Abstract
Exposure of eukaryotic cells to UV light induces a checkpoint response that delays cell-cycle progression after cells enter S phase. It has been hypothesized that this checkpoint response provides time for repair by signaling the presence of structures generated when the replication fork encounters UV-induced DNA damage. To gain insight into the nature of the signaling structures, we used time-lapse microscopy to determine the effects of deficiencies in translesion DNA polymerases on the checkpoint response of the fission yeast Schizosaccharomyces pombe. We found that disruption of the genes encoding translesion DNA polymerases Polκ and Polη significantly prolonged the checkpoint response, indicating that the substrates of these enzymes are signals for checkpoint activation. Surprisingly, we found no evidence that the translesion polymerases Rev1 and Polζ repair structures that are recognized by the checkpoint despite their role in maintaining viability after UV irradiation. Quantitative flow cytometry revealed that cells lacking translesion polymerases replicate UV-damaged DNA at the same rate at WT cells, indicating that the enhanced checkpoint response of cells lacking Polκ and Polη is not the result of stalled replication forks. These observations support a model in which postreplication DNA gaps with unrepaired UV lesions in the template strand act both as substrates for translesion polymerases and as signals for checkpoint activation.
Keywords: DNA damage checkpoint, Polη, Polκ, Polζ, Rev1
DNA damage produced by the UV component of sunlight presents a constant challenge for the survival of organisms on the earth's surface. In response to this challenge, eukaryotic cells have evolved excision repair processes that remove the damage, postreplication repair processes that facilitate the replication of damaged DNA, and checkpoint mechanisms that delay cell-cycle progression to make time for repair (1–3). Because the genes that mediate these processes are conserved in eukaryotes, model systems such as yeast have provided valuable insights that are applicable to the DNA damage responses of higher organisms (4). In previous studies, we used time-lapse microscopy to measure the cell-cycle dynamics of UV-irradiated fission yeast cells and observed two distinct DNA damage checkpoint responses: the previously characterized G2/M checkpoint response that delays cell division when cells are irradiated in G2 phase and a postreplication checkpoint response that delays division when cells irradiated in any stage of the cell cycle carry lesions into S phase (5). Only the latter response occurs after moderate UV doses comparable to sunlight exposure, so it is likely to be particularly important in nature (3). The postreplication checkpoint response is activated following the encounter of replication forks with UV-induced DNA damage and requires the activity of checkpoint proteins that recognize structures containing transitions between double-stranded and single-stranded DNA (3, 6–11). The nature and origin of the signaling structures that determine the Schizosaccharomyces pombe postreplication checkpoint response are currently unknown.
The DNA lesions produced with the greatest efficiency by UV light are cyclobutane pyrimidine dimers (CPDs) and (6–4) photoproducts (12). Several distinct cellular mechanisms repair structures created by the replication of DNA containing these lesions. The repair mechanisms have been best characterized in budding yeast and include translesion DNA synthesis (TLS), homology-directed repair (HR), and Rad5-dependent error-free postreplication repair (13–16). It is not known which of these processes, if any, repairs structures that elicit the fission yeast postreplication checkpoint response. The S. pombe genome encodes four specialized translesion polymerases that are potentially involved in TLS through UV-damaged DNA: Polζ, Rev1, Polκ, and Polη (17). All four of these polymerases are conserved in most eukaryotes, including humans. Polη can accurately copy template DNA containing UV-induced CPDs (18, 19). Polκ is required for full UV resistance in mammalian cells, but the lesions that it bypasses are unknown (17, 20, 21). The exact roles of Polζ and Rev1 in the UV response are enigmatic, but these polymerases are required for UV resistance and UV-induced mutagenesis in a number of species (17, 22). Human cells have an additional translesion polymerase, Polι, whose mechanism of action is under investigation (23).
It has been hypothesized that translesion polymerases function at the replication fork, where they may be exchanged dynamically with the replicative polymerases in a polymerase-switch mechanism (2, 24). Alternatively, translesion polymerases may function to fill in gaps containing lesions that are left behind by replication forks (25). These two models posit different DNA intermediates containing single-strand to double-strand transitions that could elicit a checkpoint response. In the former case, the transitions would be associated with stalled replication forks, whereas in the latter case, they would be associated with postreplication gaps.
To gain insight into the structures recognized by the DNA damage checkpoint pathway, we analyzed the UV-induced postreplication checkpoint response of TLS mutants using time-lapse microscopy. Our data indicate that defects in Polκ and Polη cause enhanced checkpoint delays, suggesting that these translesion polymerases are involved in the repair of structures that signal to the checkpoint when replication forks encounter UV lesions. To determine whether these structures are stalled replication forks, we measured the kinetics of DNA replication after UV irradiation using quantitative flow cytometry. We found that the elimination of translesion polymerases did not detectably reduce the rate of replication through dimeric photoproducts over a wide range of lesion densities. These observations suggest a model in which the DNA damage checkpoint monitors the presence of postreplication gaps to prevent cell-cycle progression until the gaps are removed by TLS and perhaps other mechanisms.
