C-terminal, so-called BRX domains. Genotyping of 42 additional accessions also found this deletion in Kz-1, Pu2-7, and Ws-0. In segregating recombinant inbred lines, the Lc-0 allele (AtBRXLc-0) conferred significantly enhanced root growth. Moreover, when constitutively expressed in the same regulatory context, AtBRXLc-0 complemented brx mutants more efficiently than an allele without deletion. The same was observed for AtBRXL1, which compared with AtBRX carries a 13 amino acid deletion that encompasses the deletion found in AtBRXLc-0. Thus, the AtBRXLc-0 allele seems to contribute to natural variation in root growth vigor and provides a rare example of an experimentally confirmed, hyperactive allelic variant.
Keywords: Brachypodium, BREVIS RADIX, adaptation
Natural genetic variation, together with environmental factors, is principally responsible for the observed intraspecific variation in plant and animal morphology. This variation is increasingly recognized as a valuable resource to isolate either novel alleles or previously uncharacterized genes (1). Generally, morphologic variation in multicellular organisms can reflect differences in the extent of cellular growth, localized cell death, cell migrations, or differential growth patterns. In plants, cell walls largely prohibit cell migration and keep cells in their relative positions to each other throughout development. Thus, local acceleration or slowdown of cell proliferation or expansion might have a central role in shaping plant morphology (2–5). Indeed, even very local spatiotemporal constraints on growth can significantly affect organ shape. For instance, developmentally controlled formation of growth-promoting local maxima of the plant hormone auxin along the proximo-distal axis of outgrowing leaves determines leaf serration (6). The timing and amplitude of this phenomenon can even explain the transition from simple to compound leaves (2, 6, 7) and impinges significantly on final organ size (3, 5, 8). In summary, quantitative differences in localized cellular growth and differentiation seem to play a major role in morphologic variation in plants.
Compared with mutant analyses, dissecting the natural basis of growth phenomena is often complicated by the complexity of the associated genetics. Studies that aim to clone quantitative trait loci (QTL) underlying growth rate are limited by genetic marker density, recombinant inbred line (RIL) population size, and the resolution of the growth measurement (9). Moreover, plant organ formation displays remarkable plasticity, and environmental factors often play an important role in trait expression, resulting in low heritability. Combined with multigenic control, this can render identification of the molecular polymorphisms underlying individual QTL practically impossible. By concentrating on growth phenomena that display an easy to measure, developmentally robust, and wide quantitative range, these problems can be minimized. These features apply to elongation growth of plant roots, which is driven by apical meristems and can be treated as a one-dimensional process.
The model plant, Arabidopsis thaliana (Arabidopsis), displays a typical dicotyledonous, allorhiz root system, dominated by a primary root (10). Significant natural variation between isogenized Arabidopsis strains, so-called accessions, has been observed in root growth vigor, and in a few cases the underlying allelic variation could be resolved at the molecular level (11–13). We have isolated a large effect root growth QTL from a cross between the accessions Slavice-0 (Sav-0) and Umkirch-1 (Uk-1), which we named BREVIS RADIX (BRX) (11). The short root phenotype of Uk-1 results from loss of function of the BRX gene, which was identified by map-based cloning. The gene represented the first characterized member of a plant-specific gene family consisting of five paralogs in Arabidopsis, termed BRX and BRX-LIKE 1–4 (BRXL1-4).
BRX family proteins contain four highly conserved domains of initially unknown function; most characteristically, a tandem repeat of 55 amino acids termed the BRX domain, which likely mediates protein–protein interaction (11, 14). Despite the high degree of amino acid similarity between BRX family proteins, molecular and genetic analyses revealed that only BRX is involved in root growth, indicating functional diversification (14). Moreover, BRX is also needed for optimal growth in the radial dimension of the root (15). Finally, physiologic and gene expression analyses are consistent with a role of BRX in mediating crosstalk between the auxin and brassinosteroid plant hormone pathways (16), which seems to involve auxin-responsive plasma membrane to nucleus signaling of BRX protein (17). These results have been mostly achieved by functional analyses involving the Uk-1 allele, which carries an early stop codon and can be considered a null allele (11, 14).
Similar to BRX, the vast majority of loci isolated in Arabidopsis through the natural variation approach to date are often drastic loss-of-function mutations associated with large effect QTL (1). Although these types of mutations have greatly improved our understanding of molecular mechanisms affecting plant traits, they are rarely found to be relevant in nature, because their effects on plant fitness are likely too deleterious and rejected by purifying selection. However, isolation of differentially active, functional alleles remains the elusive goal of studies that aim to identify polymorphisms underlying QTL that are relevant for the evolutionary life history of plants. So far, few convincing, experimentally verified examples of hypo- or hyperactive alleles that might mediate adaptive developmental variation have been reported, such as the amino acid replacements in photoreceptors, which have been shown to differentially affect physiologic responses to light quality and intensity (18–21).
