Abstract
Mouse and rat embryonic stem cells can be sustained in defined medium by dual inhibition (2i) of the mitogen-activated protein kinase (Erk1/2) cascade and of glycogen synthase kinase 3. The inhibitors suppress differentiation and enable self-renewal of pluripotent cells that are ex vivo counterparts of naïve epiblast cells in the mature blastocyst. Pluripotent stem cell lines can also be derived from unipotent primordial germ cells via a poorly understood process of epigenetic reprogramming. These are termed embryonic germ (EG) cells to denote their distinct origin. Here we investigate whether EG cell self-renewal and derivation are supported by 2i. We report that mouse EG cells can be established with high efficiency using 2i in combination with the cytokine leukaemia inhibitory factor (LIF). Furthermore, addition of fibroblast growth factor or stem cell factor is unnecessary using 2i-LIF. The derived EG cells contribute extensively to healthy chimaeric mice, including to the germline. Using the same conditions, we describe the first derivations of EG cells from the rat. Rat EG cells express a similar marker profile to rat and mouse ES cells. They have a diploid karyotype, can be clonally expanded and genetically manipulated, and are competent for multilineage colonisation of chimaeras. These findings lend support to the postulate of a conserved molecular ground state in pluripotent rodent cells. Future research will determine the extent to which this is maintained in other mammals and whether, in some species, primordial germ cells might be a more tractable source than epiblast for the capture of naïve pluripotent stem cells.
Keywords: EG cells, PGC, Pluripotent stem cell, Rat, Mouse, Reprogramming
INTRODUCTION
Pluripotency can be defined as the ability at the single cell level to initiate formation of all somatic cell lineages and the germ cells. In the mammalian embryo, pluripotency is an emergent property first established in the epiblast of the late blastocyst (Selwood and Johnson, 2006). Cultured epiblast gives rise to pluripotent embryonic stem (ES) cells (Batlle-Morera et al., 2008; Brook and Gardner, 1997; Evans and Kaufman, 1981; Martin, 1981; Nichols et al., 2009b). ES cells can self-renew indefinitely in vitro while maintaining an undifferentiated pluripotent state, demonstrated by their ability to colonise all cell lineages, including the germline, when reintroduced to the blastocyst (Beddington and Robertson, 1989; Bradley et al., 1984). Identification of the transcriptional organisers that determine and maintain pluripotency in the embryo and in ES cells has enabled the induction of pluripotent stem (iPS) cells from somatic cells by molecular reprogramming (Takahashi and Yamanaka, 2006). iPS cells share the defining properties of ES cells, including contribution to germline-competent chimaeras following blastocyst injection (Okita et al., 2007; Wernig et al., 2007). A third type of pluripotent stem cell is obtained by epigenetic reprogramming. These are embryonic germ (EG) cells which are derived by culturing primordial germ cells (PGCs) from embryonic day (E) 8.5-E12.5 mouse embryos in the presence of leukaemia inhibitory factor (LIF), basic fibroblast growth factor (bFGF), stem cell factor (SCF) and serum (Durcova-Hills et al., 2001; Matsui et al., 1992; Resnick et al., 1992; Tada et al., 1998). Like ES and iPS cells, EG cells can contribute to chimaeras and, on occasion, give rise to germline transmission (Labosky et al., 1994; Stewart et al., 1994).
PGCs are specified prior to gastrulation at E6.25 in mice (McLaren and Lawson, 2005; Ohinata et al., 2005). Soon after, their specification transcription of many pluripotency-associated genes, including Sox2, Nanog, Klf2 and Stella, is upregulated in PGCs (Kurimoto et al., 2008). However, PGCs are unipotent, making only sperm or eggs, and they do not contribute to chimaeras following blastocyst injection (Durcova-Hills and Surani, 2008; Matsui, 1998). Therefore the conversion of unipotent PGCs to pluripotent EG cells is considered a reprogramming event (McLaren, 1992). Attempts to understand this conversion have been hampered by the inconsistency of EG cell derivation and, as a result, the process and mechanism remain elusive (Durcova-Hills and Surani, 2008; Shamblott et al., 2004). Derivation of human cells with features of EG cells has also been reported (Shamblott et al., 1998; Turnpenny et al., 2003). However, although these cultures express pluripotency marker genes and demonstrate broad differentiation, undifferentiated cells have not been clonally propagated or maintained during long-term culture (Turnpenny et al., 2006). Thus, whether the ability to reprogram to pluripotent stem cells in vitro is a phenomenon specific to mice or is a general property of mammalian PGCs remains an open question.