Results
Disruption of Polη or Polκ Activity Prolongs Checkpoint-Dependent Cell-Cycle Delays in Response to UV Damage.
To determine whether translesion polymerases function to remove UV-induced structures that signal to the DNA damage checkpoint, we analyzed TLS-defective cells by time-lapse microscopy. The fission yeast gene eso1 encodes a fusion protein with the translesion polymerase Polη at the N terminus and the essential sister chromatid cohesion protein Ctf7 at the C terminus (26). Our attempts to delete the entire Polη subregion of eso1 yielded mutants with highly abnormal cell cycles and high rates of spontaneous cell death, possibly because the deletion affected the activity of Ctf7. Thus, we eliminated Polη activity by introducing a missense mutation that changed the highly conserved aspartate 147 to asparagine. Aspartate 147 is critical for coordinating magnesium at the active site of the enzyme, and the structurally analogous D882N mutation in Escherichia coli DNA polymerase I eliminates its catalytic activity and reduces its affinity for DNA (27). An analogous D155A mutant in budding yeast Polη has the UV sensitivity of a null mutant (28).
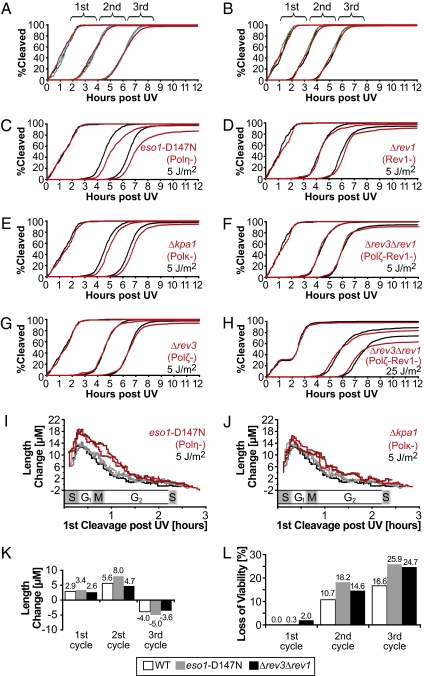
The eso1-D147N mutant and cells lacking Polζ, Rev1, or Polκ were observed by time-lapse microscopy. For each strain, an asynchronous population of 300 cells was followed for three divisions after mock irradiation or irradiation with 5 J/m2 of UV. In all cases, the kinetics of division in the absence of irradiation were unaffected by the lack of TLS activity (Fig. 1 A and B). Consistent with our previous studies, WT S. pombe cells exposed to 5 J/m2 delayed cell division during the second cycle after irradiation. This reflects the fact that the DNA damage checkpoint response at moderate UV doses is entirely dependent on entry of the cells into S phase, which in S. pombe occurs just before cell division (5). In each of the mutant strains analyzed, the kinetics of the first division after irradiation were normal (Fig. 1 C–G); however, after 5 J/m2 of UV, the second cell cycle was delayed in cells lacking Polη and Polκ relative to WT controls (population averages of 43 min and 13 min, respectively, above the WT delay of 59 min). Cells lacking Polη and Polκ also showed greater length increases compared with WT cells during the second cycle after irradiation, indicating that the enhanced delay was due not to the direct inhibition of growth by UV damage, but rather to a checkpoint-mediated inhibition of cell division (Fig. 1 I and J). Because the average duration of cell-cycle phases is known for S. pombe, the position of a given cell in the cell cycle can be deduced from its time of division (5). It is apparent from this analysis that cells lacking Polη and Polκ had enhanced checkpoint responses in cells irradiated at the beginning of S phase (Fig. 1 I and J), consistent with a role for these polymerases in removing signaling structures generated during this phase of the cell cycle. The mutant phenotype of eso1-D147N cells was less pronounced in cells irradiated in G2 phase, which was expected because these cells had more time before S phase to excise the CPDs that Polη normally bypasses (Fig. 1I). Because the phenotype of cells lacking Polκ is more subtle, it is unclear if the lesions bypassed by Polκ are subject to the same time-dependent repair (Fig. 1J). These results indicate that after UV lesions are carried into S phase, substrates normally repaired by Polη and Polκ trigger a checkpoint response that delays cell-cycle progression.
Fig. 1.