In this study, we present results of a combined functional and molecular evolutionary analysis of BRX family genes in monocotyledons and dicotyledons. We show that evolutionary rates in dicotyledons have accelerated relative to those of monocotyledons, resulting in higher levels of functional diversification. Population genetic data are consistent with a possible adaptive role of BRX alleles. Moreover, we could identify a polymorphism in functional BRX alleles of a few accessions, which seems to confer enhanced primary root growth and might represent local adaptation.
Results and Discussion
Identification of BRX Family Genes in Brachypodium distachyon.
Previously, BRX family genes have been characterized in the dicotyledons Arabidopsis (Arabidopsis thaliana, At) and poplar (Populus tremuloides, Pt), and in the monocotyledon rice (Oryza sativa, Os) (14); they are also present in Arabidopsis lyrata (Al). Each of those genomes contains five paralogous gene family members, which are highly conserved in amino acid sequence and exon structure. An exception is OsBRXL5, which seems to be a retrotransposed pseudogene (14). To describe the molecular evolution and functional constraint on BRX family genes, we sought to identify additional family members from a monocotyledon species roughly equidistant to the dicotyledon At-Pt pair in terms of genetic distance, to conduct within monocotyledon and between monocotyledon and dicotyledon comparisons. The monocotyledon Brachypodium (Brachypodium distachyon, Bd), for which genome sequence data have become available recently, met this criterion (22).
Four bona fide Brachypodium BRX family members were identified, and all of them share the characteristic features observed previously (11, 14). Expression analyses by RT-PCR confirmed that most of the monocotyledon genes were expressed in young seedlings and/or leaves. The exceptions are OsBRXL4 and OsBRXL5, for which no transcript could be detected in repeated attempts. Finally, we could confirm that OsBRXL3 encodes a bona fide, full-length BRX family protein. An alternative OsBRXL3 transcript lacking two exons reported as an expressed sequence tag (14) was not observed in our study.
Phylogenetic Analysis of BRX Family Genes.
The high level of amino acid conservation among BRX family proteins described previously (14) is consistent with the importance of each paralog for plant fitness, and thus a role for purifying selection to maintain sequence functionality. We included all BRX family genes from two monocotyledons and three dicotyledons, except the too divergent OsBRXL5, to conduct a detailed sequence survey to evaluate this notion. We obtained robust phylogenies when the genomic sequences of BRX family genes were analyzed using Bayesian inference and assuming the generalized time reversible evolutionary model (23) (Fig. 1A). The separate grouping of the dicotyledon and monocotyledon paralogs was supported by the phylogenetic analysis. To account for possible ambiguities in ortholog relationships, all pairwise comparisons between genes were considered during subsequent molecular evolutionary analyses.
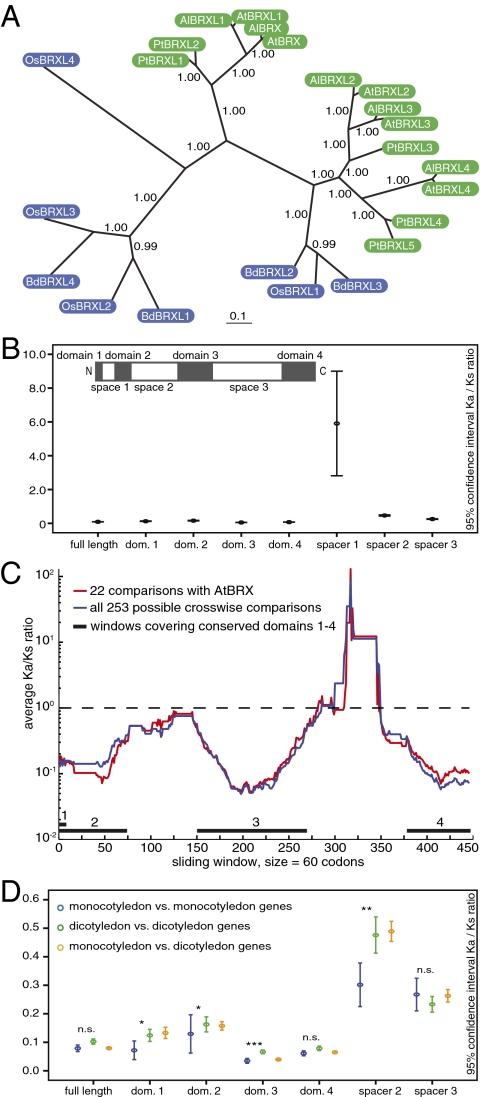
Fig. 1.
Phylogenetic and Ka/Ks analyses of BRX family genes. (A) Phylogenetic tree of BRX family genes from A. thaliana (At), A. lyrata (Al), poplar (Pt), rice (Os), and Brachypodium (Bd). Dicotyledon genes are depicted in green, monocotyledon genes in blue. (B) Ninety-five percent confidence interval Ka/Ks for all pairwise combinations of BRX family proteins, calculated for full-length coding sequence or individual conserved domains or spacer regions (see Inset schematic of BRX family protein structure, drawn to scale for AtBRX). (C) Ka/Ks calculated for pairwise combinations of BRX family proteins using a sliding window of 60 amino acids. Total length of the aligned amino acid sequences was 503. (D) Pairwise Ka/Ks calculated within mono- or dicotyledon proteins, or between the two groups. Significance is indicated for intergroup comparison between Ka/Ks of monocotyledon and dicotyledons. The outlier spacer 1 (Fig. S1) was removed for better viewing of the other ratios. dom., domain; n.s., not significant. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. Full statistics are given in Table S1.