Once established, all three pluripotent cell types can be maintained in medium containing foetal calf serum (FCS) and LIF, with the optional addition of a mouse embryonic fibroblast (MEF) feeder layer. Recently it has been shown that mouse ES cells can self-renew in basal medium if autocrine mitogen-activated protein kinase (MAPK) signalling is eliminated and glycogen synthase kinase 3 activity is reduced (Ying et al., 2008). The two inhibitor (2i) culture system, supplemented with LIF, allows efficient derivation and expansion of germline-competent ES cells from different strains of mice tested, including 129, CBA and NOD (Nichols et al., 2009a; Ying et al., 2008), and, for the first time, from the rat (Buehr et al., 2008; Li et al., 2008). Reprogramming of mouse somatic cells to induced pluripotency is also enhanced using 2i-LIF (Silva et al., 2008). Furthermore, application of 2i to cleavage-stage embryos suppresses formation of the hypoblast, resulting in the entire inner cell mass becoming pluripotent epiblast (Nichols et al., 2009b). These findings have led to the hypothesis that 2i anchors pluripotent cells in a self-renewing ground state by eliminating extrinsic stimuli that initiate differentiation (Silva and Smith, 2008; Smith, 2009; Ying et al., 2008). Significantly, epiblast stem cells (EpiSCs) derived from postimplantation epiblast cannot be propagated in 2i (Guo et al., 2009; Nichols and Smith, 2009). EpiSCs exhibit some features of pluripotency but are transcriptionally and epigenetically differentiated and are unable to colonise chimaeras (Brons et al., 2007; Tesar et al., 2007). Sustained proliferation in 2i-LIF is thus a specific attribute of ground state pluripotent cells.
Here we evaluate whether, despite their distinct origins, EG cells are in a similar molecular ground state to ES cells and investigate the application of 2i during conversion of mouse and rat PGCs into pluripotent EG cells.
MATERIALS AND METHODS
Mouse EG cell derivation and culture
Cell lines were established from embryos produced by crossing mixed background Oct4ΔPE-GFP transgenic males (Yoshimizu et al., 1999) with strain 129 female mice. GFP fluorescence was monitored either by fluorescence microscopy or flow cytometry. EG cells were derived from E8.5 mouse embryos as described (Durcova-Hills and McLaren, 2004). In brief, the posterior fragment of the embryo containing PGCs was dissected free of the extraembryonic membranes and trypsinised to a single cell suspension. Cells were collected by centrifugation, resuspended in FCS-LIF medium (see below) supplemented with bFGF (25 ng/ml; Invitrogen) and plated on stem cell factor (SCF)-producing Sl4-m220 feeders (Majumdar et al., 1994) in two 2-cm2 wells per starting embryo. After 2 days, 50% of the medium was replaced with fresh medium lacking bFGF. Subsequently, medium was changed every 24-48 hours without bFGF supplementation. After 10-14 days, macroscopic colonies were individually picked, replated on a MEF feeder layer and expanded into EG cell lines. Derivations using 2i-LIF followed the same schedule but, on day 3 of derivation (first medium change), the cultures were washed with PBS and the medium replaced with serum-free N2B27 supplemented with 2i-LIF without bFGF. Cultures were then maintained continuously in 2i-LIF. Picked colonies were expanded on gelatin-coated plates without feeders. FCS-LIF conditions comprise DMEM-F12 medium supplemented with 15% FCS, 0.1% MEM non-essential amino acids, 4 mM glutamate, 2 mM sodium pyruvate, 0.1 mM 2-mecaptanethanol and recombinant mouse LIF (1200 U/ml; ESGRO, Chemicon) or human recombinant LIF generated in-house by transient transfection of Cos7 cells with plasmid encoding human LIF. 2i-LIF conditions comprise the MEK inhibitor PD0325901 (1 μM), the GSK3 inhibitor CHIR99021 (3 μM) and LIF (1200 U/ml) in N2B27 medium (Ying et al., 2003a). After colony picking, cells were expanded by dissociation with trypsin and replating every 2-4 days.
Rat EG cell derivation and culture
Rat EG cells were derived using the 2i-LIF protocol above with the following modifications. Sprague Dawley (SD) rat embryos were collected and dissected early on the morning of E10. At this time they are at late head fold/early somite stage, prior to turning, equivalent to E8.5 in mouse (Theiler stage 12). After 10-14 days of primary culture, each well was passaged into 1 well of a 6-well plate, on a MEF feeder layer, in 2i-LIF. After 7-15 days of further culture, macroscopic colonies were picked and expanded. Cells were maintained in 2i-LIF on MEFs and were expanded by dissociation with trypsin and replating every 2-4 days. Cells were cryopreserved in 2i-LIF medium supplemented with 10% DMSO and 10% FCS. They survived multiple freeze thaw cycles with transfer into serum-free 2i-LIF immediately upon thawing.