Cells lacking Polκ and Polη exhibit enhanced UV-induced delays during the second cell cycle after irradiation, but cells lacking Polζ and Rev1 have no phenotype until the third cycle. Asynchronous populations of mutant and WT S. pombe cells were analyzed by time-lapse microscopy. Some of the strains were ura4− (Left), and others were ura4+ (Right). Because the former have doubling times 15 min longer than the latter, two sets of WT controls were analyzed. (A) Division kinetics after mock irradiation. Black, WT (smt0, ura4-D18, leu1-32); green, eso1-D147N::kanMX; blue, kpa1::kanMX; red, rev3::kanMX. (B) Division kinetics after mock irradiation. Black, WT (smt0, leu1-32); green, rev1::ura4+; red, TLS− (lacking all four translesion polymerases). (C–H) Comparison of the cleavage kinetics of TLS mutants (red) with the appropriate WT control (black) irradiated at the same UV dose. (I and J) Cell length changes induced by UV irradiation of WT controls (gray) and TLS mutants (red). Three independent experiments were performed with WT controls, and two experiments were performed for each TLS mutant. The length change, calculated as the length of a cell just before its second cleavage after UV exposure minus the same measurement performed on its mother, is plotted as a function of first cleavage time post-UV (n = 600). To smooth the data, the running median value of 40 cells is plotted. The approximate cell-cycle stage at the time of irradiation is indicated on the abscissa. (K and L) Length change and loss of viability of WT and mutant cells. The indicated strains were irradiated with 25 J/m2, and the population averages of length change and loss of viability were determined (n = 300, 600, and 1,200 for the first, second, and third cycles, respectively). Cells that degraded or failed to divide during the ~18-h duration of the time-lapse series were scored as inviable.
Cells Lacking Polζ or Rev1 Do Not Have Extended Checkpoint-Mediated Cell-Cycle Delays.
Surprisingly, the division kinetics of cells lacking Polζ and Rev1 were identical to those of WT cells after exposure to 5 J/m2 of UV (Fig. 1 D, G, and F). These polymerases have an established role in bypassing (6–4) photoproducts (21, 29), a class of pyrimidine dimers expected to be abundant after a dose of 5 J/m2. To determine whether the phenotype was too small to be detected at 5 J/m2, or whether these polymerases function only when other repair mechanisms have been exhausted, the UV dose was raised to 25 J/m2 (75% viability in WT cells). Even at this dose, the checkpoint response of cells lacking Polζ and Rev1 was the same as that in WT (Fig. 1 H and K). In contrast, the loss of viability during the third cycle after irradiation in these cells was comparable to that of the eso1-D147N mutant (Fig. 1L). These results indicate that Polζ and Rev1 do not repair structures recognized by the postreplication checkpoint system and suggest that the initial bypass of (6–4) photoproducts may be independent of these polymerases.
Translesion Polymerases Do Not Facilitate Replication Fork Movement Through CPDs or (6–4) Photoproducts.
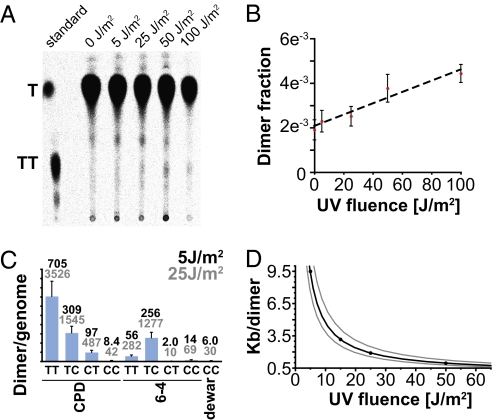
The enhanced checkpoint response in cells lacking Polη and Polκ could result from the accumulation of stalled replication forks that are unable to bypass lesions via a putative polymerase-switch mechanism or, alternatively, from the accumulation of postreplication gaps that are repaired more slowly in the absence of these polymerases (17). A strong prediction of the stalled fork model is that on introduction of a sufficient number of UV lesions, cells lacking translesion polymerases should have a slower rate of bulk DNA synthesis than WT cells. To test this prediction, we first measured the CPD yield in fission yeast cells irradiated on the surface of agar plates under conditions identical to our time-lapse and flow cytometry experiments. Thymine monomers and dimers, generated by acid hydrolysis of DNA from cells labeled with 14C-thymidine, were resolved by polyethylenimine (PEI)-cellulose TLC (Fig. 2A). The fraction of CPDs in the DNA was calculated for multiple UV doses in three independent experiments, and linear regression analysis was performed to estimate the efficiency of dimer production (Fig. 2B). The data indicate that 224 ± 52 CPDs are generated per duplex haploid genome per J/m2 of UV in our experiments. In addition to CPDs, UV exposure is known to create (6–4) photoproducts and Dewar photoproducts that would not be detected by our assay; however, the precise mole ratios of several photoproducts generated in calf-thymus DNA have been determined using calibrated HPLC (30). We adjusted these ratios for the guanine-cytosine content of fission yeast DNA to calculate the expected frequencies of the various UV photoproducts (Fig. 2C). When all known dimeric photoproducts are taken into account, the total yield under the conditions of our experiments is 291 ± 67 lesions per genome per J/m2. Therefore, a dose of 25 J/m2 introduces 10 or more lesions per replicon in >75% of the S. pombe genome; the remaining portion of the genome is duplicated by small replicons with fewer lesions per replicon (see SI Materials and Methods for detailed calculations). Because the half-life of dimeric photoproducts in S. pombe cells is ~20 min (31, 32), the majority of replication forks will encounter a UV lesion in cells irradiated during S phase and in cells in which several repair half-lives elapse before S phase. Mutations that result in the replication fork stalling at dimeric photoproducts should exhibit a slowing or cessation of bulk DNA synthesis that could be easily detected in a substantial portion of an asynchronous population after 25 J/m2.