Molecular Evolutionary Analysis of BRX Family Genes.
We estimated the ratio of the rate of nonsynonymous substitutions per nonsynonymous site (Ka) over the rate of synonymous substitutions per synonymous site (Ks), between all monocotyledon and dicotyledon BRX orthologs. We applied the maximum likelihood model of sequence evolution as implemented in PAML software (24). Overall, BRX family sequences display averaged Ka/Ks = 0.089 ± 0.036 (Fig. 1B and Table S1), which is much below the neutral expectation of 1.0. These ratios are indicative of intense selective constraint on nonsynonymous nucleotide sites and are consistent with functional importance of the genes.
The latter conclusion remains valid when Ka/Ks was calculated after the sequences were split into the conserved domains 1–4 and spacer regions 1–3 (11) (Fig. 1B and Table S1). Spacer 1 is an exception, displaying Ka/Ks above the neutral expectation. Domains displayed lower average Ka/Ks (0.050–0.157) than spacer regions (0.251–5.905). To account for stochastic fluctuations due to short sequence (i.e., spacer 1) and loss of resolution due to averaging of Ka/Ks over long sequences, we conducted sliding-window analyses considering 60–90 codons at a time, moving the window one codon at a time (Fig. 1C). These analyses largely supported our qualitative conclusions based on the whole coding region and domain analyses. However, results for spacers were affected. First, spacer 1 displayed a Ka/Ks < 1.0. Second, peaks of Ka/Ks near and above 1.0 were identified within spacer 2 and 3, respectively. Overall, despite differences between domains and spacers (all P < 0.001, two-tailed Mann-Whitney U tests), the estimated Ka/Ks are consistent with selective constraint on the entire region as well as individual domains and spacers of the BRX family proteins. However, within the spacers, short regions experience reduced constraint (spacers 1 and 2), and/or positive selection has acted upon them (spacer 3). Although these results point to the spacer regions as the source for diversification among BRX family genes, analysis of Ka/Ks in dicotyledons when compared with monocotyledons revealed that dicotyledon BRX family genes diverged at significantly different rates in domain 1 (P = 0.029, Mann-Whitney U test, two-tailed), domain 2 (P = 0.039), and domain 3 (P < 0.001), as well as spacer 2 (P = 0.014) (Fig. 1D).
Experimental Verification of AtBRX Activity in Monocotyledon BRX Family Genes.
The evolutionary rate acceleration in dicotyledon BRX family genes is partly consistent with previous experimental analyses of Arabidopsis BRX family proteins that found functional diversification (14). This was demonstrated by transgenic complementation of the Arabidopsis brx mutant short root phenotype through ectopic overexpression of AtBRX-LIKE genes in the root under control of the cauliflower mosaic virus 35S promoter. In these experiments, AtBRXL1 could fully rescue the brx mutant, unlike AtBRXL2 and AtBRXL4.
To determine which monocotyledon genes are functionally equivalent to AtBRX, we used the same approach. We restricted ourselves to genes for which transcripts could be detected (i.e., OsBRXL1-3 and BdBRXL1-4). As previously reported, both AtBRX and AtBRXL1 could restore root growth to wild-type levels, whereas AtBRXL2 and AtBRXL4 could not (Fig. 2A). Moreover, AtBRXL3 also could not complement the brx mutant. These results confirm that it is not only differential expression that is responsible for a lack of redundancy between Arabidopsis BRX family genes, as in the case of AtBRXL1 (17), but mostly differential protein activity.
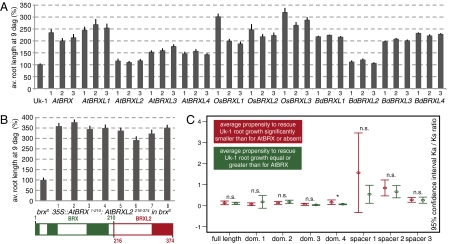
Fig. 2.
Functional analyses of AtBRX homologs in Arabidopsis. (A) Propensity of BRX family genes to complement the Arabidopsis brx null mutant short root phenotype when ectopically overexpressed under control of the 35S promoter in transgenic brx seedlings, scored at 9 days after germination (dag). The three independent transgenic lines for each construct are shown. (B) Same assay as in A, with a transgenic fusion of the N-terminal half of AtBRX and the C-terminal half of AtBRXL2. (C) Ninety-five percent confidence interval Ka/Ks for all pairwise combinations between BRX family proteins that could rescue the brx mutant if expressed constitutively and those that could not, calculated for full-length or individual conserved domains or spacer regions (see Inset schematic in Fig. 1B). dom., domain; n.s., not significant. *P ≤ 0.05. Full statistics are given in Table S1.