Colony forming assays
Medium containing the appropriate cytokines was placed in gelatinised 6-well plates, which were placed in the incubator to pre-equilibrate. Cells were harvested by trypsinisation, triturated to a single-cell suspension, counted using a haemocytometer and diluted to a final cell density of 1000 cells/ml. Six hundred microlitres of this cell suspension was added to each well. Cells were cultured for 6-7 days then fixed, stained for alkaline phosphatase (AP) (Durcova-Hills and McLaren, 2004) and scored for the frequency of undifferentiated (AP-positive), mixed and differentiated (AP-negative) cells. For rat EG cells, 800 cells/well were plated onto laminin coated plates and resultant colonies were called either AP-positive or AP-negative.
Immunostaining
Immunostaining was performed using standard protocols. Primary antibodies were: Nanog (Abcam, 1:200; Cosmo Bio, 1:500), Oct4 (BD, 1:200; Santa Cruz, 1:200), Sox2 (Abcam, 1:100), βIII tubulin (Sigma, 1:200), myosin (MF-20, 1:5) and DsRed (Clontech, 1:1000). Nuclei were stained with DAPI. Alexa Fluor secondary antibodies (Invitrogen) were used at 1:500 dilution.
RT-PCR
Total RNA was prepared using the RNeasy Mini Kit (Qiagen) with DNaseI treatment. Eluted RNA concentration was determined by spectrophotometry. cDNA was synthesized using the Invitrogen Superscript III First-Strand Synthesis Kit (Invitrogen) according to the manufacturer's protocol using oligo-dT primers. Two micrograms RNA was used as template. cDNA was diluted 1:10 in sterile water and used for PCR. Primer sequences are as described previously (Buehr et al., 2008), with the following additions: Blimp1, 5′-AGTGCAATGTCTGTGCCAAG-3′ and 5′-ATGTCCTCAAGACGGTCAGC-3′; Rex1 (Zfp42 – Mouse Genome Informatics), 5′-CGAAACTAAAGCGGCACTTC-3′ and 5′-AGCATTTCTTCCCTGCCTTT-3′; Flk1 (Kdr – Mouse Genome Informatics), 5′-ATACACCTGCACAGCGTACAG-3′ and 5′-TCCCGCATCTCTTTCACTCAC-3′; Afp, 5′-GTCCCACCCTTCCACTTT-3′ and 5′-CCATCCTGTAGGCACTCC-3′; Gata4, 5′-GCATCCATTTCCACCTCTT-3′ and 5′-TCCATCACCCTTGTCCTTT-3′; nestin, 5′-AGCCATTGTGGTCTACTGA-3′ and 5′-TGCAACTCTGCCTTATCC-3′; Olig2, 5′-ACCCGATGATCTTTTTCTGC-3′ and 5′-GGGCTCAGTCATCTGCTTCT-3′.
In vitro differentiation of rat EG cells
To induce embryoid body formation, rat EG cells were collected by trypsinisation. Feeders were removed by culturing, on untreated tissue culture, for 55 minutes and the purity of the EG cell suspension observed by microscopy. The cell suspension was centrifuged and resuspended in DMEM-F12 medium supplemented with 20% serum, 0.1 mM NEAA, 2 mM L-glutamine, 0.1 mM 2-mercaptoethanol and penicillin or streptomycin. Fifteen hundred cells were deposited per well in two untreated round bottom 96-well plates in 100 μl of medium. An extra 100 μl of medium was added to each well after four days. Sixty similar-sized embryoid bodies were recovered (1 per well) at day 4 and day 8 for RT-PCR analysis. To induce cardiac differentiation, day-4 embryoid bodies were picked and plated on gelatine-coated 6-well plates (6 per well) in the same medium, with gentle medium changes every 3-4 days. Monolayer neuronal differentiation was performed as described (Ying et al., 2003a), with the following modifications: cells were grown on laminin in the absence of MEFs for 3 passages and then dissociated, pelleted, resuspended in 2i-LIF and plated at 0.75, 1.5 or 3.0×104 cells/cm2 in 24-well (2 cm2/well) tissue culture plates coated with laminin. The next day, 2i-LIF was removed and cells were cultured in N2B27 for 10 days before fixation and staining.
PiggyBac vector transposition
1×106 cells were transfected using FuGENE (Roche) with 2 μg pGG131 vector (CAG-DsRed-IRES-Hygro) (Guo et al., 2009) plus 2 μg pCAGPBase (Wang et al., 2008). Transfection was performed in 2i-LIF medium for 7 hours. To select for stable transfectants, hygromycin (200 μg/ml) was applied for at least 7 days. Cells were grown on multi-drug-resistant DR-4 feeders (Tucker et al., 1997) throughout.