Fig. 2.
Pyrimidine dimer yields in UV-irradiated S. pombe cells. (A) 14C-labeled thymine monomers and dimers were separated by PEI-cellulose TLC and visualized with a phosphorimager. The positions of thymine and cis-syn TT dimers are indicated by the standard in the leftmost lane. Note that fewer radiolabeled cells were used for the sample irradiated at 100 J/m2 to conserve material. (B) The fraction of radioactivity found in the dimer region is plotted as a function of UV dose. Values are the average of three samples, with error bars representing the SD. (C) The average dimer yields after 5 and 25 J/m2 UV are shown. Values are calculated from empirically derived dimer distributions in irradiated calf thymus DNA (41), adjusted to reflect the dinucleotide probabilities in the S. pombe genome; for instance, the probability of a TC CPD is calculated as Pr(TCCPD pombe) = Pr(TCCPD calf) × Pr(TC pombe)/Pr(TC calf). (D) The average number of kilobases that a replication fork would travel before encountering a pyrimidine dimer on either the leading-strand or lagging-strand template is plotted as a function of UV dose. The gray lines indicate SD.
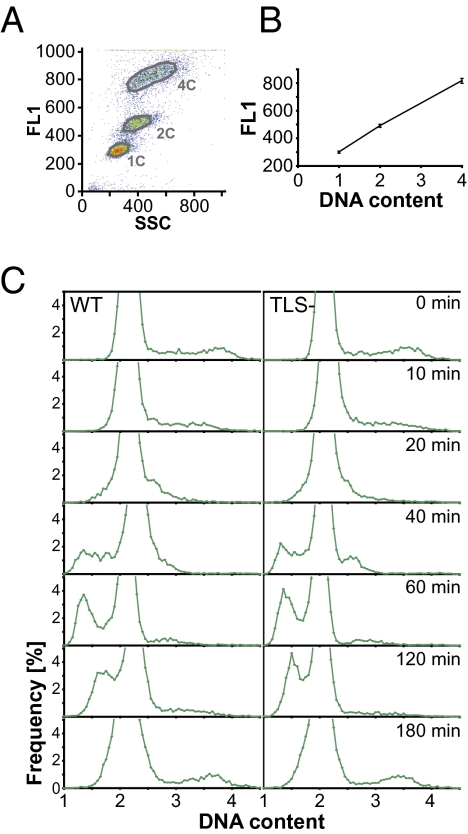
To determine whether TLS− cells are defective in bulk DNA synthesis after DNA damage, we used quantitative flow cytometry to compare the DNA content in WT cells and mutant cells lacking all four translesion polymerases. We developed a protocol for staining Rnase A–treated S. pombe with SYTOX Green that gave a strict linear relationship between cellular fluorescence and DNA content (Fig. 3 A and B). Using haploid and diploid S. pombe as calibration standards, we were able to use this relationship to obtain accurate estimates of absolute cellular DNA content. These estimates included a correction for the component of background fluorescence that is dependent on cell size rather than on DNA content (see Materials and Methods for details). Asynchronous WT and TLS− cells irradiated with 25 J/m2 UV were harvested for flow cytometry at various times after irradiation, and the distributions of cellular DNA content were calculated (Fig. 3C). In fission yeast, cytokinesis does not occur until late S phase, so the majority of cells in the population had 2C DNA contents before irradiation, and the small subset of cells in S phase had DNA contents between 2C and 4C. Because DNA synthesis slowed significantly after UV irradiation, many cells divided before completing DNA replication. The cells with less than 2C DNA contents resulting from this slow replication went on to synthesize DNA with the same kinetics in WT and TLS− cells, achieving a 2C DNA content between 2 and 3 h after irradiation. Whereas bulk DNA synthesis was completed by 3 h, the enhanced checkpoint response of TLS mutants delayed division until 4–8 h after irradiation (Fig. 1 C and E), suggesting that TLS normally occurs after S phase.
Fig. 3.