Most importantly, we observed that nearly all monocotyledon genes tested complemented the mutant, with the exception of BdBRXL2 (Fig. 2A). This was unexpected, because monocotyledon BRX family genes generally group apart from the dicotyledon homologs in phylogenetic analysis (Fig. 1A) and are more distantly related to AtBRX than the dicotyledon counterparts. In summary, our results demonstrate that most monocotyledon BRX family genes are functional paralogs of the dicotyledon gene AtBRX.
BRX Family Protein Diversification Is Likely Due to Variable N-Terminal Regions.
The Ka/Ks analysis indicates that BRX family genes might have diversified because of variation in the N-terminal region, including domains 1–3 and spacers 1 and 2. To examine whether the molecular evolutionary analysis and the results of complementation assays correspond, we generated a transgene encoding a fusion between the N terminus of AtBRX, up to the end of domain 3, and the C terminus of AtBRXL2, including spacer 3 and domain 4. This hybrid protein was able to rescue the brx mutant phenotype (Fig. 2B). Interestingly, domain 3 is largely identical in AtBRX and AtBRXL2, and similar to domains 1 and 2, which are highly conserved across all BRX family proteins, most amino acid changes are conservative. Importantly, considering all BRX family proteins for which functional data are available, there is no single substitution in the conserved domains that would exclusively occur within proteins that were able to complement the brx mutant when ectopically overexpressed as compared with those that were not. Finally, Ka/Ks analysis comparing those two groups of proteins indicates that the conserved domains and spacer 3 are equally well evolutionarily constrained (Ka/Ks similarly low) in both groups (Mann-Whitney test, P > 0.05). A higher evolutionary rate in the noncomplementing group was observed exclusively for spacer 1 and, to some degree, spacer 2 (Fig. 2C).
Collectively, the combined analyses of molecular evolutionary rates and protein function are consistent with the idea that variation in N-terminal regions has contributed to functional diversification among BRX family proteins. However, our ability to pinpoint functional changes even further is limited by lack of information on BRX family protein structure, and the considerable number of nucleotide substitutions between orthologs in the spacers, such that site-directed mutagenesis analysis of AtBRX residues combined with functional analysis will require long-term, comprehensive efforts.
Haplotyping of AtBRX and AtBRXL1 Is Consistent with an Adaptive Role of AtBRX Alleles.
Evaluation of intraspecific genetic polymorphisms is a complementary approach to narrow down functionally relevant amino acid residues. We concentrated our analyses on Arabidopsis, for which isogenized accessions are easily available, and the two genes for which positive functional data have been generated, AtBRX and AtBRXL1. We used AlBRX and AlBRXL1 as outgroup sequences to categorize SNPs as either ancestral or derived.
To determine the amount of genetic variation of AtBRX, we amplified the genomic sequence that encompasses the coding sequence (i.e., exon 3 through 7) (11). Corresponding DNA fragments were obtained for 20 worldwide accessions, as well as for 9 accessions from Umkirch, Germany, around the collection site of the original brx loss-of-function line, Uk-1 (25). It is noteworthy that the premature stop codon in the Uk-1 allele (AtbrxUk-1) is found in all corresponding stock center material available [i.e., in all individuals (>20 per seed pool) genotyped both from the bulk as well as the single seed descent lines]. Thus, the loss-of-function mutation was likely present in the original isolate, and reproductive isolation of Uk-1 possibly contributed to its fixation (26). In total, 1,032 bp of coding sequence and 1,215 bp of intron sequence as referred to the reference accession, Col-0, were surveyed.
The haplotyping revealed a low level of variation in AtBRX, with 91 variable nucleotide positions observed across the entire sample, 40 of them within the coding sequence (Table S2). None of the SNPs, except the known one in Uk-1, leads to a premature stop codon. In addition, eight indels were identified, most of them (seven) in introns. Interestingly, a recently collected isolate from Umkirch, UkD-1, shares all of the unique features of the Uk-1 haplotype, except the premature stop codon, confirming the Uk-1 geographic origin.
Only 10 of the 43 exonic SNPs result in amino acid polymorphisms when compared with the Col-0 reference allele (Fig. S2A). Among them, seven lead to nonconservative amino acid replacements, but only two of them (I151T in AtBRXLz-0 and E312G in AtBRXSpr1-6) map within the conserved domains. Assuming that loss of AtBRX activity is not masked by redundancy with AtBRXL1 in any of the accessions investigated, the observed amino acid polymorphisms should not compromise protein function, because all lines (except Uk-1) displayed normal root growth vigor.