Chimaera production
Mouse chimaeras were produced by micro-injection of Oct4ΔPE-GFP EG cells (agouti) into E3.5 C57BL/6 blastocysts. Chimaerism was assessed by agouti coat colour. Rat chimaeras were produced by micro-injection of SD-derived fluorescent rat EG cells into E4.5 SD blastocysts produced by natural matings. Injected blastocysts were transferred into pseudopregnant SD recipients. Chimaerism was assessed by DsRed fluorescence. Animal studies were authorised by a UK Home Office Project Licence and carried out in a Home Office designated facility.
RESULTS
Mouse EG cells can be propagated and remain pluripotent in 2i-LIF
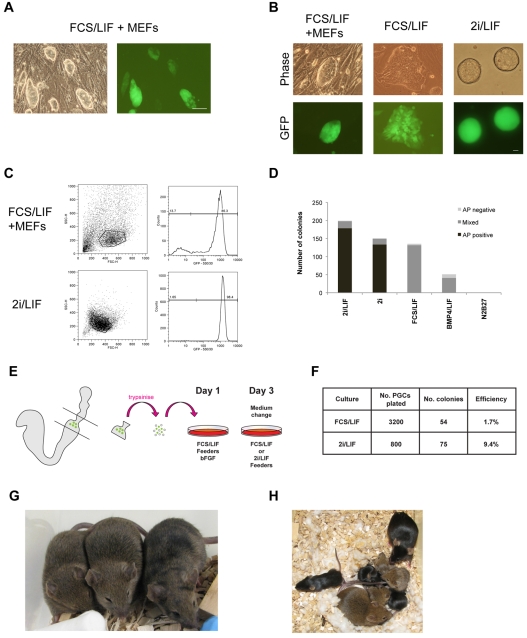
We derived EG cells from E8.5 mouse embryos carrying the Oct4ΔPE-GFP transgene (Yoshimizu et al., 1999). This reporter is expressed in preimplantation embryos, primordial germ cells and ES cells, but not in postimplantation epiblast. It has been used as a marker for pluripotent cells in reprogramming protocols (Bao et al., 2009; Ko et al., 2009). EG cell lines were derived as previously described (Durcova-Hills and McLaren, 2004). The posterior fragment of the embryo containing PGCs was dissected free of extraembryonic membranes, trypsinised to a single-cell suspension and plated on an Sl4-m220 feeder line (Majumdar et al., 1994) in medium supplemented with FCS and LIF. bFGF was added for the first 2 days of primary culture. Colonies emerged after 10 days and were picked and expanded on a MEF feeder layer in medium containing FCS and LIF. The majority, although not all, cells in these passaged cultures expressed GFP (Fig. 1A), consistent with observations on ES cells carrying this Oct4ΔPE-GFP transgene (H.G.L. and M.A.S., unpublished observations). We plated these established EG cells without feeders, either in FCS-LIF or in 2i-LIF. In FCS-LIF conditions without feeders, transgene expression was more heterogeneous (Fig. 1B). In 2i-LIF however, the transgene was expressed strongly in all cells (Fig. 1B,C). This indicates that, in 2i-LIF conditions, the distal Oct4 ‘enhanceosome’ is efficiently activated, presumably reflecting optimal expression of pluripotency factors (Chen et al., 2008), and the cultures appear homogenous, as described for other markers in ES cells (Wray et al., 2010).
Fig. 1.
Mouse EG cells can be maintained and derived in 2i-LIF. (A) Phase contrast and fluorescence images of Oct4-ΔPE-GFP EG cells. (B) Phase contrast and fluorescence images of Oct4-ΔPE-GFP EG cells cultured in: FCS-LIF on a MEF feeder layer; FCS-LIF after one passage without feeders; 2i-LIF without feeders. (C) Flow cytometry analysis of Oct4-ΔPE-GFP EG cells cultured in 2i-LIF compared with FCS-LIF on feeders. Gates were set to eliminate feeders and cell debris. (D) Colony-forming assay on Oct4-ΔPE-GFP EG cells. Colonies were scored for alkaline phosphatase (AP) and designated positive, negative or mixed. Data are means of two biological replicates. (E) Schematic of EG cell derivation protocol. On day 3, medium was changed to either FCS-LIF or 2i-LIF. (F) Comparison of GFP-positive EG cell colony formation following addition of 2i-LIF on day 3 compared with FCS-LIF. (G) High-contribution coat colour chimaeras generated with agouti EG cells derived in 2i-LIF injected into C57BL/6 blastocysts. (H) Chimaera, C57BL/6 mate, and mixed litter of agouti and black pups. Scale bars: 100 μm in A; 25 μm in B.