S. pombe cells lacking TLS polymerases complete bulk DNA synthesis at the same rate as WT cells. (A) The linear relationship between fluorescence and DNA content in fission yeast cells stained with SYTOX Green is illustrated by flow cytometry analysis of a diploid strain (4C) that was mixed with a haploid strain after the latter had been held in 25 mM hydroxyurea for 2 h to block DNA synthesis (1C and 2C). The mixture was analyzed by flow cytometry. The green fluorescence (FL1), a measure of the DNA content, is plotted as a function of the side scatter (SSC), a measure of cell size. (B) The average FL1 values of populations gated as in A are plotted as a function of their DNA content. The mean value of three different samples is displayed, with error bars representing SD. (C) Asynchronous cell populations were plated on YE6s and UV-irradiated under conditions similar to those described in Fig. 1. After irradiation with 25 J/m2, cells were harvested at multiple times post-UV and analyzed by flow cytometry. The frequency distributions of calculated DNA contents, in genome equivalents [C], are plotted for 9,000–10,000 cells after binning into 0.05 C bins. The DNA content of WT cells is compared with that of a strain lacking all translesion polymerases.
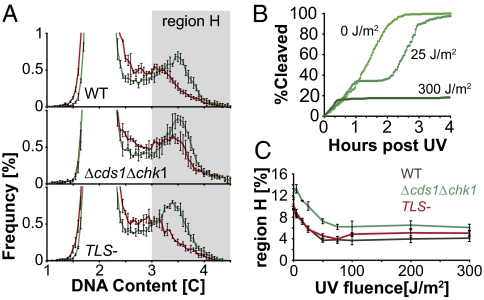
To further assess the possible role of translesion polymerases in replication fork progression through UV lesions, we studied the rate of DNA synthesis in the first 10 min after irradiation with a broad range of UV doses. In this short interval, the majority of UV lesions remain unrepaired, and the Cds1-mediated checkpoint that slows replication when DNA is damaged (33, 34) has a minimal effect on the overall rate of DNA synthesis (see below). Thus, the potential role of TLS polymerases on the rate of DNA chain elongation after UV irradiation can be assessed more directly. Figure 4A compares the DNA content distributions in asynchronous cells at 10 min after mock irradiation or irradiation with 25 J/m2 UV. The data indicate that UV irradiation caused a reduction in the number of cells with DNA contents above 3C (“region H”) and an increase in the number of cells with DNA contents below 3C. Because the rate at which cells divide and exit region H is unaffected by the UV doses used in this experiment (Fig. 4B), the loss of cells in region H must be due to a reduction in the rate at which cells enter the region by synthesizing DNA. It follows from these considerations that the abundance of cells in region H at 10 min after irradiation is a function of the average rate of DNA synthesis. At UV doses of ≥50 J/m2, the reduced number of cells in region H represents ~6% of the total cell population (Fig. 4C). Under our experimental conditions, 5.8% of an asynchronous population of S. pombe cells divided every 10 min (doubling time, 122 min); thus, the observed depletion of the population from region H at high UV doses is consistent with a near cessation of DNA synthesis along with a loss of 6% of the cells from region H by division. At lower UV exposures, there was a dose-dependent depletion of cells in region H consistent with blocked replication forks or a slower mode of DNA synthesis through UV lesions. The UV-induced depletion of region H was similar in WT cells and the Δcds1Δchk1 checkpoint mutant, indicating that it is not caused by a checkpoint response (Fig. 4C). Unexpectedly, the checkpoint mutant cells had a somewhat higher DNA content than WT cells in the absence of UV irradiation (Fig. 4C and Fig. S1). We suspect that this phenotype might be due to the firing of a greater number of replication origins by the mutant cells (35), but this interpretation will require further confirmation. Importantly, the UV-induced depletion of region H was similar in WT and TLS− mutants over a broad range of UV doses (Figs. 2D and 4C and Fig. S2), indicating that the rate at which cells replicate UV-damaged DNA is unaffected by the absence of translesion polymerases. This experiment provides strong evidence that translesion polymerases do not facilitate the movement of replication forks through damaged DNA and supports a model in which they function to repair postreplication gaps.
Fig. 4.
The synthesis of UV-damaged DNA occurs at the same rate in WT and TLS- cells over a broad range of UV fluences. (A) The distribution of DNA contents, measured as detailed in Fig. 3, is shown for cells 10 min after mock irradiation (green) or irradiation with 25 J/m2 (red). The data for three experiments, with 20,000 cells each, were binned into 0.05 C bins. The average frequency of cells in each bin is plotted, with error bars indicating SD. The shaded box labeled “region H” indicates the >3C region analyzed in C. (B) Division kinetics after irradiation with the indicated UV doses are shown (n = 300). (C) Average percentage of cells in region H at 10 min after irradiation is plotted as a function of the UV dose. The experiment was carried out in triplicate, with the three different strains coirradiated on the same plate in each case. Error bars indicate the SDs calculated from the three experiments.