Overall, in the accessions representing a worldwide sample per nucleotide diversity for AtBRX was θW = 0.0107 and expected site heterozygosity π = 0.0060. Gene flow between Uk accessions and the remaining worldwide accessions was high, with zero fixed differences between populations and all 23 polymorphisms shared, amounting to low levels of genetic differentiation among populations (FST = 0.043). Tajima's D was negative, albeit not significant (−1.31), which is consistent with an expanding population of Arabidopsis across the globe and/or selection. Fay and Wu's normalized Hn statistic was −2.84 (P = 0.01), which is consistent with a selective sweep due to positive selection on sites within the gene or nearby sites. Fu and Li's D* was 2.30 (P < 0.02), which is consistent with population structure following an island model. Only Fu and Li's D* was significant in the Umkirch site (1.90; P < 0.02). Overall, these composite population genetic measures are consistent with complex demographic scenarios affecting the worldwide accessions of Arabidopsis, yet a combination of negative Tajima's D (albeit nonsignificant) and significant Hn indicates that selection might have acted on alleles at the AtBRX locus.
For comparison, we determined the AtBRXL1 haplotypes from a sample of 17 accessions. Within 993 bp of coding sequence and 1,197 bp of intron sequence, we found only 10 polymorphisms, all of them SNPs except one single base pair deletion. Half of them, including the indel, fall into introns, whereas of the five exonic SNPs, three are silent, and one each lead to a conservative and nonconservative amino acid replacement, respectively. In the corresponding accession sample of AtBRX, 73 variable nucleotides and all eight indels are found, suggesting that genetic variation in AtBRXL1 is lower as compared with AtBRX (Fig. S2B).
Genetic variation in AtBRXL1 was θW = 0.0009 and π = 0.0009. Average genetic differentiation between the Uk and worldwide accessions was FST = 0.163. Although Fay and Wu's Hn statistic was consistent with selective sweep (−2.73; P = 0.021) this should be viewed with caution, given that Tajima's D was near zero and population structure (positive Fu and Li's D*, 1.37, P < 0.02) can lead to bias in the H statistic. None of the statistics were significant for the Umkirch sample.
In summary, the population genetic data to infer ecologic/fitness significance of developmental root growth parameters mediated by AtBRX alleles are suggestive of the presence and spread of adaptive alleles. This should motivate further detailed studies of their ecologic significance in greenhouse and/or common garden experiments. Furthermore, future population genetic studies would need to exclude the possibility of selection at nearby genes. We suggest that such efforts are merited, because the concomitant population genetic dynamics of a paralogous gene pair underlying a major QTL of ecologic relevance has rarely been documented in detail.
AtBRX Alleles from Two Northern European Accessions Carry a Seven Amino Acid Deletion in Spacer 3.
Among the eight indels identified in the AtBRX haplotypes, the only exonic deletion, observed in AtBRXLov-5 and AtBRXLc-0, is located in exon 6 and takes out 21 bp, resulting in the in-frame deletion of a stretch of seven amino acid residues in spacer 3. The AtBRXLov-5 and AtBRXLc-0 haplotypes are distinct from the bulk of other accessions during phylogenetic analysis (Fig. S2B). RT-PCR permitted amplification and sequencing of AtBRX cDNA from Lc-0, confirming that the mRNA is correctly spliced and gives rise to a transcript that should indeed result in the synthesis of variant AtBRX protein. Interestingly, in a screen for natural variation in primary root growth vigor, Lc-0 was the accession with the strongest root growth (11), suggesting that this AtBRX variant is functional. This is further supported by the analysis of the AtBRXL1 sequence, which compared with AtBRX carries an even larger deletion of 13 amino acids in spacer 3, but still can complement the brx mutant when ectopically overexpressed (see above). Notably, this deletion encompasses the seven amino acids deleted in AtBRXLov-5 and AtBRXLc-0 (Fig. 3A). In summary, these observations suggest that the AtBRXLov-5 and AtBRXLc-0 proteins are functional.
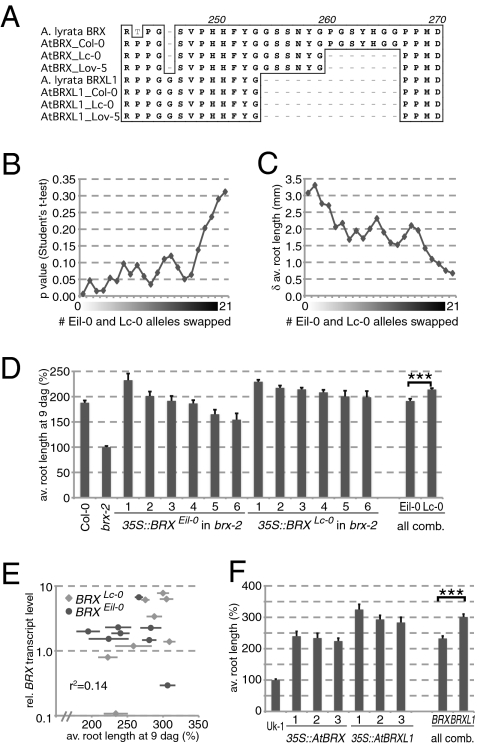
Fig. 3.