To confirm that 2i-LIF conditions are sufficient for EG cell propagation, we investigated colony formation from individual cells in a feeder-free assay (Fig. 1D). Numerous colonies grew up in FCS-LIF, with fewer in Bmp4-LIF (Ying et al., 2003b); in both of these conditions, colonies were mostly mixed, containing a core of alkaline phosphatase (AP)-positive undifferentiated cells and a skirt of differentiating AP-negative cells. By contrast, in 2i alone or 2i-LIF, colonies were almost all tightly packed, undifferentiated and highly AP-positive. Colonies were readily obtained in 2i alone but more were obtained on addition of LIF, as previously noted for mouse ES cells (Wray et al., 2010; Ying et al., 2008). We therefore used this condition in all future experiments. Undifferentiated morphology and homogenous Oct4ΔPE-GFP transgene expression were maintained during routine bulk passaging of five independent Oct4-ΔPE-GFP EG cell lines cultured in 2i-LIF (data not shown). We conclude that mouse EG cells can be propagated in the selective conditions defined for mouse ES cells in which Erk signalling is eliminated (Ying et al., 2008).
2i-LIF facilitates derivation of germline-competent mouse EG cells
We then tested whether 2i-LIF can be applied to derive EG cells. EG cell derivation protocols rely on initial provision of SCF and bFGF in addition to serum and LIF. SCF is known to improve PGC viability and proliferation (Dolci et al., 1991; Godin et al., 1991; Matsui et al., 1991) and bFGF is postulated to trigger the reprogramming process (Durcova-Hills et al., 2006). We therefore allowed PGCs to attach to Sl4-m220 feeders, a source of membrane-bound SCF, in standard conditions with serum and bFGF and transferred them into 2i-LIF from the first medium change on day 3 (Fig. 1E). We observed a significant amount of cell death shortly following transfer to 2i-LIF. This appears to be due to the rapid degeneration of Sl4-m220 feeders in serum-free 2i culture. Nonetheless, numerous Oct4ΔPE-GFP-positive colonies emerged over a 14-day period. These had the tightly packed morphology of EG cell colonies. In a side-by-side experiment, we plated approximately 100 PGCs (half an embryo) per well and medium was changed either to 2i-LIF or FCS-LIF. After 14 days, Oct4ΔPE-GFP colonies were counted. Wells were then fixed and colonies rescored by AP staining. From 24 wells in conventional FCS-LIF culture conditions, a total of 54 colonies were obtained, representing a conversion efficiency of 1.7% per PGC plated. By contrast, 8 wells in 2i-LIF yielded 75 colonies, an efficiency of 9.4% (Fig. 1F). Furthermore, this procedure has proved highly consistent, producing multiple Oct4ΔPE-GFP-positive colonies in five independent experiments, at frequencies of 2-10 colonies per 100 PGCs plated. This included occasions when the traditional protocol performed in parallel failed to produce any EG cells.
To test whether EG cells derived in this manner are pluripotent, we picked and expanded single colonies (from Experiment 2, Fig. 1F). Two of these lines were injected into C57BL/6 blastocysts. They produced very high contribution chimaeras, as shown by the predominance of agouti coat colour (Fig. 1G). Chimaeras from one of these lines were test-mated and exhibited germline transmission (Fig. 1H; see Table S1 in the supplementary material). Therefore, mouse EG cells derived and propagated in 2i-LIF display the defining functional properties of ground state pluripotency.
2i-LIF is sufficient to convert PGCs into EG cells without serum, bFGF or SCF
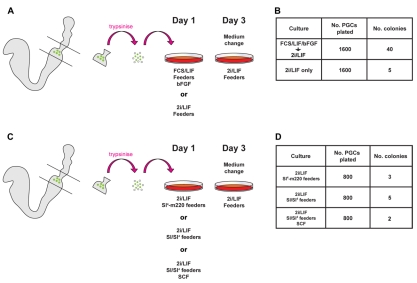
We then tested whether 2i-LIF acts only to expand reprogrammed PGCs or might be sufficient to induce reprogramming. We plated E8.5 posterior populations directly into 2i-LIF with no exposure to serum or bFGF (Fig. 2A). Despite the degeneration of Sl4-m220 feeders in serum-free 2i culture, AP-positive undifferentiated colonies were generated (Fig. 2B). The yield was lower than from parallel cultures initiated in serum and bFGF, but the colonies could readily be expanded into EG cell lines after picking. We also assessed the requirement for the membrane-bound SCF supplied by Sl4-m220 feeders. We plated cells side by side in 2i-LIF onto Sl4-m220 feeders or onto the parental Sl4 feeders, which are null for SCF (Matsui et al., 1991), with or without the addition of soluble SCF (Fig. 2C). AP-positive colonies were recovered in all three conditions at similar frequencies, indicating that SCF stimulation is not a requirement for derivation of EG cells (Fig. 2D). These results demonstrate that addition of neither bFGF and serum, nor SCF, are necessary for the generation of EG cells in 2i-LIF. We conclude that serum-free culture in 2i-LIF might be sufficient for PGC reprogramming without stimulation by additional factors.