Discussion
The fission yeast checkpoint response to UV requires several components, including the Rad3 kinase that binds to RPA-coated single-stranded DNA via its Rad26 subunit and the Rad17-RFC complex that loads the heterotrimeric 9-1-1 clamp onto DNA at double-strand to single-strand transitions. It has been suggested that single-stranded regions transiently generated during nucleotide excision repair (NER) are recognized by these proteins, leading to checkpoint activation (36–38). However, our previous studies of the response to a UV dose of 5 J/m2 demonstrated that checkpoint activation does not occur in G1 or G2 phase yeast cells in which active NER is ongoing, challenging the notion that NER intermediates represent important signals for checkpoint activation outside of S phase. It is unlikely that NER intermediates fail to elicit a checkpoint response because they are too few in number at a UV dose of 5 J/m2. We have shown that under our experimental conditions, this dose introduces 1,037 ± 240 pyrimidine dimers and 304 ± 70 (6–4) photoproducts per genome. This indicates that a large number of single-strand to double-strand transitions are formed by NER even at physiological UV doses but simply are not recognized by the DNA damage checkpoint. It is now clear that the relevant UV-induced structures monitored by the checkpoint system are formed during S phase.
Electron microscopy has revealed that the replication of UV-irradiated budding yeast DNA creates extended regions of single-stranded DNA at replication forks and single-stranded gaps behind forks (25). The former structures are suggestive of replication forks that have been stalled at UV lesions, and the latter are consistent with postreplication gaps in the nascent DNA strands. Because UV-induced checkpoint delay occurs hours after the completion of bulk DNA synthesis, we have been particularly interested in determining whether structures left behind after DNA replication, such as postreplication gaps, can be recognized by the DNA damage checkpoint. Postreplication gaps behind the fork can be formed when Okazaki fragment synthesis is interrupted by UV lesions in the lagging strand template (39). It also has been suggested that these gaps might form when the replisome reprimes DNA synthesis downstream from UV lesions in the leading strand template, leaving an unreplicated segment behind (25). A UV lesion on the leading strand template also might lead to a postreplication gap if two converging forks were to terminate at the site of the lesion. We reasoned that if postreplication gaps were recognized by the checkpoint system, then perturbations that reduce the rate of postreplication gap repair would increase the checkpoint response. On the other hand, if these structures, like NER intermediates, were invisible to the checkpoint surveillance mechanism, then the duration of the checkpoint response would be unaffected. The disruption of translesion polymerases in budding yeast cells leads to an accumulation of postreplication gap-like structures without increasing the number of single-stranded regions at forks (25). This observation would suggest that TLS mutants specifically accumulate postreplication gaps after UV irradiation. However, there is also evidence suggesting that translesion polymerases may facilitate replication fork movement through damaged DNA, and that their disruption leads to increased fork stalling (2, 24). Our finding that TLS− cells synthesize UV-irradiated DNA at a rate similar to WT cells over a wide range of UV doses indicates that S. pombe translesion polymerases do not promote replication fork progression through dimeric photoproducts, consistent with qualitative results from budding yeast obtained using 2D gel analysis (25). Therefore, the primary effect of disrupting S. pombe TLS genes is to reduce the rate at which postreplication gaps are repaired after UV irradiation.
Our time-lapse analysis of cells lacking Polη indicated that these cells respond to slower gap repair with an extended checkpoint-mediated delay in cell-cycle progression. This result is consistent with the observation that human cells lacking Polη exhibit enhanced markers of checkpoint activation after UV irradiation (40), and strongly suggests that postreplication gaps at CPDs are signals for checkpoint activation. Cells lacking Polκ also had an enhanced postreplication checkpoint response. By analogy to Polη, we suggest that this phenotype might reflect the attenuated repair of postreplication gaps.
Because Polζ or Rev1 are thought to mediate TLS through (6–4) photoproducts (17), we were surprised to find that the checkpoint response in cells lacking these polymerases was normal after the introduction of 1,518 ± 352 (6–4) photoproducts per genome. Thus, it seems likely that Polζ and Rev1 do not repair gaps containing (6–4) photoproducts under our experimental conditions. This interpretation is consistent with the failure to observe a defect in nascent strand joining in UV-irradiated budding yeast cells lacking Polζ (13, 14). Interestingly, the diminished viability of cells lacking Polζ and Rev1 during the third cell cycle after UV was comparable to that of the Polη mutant. The simplest explanation for this phenotype is that Polζ and Rev1 function only after lesions have been carried into the third cycle. For instance, if cells were to eventually adapt to the checkpoint response and continue dividing, then these polymerases might participate in a mutagenic process that restores a replication fork after it encounters a gap containing an unrepaired lesion.