Quantitative functional analyses of the AtBRXLc-0 allele. (A) Alignment of the homologous spacer 3 regions of AlBRX, AlBRXL1, AtBRX, and AtBRXL1, the latter including the Col-0, Eil-0, and Lc-0 alleles. (B) Analysis of recombinant inbred lines derived from an Eil-0 × Lc-0 cross. Significance of average root length difference between 49 lines that carry the Lc-0 allele of AtBRX vs. 57 lines that carry the Eil-0 allele (at zero on x axis), and increasing permutation by swapping lines between the two groups. (C) As in B, for absolute difference in root length. (D) Average propensity of the AtBRXEil-0 and AtBRXLc-0 alleles to complement the Arabidopsis brx-2 null mutant short root phenotype when expressed under control of the 35S promoter in transgenic brx seedlings, scored at 9 dag. The six best rescuing independent transgenic lines for each construct are shown, as well as all data combined. (E) Correlation graph of BRX expression level (log) vs. root length (percentage), determined in a replicate experiment similar to D, with all eight independent transgenic lines. BRX mRNA levels were determined by quantitative PCR, expressed as relative expression level as compared with the housekeeping gene, EF1 (15). For reference, expression level in Col-0 wild-type control was 0.0021 ± 0.0010. (F) As in D, for AtBRX and AtBRXL1 expressed in the brx null mutant background, Uk-1, three independent transgenic lines each.
The deletion could also be identified by a length polymorphism PCR genotyping. In a survey of 42 additional accessions (Table S3), the same deletion was thus found in Kz-1, Pu2-7, and Ws-0 (confirmed by sequencing). Thus, the deletion has been retained in ca. 7% of accessions examined. Combined with the above finding of positive selection on spacer 3, this could mean that these alleles might be adaptive. Finally, the deletion is not present in the outgroup BRX homolog of A. lyrata (Fig. 3A), suggesting that it is derived.
AtBRXLc-0 Allele Is Associated with Enhanced Primary Root Growth Vigor in a RIL Population.
To determine whether the observed deletion in AtBRXLc-0 protein in any way affects its activity quantitatively, we took advantage of a previously established RIL population derived from a cross between the Lc-0 and Eil-0 accessions (27). Compared with the AtBRXCol-0 allele, AtBRXEil-0 only carries very few polymorphisms (Table S2). It also does not carry the seven amino acid deletion observed in AtBRXLc-0, allowing us to easily score the AtBRX genotype of the RILs. In parallel, their root growth was assayed in tissue culture, for ≥18 seedlings per line grown in two independent plate sets (Table S4).
Comparison of the group of 49 RILs assayed that carry the Lc-0 allele vs. the 57 RILs that carry the Eil-0 allele revealed a statistically significant difference in average primary root length for both plate sets and the combined data (t test, P < 0.006) (Fig. 3B), suggesting that AtBRXLc-0 is associated with enhanced root growth. This difference accounted for ca. 3 mm or 10% of overall root growth (Fig. 3C). Increasing permutation of the two RIL groups resulted in less, and less significant, average root length difference (Fig. 3 B and C), confirming the influence of the differential AtBRX alleles on this trait. Notably, the association of AtBRX with root growth vigor seems not to be due to linkage to other root growth QTL segregating in this RIL population, because linkage disequilibrium decayed rapidly around the AtBRX locus, and linkage disequilibrium toward other, more distant loci was not statistically supported.
AtBRXLc-0 Is a Differentially Active AtBRX Variant.
However, the effect of AtBRXLc-0 could still be due to the action of a very tightly linked QTL. Moreover, the RIL approach did not allow us to distinguish whether the enhancement of root growth by AtBRXLc-0 was due to protein polymorphisms or regulatory influences. We therefore sought to remove the alleles from their native regulatory context and monitor their activity in an unrelated, isogenized genetic background. To this end, we introduced the coding sequences of AtBRXLc-0 and AtBRXEil-0, expressed under control of the 35S promoter and the same 5′ and 3′ UTRs, into a brx loss-of-function mutant in the Col-0 background (28). We then assayed their propensity to complement the root phenotype in a set of independent transgenic lines.
Both AtBRXEil-0 and AtBRXLc-0 complemented the brx phenotype. Typically, some variation in the extent of rescue was observed between individual lines. This variation notwithstanding, across all independent transgenic lines, complementation by AtBRXLc-0 was generally more efficient than complementation by AtBRXEil-0, suggesting overall weaker activity of the Eil-0 allele (Fig. 3D). Altogether, the average root length of brx seedlings complemented with the Eil-0 variant was significantly different from the average root length of brx seedlings complemented with the Lc-0 variant (t test, P < 1.4e-6). Again, the AtBRXLc-0 allele accounted for a ca. 10% increase in root length.
The differential activity of AtBRXEil-0 and AtBRXLc-0 could result from different activity or stability of the encoded proteins, or different transcript stability or translation efficiency. The latter possibilities appear unlikely, however, because transcript levels in the transgenics, as determined by qPCR, were typically 2 to 3 orders of magnitude higher than the native AtBRX expression level (11, 14) (Fig. 3E). Moreover, a replicate experiment including all eight expressing transgenic lines obtained for each construct confirmed the better rescue by lines complemented by AtBRXLc-0, and although they exhibited on average higher expression levels, this difference was not significant (t test, P = 0.32), and there was little correlation between transgene expression level and extent of rescue (r2 = 0.14) (Fig. 3E).