Fig. 2.
Initial provision of serum, bFGF or SCF is not necessary for conversion of mouse PGCs into EG cells in presence of 2i-LIF. (A) Schematic of derivation protocol to test requirement for FCS and bFGF. Primordial germ cells (PGCs) are plated either directly into 2i-LIF only or into FCS-LIF plus bFGF. (B) Quantitation of EG cell colony formation following derivation directly into 2i-LIF or transfer to 2i-LIF after the first medium change. (C) Schematic of derivation protocol to test requirement for stem cell factor (SCF). PGCs were plated directly into 2i-LIF either onto Sl4-m220 feeders, Sl/Sl4 feeders or Sl/Sl4 feeders with the addition of soluble SCF. (D) Quantitation of EG cell colony formation following derivation directly into 2i-LIF in the presence of membrane-bound SCF, no SCF or soluble SCF.
Oct4 expression is restricted to Blimp1-positive pre-migratory primordial germ cells in the E10 rat embryo
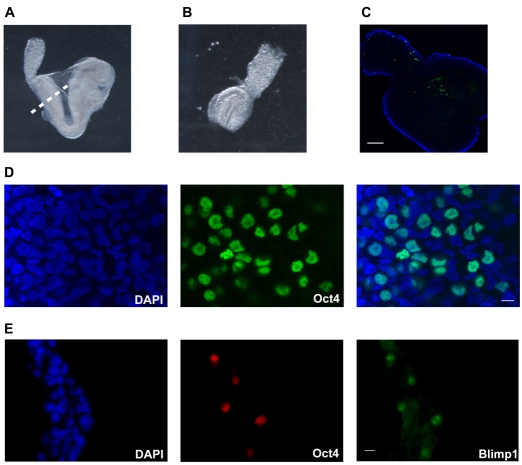
In order to test whether derivation of EG cells with 2i-LIF could be extended to the rat, we first assessed the localisation of pre-migratory PGCs in rat embryos. We found that SD rat embryos at E10 were at late head fold/early somite stage and had not commenced turning, which is therefore equivalent to E8.5 mouse embryos (Fig. 3A). The posterior fragment was isolated (Fig. 3B) and immunostained in wholemount for Oct4, a specific PGC marker at this developmental stage in mice (Yeom et al., 1996). A small cluster of Oct4-positive cells was visible at the base of the allantois (Fig. 3C,D), in the same region as PGCs are found in E8.5 mouse embryos. Double immunostaining revealed that all Oct4-positive cells also exhibited nuclear staining for Blimp1, the earliest known marker of lineage-restricted PGCs in the mouse (Ohinata et al., 2005). We therefore conclude that pre-migratory rat PGCs are similarly localised to, and express key marker genes known to be crucial for, development of PGCs in the mouse (Kehler et al., 2004; Ohinata et al., 2005). Furthermore, the restriction of Oct4 to Blimp1-positive cells indicates that there are no persistent undifferentiated pluripotent epiblast cells.
Fig. 3.
Pre-migratory rat PGCs co-express Oct4 and Blimp1. (A) Bright-field image of an E10 rat embryo. Dashed line depicts line of incision to isolate the posterior fragment. (B) Bright-field image of an isolated posterior fragment of E10 rat embryo. (C) Confocal section through the posterior fragment of an E10 rat embryo immunostained in wholemount for Oct4 (green) and co-stained with DAPI (blue outline). (D) Higher magnification confocal section through a posterior embryo fragment showing nuclear staining of Oct4 in a cluster of cells at the base of the allantois. (E) Double immunostaining for Oct4 and Blimp1 on a cryosection through the posterior fragment. Scale bars: 100 μm in C, 10 μm in D,E.