Surveillance of postreplication gaps by the DNA damage checkpoint might be critical to the cell for several reasons. In the absence of checkpoint delay, chromosomes with postreplication gaps would be carried into mitosis, and the resulting separation of sister chromatids would remove the last possibility for error-free repair by HR, potentially causing a lethal loss of genetic information. In addition, the propagation of DNA containing postreplication gaps into the subsequent S phase could lead to genomic instability or mutagenesis if the replication fork were to convert the gap into a double-strand break (DSB). A DSB generated by such an event could not be repaired by HR unless the lesion were subsequently bypassed by TLS or other postreplication repair processes. These dangers may explain why the checkpoint system has evolved the ability to ensure that cell-cycle progression is coordinated with the repair of postreplication gaps.
Materials and Methods
Time-Lapse Microscopy.
S. pombe cells, grown on agar plates containing YE6s-rich medium and maintained at 30 °C, were UV-irradiated and imaged every 2 min, as described previously (5). See Table S1 for a list of strains used in this study.
Pyrimidine Dimer Quantitation.
An S. pombe strain engineered to uptake thymidine (41) was labeled with 14C-thymidine, UV-irradiated, and then rapidly fixed in 70% ethanol. Cells were digested with RNase A, extracted with detergent, and then hydrolyzed in acid to release thymine monomers and dimers. The hydrolysates were spotted onto PEI- cellulose sheets and developed in 85% 2-propanol. A phorphorimager was used to determine the locations of thymine monomers and dimers, and these regions were cut from the TLC sheets. The recovered radioactivity was measured by scintillation counting. The protocol is described in detail in SI Materials and Methods.
Flow Cytometry.
Flow cytometry was performed as described previously (5), except that 5 μM SYTOX Green was used instead of DAPI to stain DNA. DNA content was calculated using the following equation: FL1 = a + b[DNA] + c[SSC]. The a, b, and c parameters were determined from haploid and diploid calibration standards as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Olaf Nielsen, Susan Forsburg, and Theresa Wang for sharing S. pombe strains. This work was supported by National Institutes of Health Grant 5R01GM050806-16.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1003449107/DCSupplemental.
References
- 1.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 2.Friedberg EC. Suffering in silence: The tolerance of DNA damage. Nat Rev Mol Cell Biol. 2005;6:943–953. doi: 10.1038/nrm1781. [DOI] [PubMed] [Google Scholar]
- 3.Callegari AJ, Kelly TJ. Shedding light on the DNA damage checkpoint. Cell Cycle. 2007;6:660–666. doi: 10.4161/cc.6.6.3984. [DOI] [PubMed] [Google Scholar]
- 4.Hartwell LH. Nobel lecture: Yeast and cancer. Biosci Rep. 2002;22:373–394. doi: 10.1023/a:1020918107706. [DOI] [PubMed] [Google Scholar]
- 5.Callegari AJ, Kelly TJ. UV irradiation induces a postreplication DNA damage checkpoint. Proc Natl Acad Sci USA. 2006;103:15877–15882. doi: 10.1073/pnas.0607343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffiths DJ, Barbet NC, McCready S, Lehmann AR, Carr AM. Fission yeast rad17: A homologue of budding yeast RAD24 that shares regions of sequence similarity with DNA polymerase accessory proteins. EMBO J. 1995;14:5812–5823. doi: 10.1002/j.1460-2075.1995.tb00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Majka J, Burgers PM. Yeast Rad17/Mec3/Ddc1: A sliding clamp for the DNA damage checkpoint. Proc Natl Acad Sci USA. 2003;100:2249–2254. doi: 10.1073/pnas.0437148100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thelen MP, Venclovas C, Fidelis K. A sliding clamp model for the Rad1 family of cell cycle checkpoint proteins. Cell. 1999;96:769–770. doi: 10.1016/s0092-8674(00)80587-5. [DOI] [PubMed] [Google Scholar]
- 9.Bermudez VP, et al. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc Natl Acad Sci USA. 2003;100:1633–1638. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: Clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 2003;1:e33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 12.Cadet J, Sage E, Douki T. Ultraviolet radiation–mediated damage to cellular DNA. Mutat Res. 2005;571:3–17. doi: 10.1016/j.mrfmmm.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Prakash L. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol Gen Genet. 1981;184:471–478. doi: 10.1007/BF00352525. [DOI] [PubMed] [Google Scholar]