The consequent notion that the hyperactivity of the AtBRXLc-0 allele results from the variant protein is also supported by complementation experiments that compared the Col-0 alleles of AtBRX and AtBRXL1. We had previously noticed a tendency for AtBRXL1 to more efficiently complement brx than AtBRX (14). This effect could, however, potentially be obscured by the segregation of a genetic modifier, because the complemented brxS line was obtained from an introgression of the AtbrxUk-1 allele (11). Introduction of 35S-driven AtBRX and AtBRXL1 into the isogenic Uk-1 brx mutant background confirmed elevated capacity of AtBRXL1 to compensate the short root phenotype, however (Fig. 3F). The overall difference in root growth vigor between AtBRX- and AtBRXL1-complemented lines was highly significant (t test, P < 8e-8), and enhancement of root growth by AtBRXL1 was again in the range of 10%.
Beyond the seven amino acid deletion, the Lc-0 protein has two amino acid replacements in spacer 2 as compared with the Eil-0 variant (Fig. S2A). Both residues are polymorphic among BRX family proteins, however, and not conserved between AtBRX and AtBRXL1. Thus, it seems possible that the overlapping amino acid deletions in spacer 3 are responsible for the higher activity of AtBRXLc-0 and AtBRXL1. However, because the two amino acid replacements are so far always found in conjunction with the seven amino acid deletion in all other accessions investigated, a contribution of these two amino acid polymorphisms to the observed allele hyperactivity cannot be strictly ruled out at this point.
Variation Among Functional Alleles of AtBRX Contributes to Natural Variation in Root Growth Vigor.
In summary, our data suggest that BRX family genes in general, and AtBRX and AtBRXL1 in particular, are under considerable evolutionary constraint that has maintained protein function over timescales as long as the split between monocotyledons and dicotyledons. However, diversification in sequence and function of BRX family genes can be observed and seems to have mainly occurred in dicotyledons. We showed for the N-terminal regions of the respective proteins that diversification in sequence is correlated with this sub- or neofunctionalization. However, further detailed analyses at the resolution of individual domains and spacers are required. The low intraspecific variability of AtBRX and AtBRXL1 notwithstanding, hyperactive AtBRX alleles seem to exist at medium frequency and contribute to natural variation in root growth vigor of Arabidopsis accessions. Genetic polymorphism data for the worldwide sample are partly consistent with the fitness relevance of alleles at these loci. Whether the hyperactive AtBRX alleles identified reflect a microevolutionary adaptation remains to be determined. Establishing their adaptive role in nature in more detail is a promising subject for future ecology and evolution studies.
Materials and Methods
Plant Materials.
Arabidopsis accessions and brx loss-of-function lines have been described previously (11, 25, 26). Transgenic plants were generated according to standard procedures (11, 29). Only lines expressing the transgene (routinely monitored by RT-PCR) were included in phenotypic analyses.
Molecular Biology.
Molecular biology procedures were carried out according to standard procedures (15).
Phylogenetics, Molecular Evolution, and Population Genetics.
Multiple sequence alignments of genomic or protein sequences were conducted in MUSCLE 3.7 with default settings (30). Phylogenetic trees of genomic and protein sequences were estimated using Bayesian inference (BI) using the MrBayes 3.1.2 program (23). Appropriate substitution models were selected using Modeltest ver. 3.7 (31). The haplotype sequences of AtBRX and AtBRXL1 were aligned in ClustalW 2.0.9, and their BI phylogeny was estimated according to the Hasegawa-Kishino-Yano (Nst = 2) evolutionary model. Estimators of polymorphism were calculated as two sequence groups (worldwide and Umkirch) in SITES software (32). Ka/Ks were estimated with the maximum likelihood model of sequence evolution as implemented in PAML software (24). A detailed description of materials and methods is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank A.-M. Amiguet-Vercher, C. Wyser, T. Lefevre, and S. Plantegenet for technical assistance. This work was supported by Swiss National Science Foundation Grant 3100A0-107631, a Wray-Todd graduate student fellowship (to S.L.), and the University of Lausanne (J.B.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913207107/DCSupplemental.