2i-LIF enables derivation of rat EG cell lines
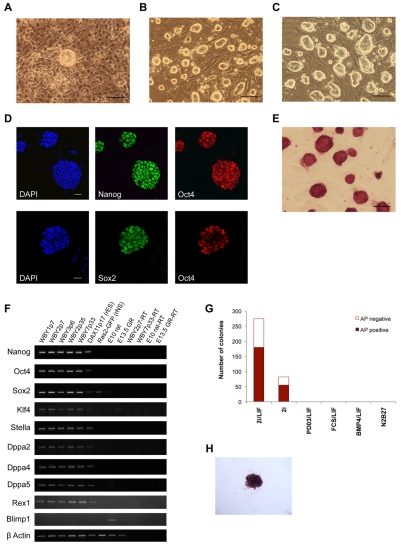
Derivation of rat EG cells has not previously been reported. We therefore tested whether 2i-LIF conditions might enable the establishment of rat EG cells. We isolated the posterior fragment containing PGCs from rat embryos (Fig. 3). After addition of 2i-LIF on day 3, cell death was observed consistent with the toxic effect of 2i on differentiated cells and Sl4-m220 feeders. However, underneath the cell debris, proliferating cells persisted. After 14 days, whole wells were passaged onto a MEF feeder layer. After a further 7-10 days, colonies with rat ES cell-like morphology were observed (Fig. 4A). These could be picked and expanded on a MEF feeder layer (Fig. 4B). They maintained an undifferentiated morphology even after extended passaging (Fig. 4C). These colonies were immunoreactive for Oct4, Sox2 and Nanog (Fig. 4D), the ‘trinity’ of transcription factors central to pluripotency (Niwa, 2007). They also stained for AP (Fig. 4E). Gene expression analysis by RT-PCR detected transcripts for all pluripotency markers tested, including Klf4 and Rex1, that are specific to ES cells and absent from EpiSCs in mice (Fig. 4F). The PGC marker Blimp1 could be detected in posterior fragments of E10 rat embryos and at a lower level in E13.5 genital ridges but not in rat EG cells (Fig. 4F). This is consistent with the previous observation that Blimp1 is rapidly downregulated during mouse EG cell derivation (Durcova-Hills et al., 2008). We performed a feeder-free colony-forming assay to assess responsiveness of rat EG cells to a range of different culture conditions (Fig. 4G). Numerous colonies grew up in 2i-LIF on laminin-coated dishes. When fixed and stained after 7 days, two thirds of these contained a central mass of AP-positive cells (Fig. 4H). A few very small AP-positive colonies formed in 2i conditions without LIF. No colonies were recovered in either FCS-LIF or Bmp4-LIF conditions. These results mirror those previously obtained with rat ES cells (Buehr et al., 2008).
Fig. 4.
Rat EG cells can be derived and expanded in 2i-LIF and express markers of pluripotency. (A) Phase contrast image of a primary rat EG cell colony. (B,C) Phase contrast images of rat EG cells after 5 (B) and 35 (C) passages. (D) Oct4, Sox2 and Nanog immunostaining of rat EG cells. (E) Alkaline phosphatase (AP) staining of rat EG cells. (F) RT-PCR analysis of pluripotency markers in three early-passage rat EG cell lines (WBY1 P7, WBY2 P7, WBY3 P6) and two late-passage rat EG cell lines (WBY2 P35, WBY7 P33). Controls are rat ES cells (DAX11), rat neural stem (rNS) cells (Ras2-GFP), posterior fragments of E10 embryos and whole E13.5 genital ridges. (G) Colony-forming assay on dissociated rat EG cells plated on laminin. Colonies were scored for AP and designated positive or negative. Data are means of two biological replicates. (H) Typical AP-positive colony. Scale bars: 100 μm in A-C,E; 25 μm in D.
In vitro differentiation of rat EG cells
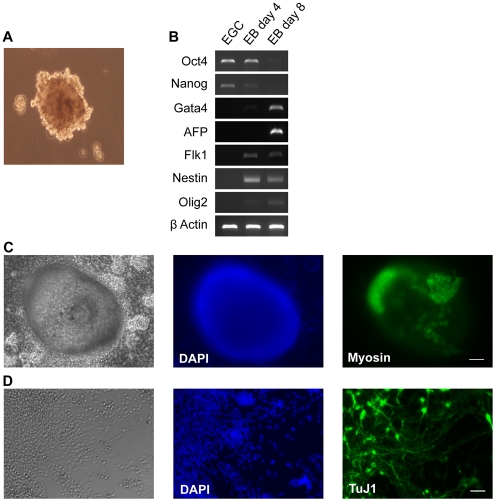
We then assessed the in vitro differentiation capacity of rat EG cells to determine whether they might be pluripotent. We induced embryoid body (EB) formation by suspension culture of rat EG cells in uncoated 96-well plates in the presence of serum (Fig. 5A). EBs were collected at day 4 and day 8 of differentiation and the gene expression profile compared with that of the parental rat EG cell line by RT-PCR. During the time course of differentiation we saw downregulation of pluripotency markers Nanog and Oct4 and upregulation of differentiation markers (Fig. 5B). We plated day-4 EBs on gelatin-coated plates and, after 12 days, beating foci were evident (see Movie 1 in the supplementary material). This continued until fixation at 21 days. The presence of cardiac myocytes was confirmed by immunostaining for myosin which colocalised with the beating foci (Fig. 5C). To further assess in vitro differentiation potential, we subjected rat EG cells to a serum-free monolayer neural differentiation protocol (Ying et al., 2003a). Cells were plated overnight in 2i-LIF then cultured in N2B27 only. After 7 days, numerous neural rosettes were visible, and after a further 3 days, cultures were dominated by cells of neuronal morphology with extended processes that were immunoreactive for TuJ1 (Fig. 5D).