- 14.Torres-Ramos CA, Prakash S, Prakash L. Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:2419–2426. doi: 10.1128/MCB.22.7.2419-2426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gangavarapu V, Prakash S, Prakash L. Requirement of RAD52 group genes for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:7758–7764. doi: 10.1128/MCB.01331-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blastyák A, et al. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waters LS, et al. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 19.Yu SL, Johnson RE, Prakash S, Prakash L. Requirement of DNA polymerase eta for error-free bypass of UV-induced CC and TC photoproducts. Mol Cell Biol. 2001;21:185–188. doi: 10.1128/MCB.21.1.185-188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schenten D, et al. DNA polymerase kappa deficiency does not affect somatic hypermutation in mice. Eur J Immunol. 2002;32:3152–3160. doi: 10.1002/1521-4141(200211)32:11<3152::AID-IMMU3152>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 21.Ziv O, Geacintov N, Nakajima S, Yasui A, Livneh Z. DNA polymerase zeta cooperates with polymerases kappa and iota in translesion DNA synthesis across pyrimidine photodimers in cells from XPV patients. Proc Natl Acad Sci USA. 2009;106:11552–11557. doi: 10.1073/pnas.0812548106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrence CW, Maher VM. Eukaryotic mutagenesis and translesion replication dependent on DNA polymerase zeta and Rev1 protein. Biochem Soc Trans. 2001;29:187–191. doi: 10.1042/0300-5127:0290187. [DOI] [PubMed] [Google Scholar]
- 23.Vidal AE, Woodgate R. Insights into the cellular role of enigmatic DNA polymerase iota. DNA Repair (Amst) 2009;8:420–423. doi: 10.1016/j.dnarep.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Edmunds CE, Simpson LJ, Sale JE. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol Cell. 2008;30:519–529. doi: 10.1016/j.molcel.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 25.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka K, et al. Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol Cell Biol. 2000;20:3459–3469. doi: 10.1128/mcb.20.10.3459-3469.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polesky AH, Steitz TA, Grindley ND, Joyce CM. Identification of residues critical for the polymerase activity of the Klenow fragment of DNA polymerase I from Escherichia coli. J Biol Chem. 1990;265:14579–14591. [PubMed] [Google Scholar]
- 28.Kondratick CM, Washington MT, Prakash S, Prakash L. Acidic residues critical for the activity and biological function of yeast DNA polymerase eta. Mol Cell Biol. 2001;21:2018–2025. doi: 10.1128/MCB.21.6.2018-2025.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibbs PE, McDonald J, Woodgate R, Lawrence CW. The relative roles in vivo of Saccharomyces cerevisiae Pol eta, Pol zeta, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics. 2005;169:575–582. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Douki T, Cadet J. Individual determination of the yield of the main UV-induced dimeric pyrimidine photoproducts in DNA suggests a high mutagenicity of CC photolesions. Biochemistry. 2001;40:2495–2501. doi: 10.1021/bi0022543. [DOI] [PubMed] [Google Scholar]
- 31.McCready S, Carr AM, Lehmann AR. Repair of cyclobutane pyrimidine dimers and 6-4 photoproducts in the fission yeast Schizosaccharomyces pombe. Mol Microbiol. 1993;10:885–890. doi: 10.1111/j.1365-2958.1993.tb00959.x. [DOI] [PubMed] [Google Scholar]
- 32.Lombaerts M, Tijsterman M, Brandsma JA, Verhage RA, Brouwer J. Removal of cyclobutane pyrimidine dimers by the UV damage repair and nucleotide excision repair pathways of Schizosaccharomyces pombe at nucleotide resolution. Nucleic Acids Res. 1999;27:2868–2874. doi: 10.1093/nar/27.14.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindsay HD, et al. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhind N, Russell P. The Schizosaccharomyces pombe S-phase checkpoint differentiates between different types of DNA damage. Genetics. 1998;149:1729–1737. doi: 10.1093/genetics/149.4.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim SM, Huberman JA. Regulation of replication timing in fission yeast. EMBO J. 2001;20:6115–6126. doi: 10.1093/emboj/20.21.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giannattasio M, Lazzaro F, Longhese MP, Plevani P, Muzi-Falconi M. Physical and functional interactions between nucleotide excision repair and DNA damage checkpoint. EMBO J. 2004;23:429–438. doi: 10.1038/sj.emboj.7600051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neecke H, Lucchini G, Longhese MP. Cell cycle progression in the presence of irreparable DNA damage is controlled by a Mec1- and Rad53-dependent checkpoint in budding yeast. EMBO J. 1999;18:4485–4497. doi: 10.1093/emboj/18.16.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouse J, Jackson SP. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297:547–551. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- 39.Nikolaishvili-Feinberg N, Cordeiro-Stone M. Bypass replication in vitro of UV-induced photoproducts blocking leading or lagging strand synthesis. Biochemistry. 2001;40:15215–15223. doi: 10.1021/bi011474t. [DOI] [PubMed] [Google Scholar]
- 40.Bomgarden RD, et al. Opposing effects of the UV lesion repair protein XPA and UV bypass polymerase η on ATR checkpoint signaling. EMBO J. 2006;25:2605–2614. doi: 10.1038/sj.emboj.7601123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hodson JA, Bailis JM, Forsburg SL. Efficient labeling of fission yeast Schizosaccharomyces pombe with thymidine and BUdR. Nucleic Acids Res. 2003;31:e134. doi: 10.1093/nar/gng134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.