References
- 1.Alonso-Blanco C, et al. What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell. 2009;21:1877–1896. doi: 10.1105/tpc.109.068114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barkoulas M, Galinha C, Grigg SP, Tsiantis M. From genes to shape: regulatory interactions in leaf development. Curr Opin Plant Biol. 2007;10:660–666. doi: 10.1016/j.pbi.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 3.Bensmihen S, et al. Mutational spaces for leaf shape and size. HFSP J. 2008;2:110–120. doi: 10.2976/1.2836738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mündermann L, Erasmus Y, Lane B, Coen E, Prusinkiewicz P. Quantitative modeling of Arabidopsis development. Plant Physiol. 2005;139:960–968. doi: 10.1104/pp.105.060483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rolland-Lagan AG, Bangham JA, Coen E. Growth dynamics underlying petal shape and asymmetry. Nature. 2003;422:161–163. doi: 10.1038/nature01443. [DOI] [PubMed] [Google Scholar]
- 6.Barkoulas M, Hay A, Kougioumoutzi E, Tsiantis M. A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat Genet. 2008;40:1136–1141. doi: 10.1038/ng.189. [DOI] [PubMed] [Google Scholar]
- 7.Hay A, Tsiantis M. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat Genet. 2006;38:942–947. doi: 10.1038/ng1835. [DOI] [PubMed] [Google Scholar]
- 8.Nath U, Crawford BC, Carpenter R, Coen E. Genetic control of surface curvature. Science. 2003;299:1404–1407. doi: 10.1126/science.1079354. [DOI] [PubMed] [Google Scholar]
- 9.Alonso-Blanco C, Koornneef M, van Ooijen JW. QTL analysis. Methods Mol Biol. 2006;323:79–99. doi: 10.1385/1-59745-003-0:79. [DOI] [PubMed] [Google Scholar]
- 10.Osmont KS, Sibout R, Hardtke CS. Hidden branches: Developments in root system architecture. Annu Rev Plant Biol. 2007;58:93–113. doi: 10.1146/annurev.arplant.58.032806.104006. [DOI] [PubMed] [Google Scholar]
- 11.Mouchel CF, Briggs GC, Hardtke CS. Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes Dev. 2004;18:700–714. doi: 10.1101/gad.1187704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sergeeva LI, et al. Vacuolar invertase regulates elongation of Arabidopsis thaliana roots as revealed by QTL and mutant analysis. Proc Natl Acad Sci USA. 2006;103:2994–2999. doi: 10.1073/pnas.0511015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Svistoonoff S, et al. Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet. 2007;39:792–796. doi: 10.1038/ng2041. [DOI] [PubMed] [Google Scholar]
- 14.Briggs GC, Mouchel CF, Hardtke CS. Characterization of the plant-specific BREVIS RADIX gene family reveals limited genetic redundancy despite high sequence conservation. Plant Physiol. 2006;140:1306–1316. doi: 10.1104/pp.105.075382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sibout R, Plantegenet S, Hardtke CS. Flowering as a condition for xylem expansion in Arabidopsis hypocotyl and root. Curr Biol. 2008;18:458–463. doi: 10.1016/j.cub.2008.02.070. [DOI] [PubMed] [Google Scholar]
- 16.Mouchel CF, Osmont KS, Hardtke CS. BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature. 2006;443:458–461. doi: 10.1038/nature05130. [DOI] [PubMed] [Google Scholar]
- 17.Scacchi E, et al. Dynamic, auxin-responsive plasma membrane-to-nucleus movement of Arabidopsis BRX. Development. 2009;136:2059–2067. doi: 10.1242/dev.035444. [DOI] [PubMed] [Google Scholar]
- 18.Balasubramanian S, et al. The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nat Genet. 2006;38:711–715. doi: 10.1038/ng1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El-Din El-Assal S, Alonso-Blanco C, Peeters AJ, Raz V, Koornneef M. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat Genet. 2001;29:435–440. doi: 10.1038/ng767. [DOI] [PubMed] [Google Scholar]
- 20.Filiault DL, et al. Amino acid polymorphisms in Arabidopsis phytochrome B cause differential responses to light. Proc Natl Acad Sci USA. 2008;105:3157–3162. doi: 10.1073/pnas.0712174105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maloof JN, et al. Natural variation in light sensitivity of Arabidopsis. Nat Genet. 2001;29:441–446. doi: 10.1038/ng777. [DOI] [PubMed] [Google Scholar]
- 22.Draper J, et al. Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol. 2001;127:1539–1555. [PMC free article] [PubMed] [Google Scholar]
- 23.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 24.Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- 25.Shindo C, Bernasconi G, Hardtke CS. Intraspecific competition reveals conditional fitness effects of single gene polymorphism at the Arabidopsis root growth regulator BRX. New Phytol. 2008;180:71–80. doi: 10.1111/j.1469-8137.2008.02553.x. [DOI] [PubMed] [Google Scholar]
- 26.Bomblies K, et al. Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 2007;5:e236. doi: 10.1371/journal.pbio.0050236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plantegenet S, et al. Comprehensive analysis of Arabidopsis expression level polymorphisms with simple inheritance. Mol Syst Biol. 2009;5:242. doi: 10.1038/msb.2008.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodrigues A, et al. The short-rooted phenotype of the brevis radix mutant partly reflects root abscisic acid hypersensitivity. Plant Physiol. 2009;149:1917–1928. doi: 10.1104/pp.108.133819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Posada D, Buckley TR. Model selection and model averaging in phylogenetics: Advantages of akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- 32.Hey J, Wakeley J. A coalescent estimator of the population recombination rate. Genetics. 1997;145:833–846. doi: 10.1093/genetics/145.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Berloo R. GGT 2.0: Versatile software for visualization and analysis of genetic data. J Hered. 2008;99:232–236. doi: 10.1093/jhered/esm109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.