Fig. 5.
Rat EG cells exhibit multilineage differentiation capacity in vitro. (A) Phase contrast image of day 4 embryoid body (EB). (B) RT-PCR analysis of undifferentiated EG cells and EBs after 4 and 8 days of differentiation. (C) Phase contrast and immunofluorescence images of an adherent EB, fixed and immunostained following 21 days of cardiac differentiation. Cardiomyocyte marker myosin colocalises with regions that exhibited spontaneous contraction prior to fixation. (D) Immunostaining for TuJ1 after 10 days of adherent monolayer neural differentiation on laminin. Scale bars: 100 μm in C; 50 μm in D.
Rat EG cells can be genetically manipulated and contribute widely to chimaeras
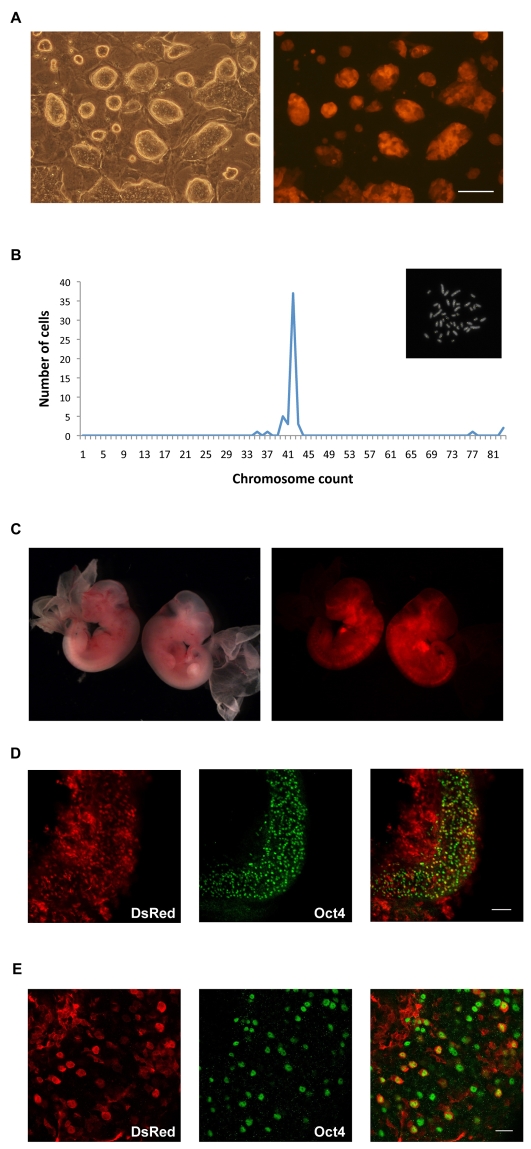
To trace cells derived from rat EG cells in vivo we introduced a transgenic reporter gene using the PiggyBac (PB) system (Wang et al., 2008). The PB vector contains a DsRed reporter with a linked hygromycin resistance gene driven by the constitutive CAG promoter (Guo et al., 2009). Rat EG cells at passage 6 were transfected and selected in hygromycin. DsRed-expressing colonies were picked, isolated and expanded into clonal lines (Fig. 6A). One of these lines, Red1.2, exhibited high transgene expression and retained a modal chromosome number of 42 (Fig. 6B). We injected these cells, expanded for 9 passages after transfection, into 75 SD blastocysts that were transferred to 6 pseudopregnant females. One of the females was sacrificed for assessment of foetal chimaerism at E13.5. Two embryos were recovered and chimaeric contribution assessed by fluorescence microscopy. Both foetuses displayed significant DsRed fluorescence throughout the whole embryo but not in the yolk sac (Fig. 6C). The genital ridges were dissected and immunostained for DsRed and Oct4 to identify germ cells. DsRed was detected throughout the genital ridge and surrounding mesonephric tissue and also in numerous Oct4-positive germ cells (Fig. 6D,E).
Fig. 6.
Contribution of rat EG cells to embryonic chimaeras including germ cells. (A) Phase contrast and fluorescence images of Red1.2 EG cell clone. (B) Graph showing chromosome number per cell in 50 metaphase spreads from line Red1.2. Inset is a karyotype of Red1.2, showing 42 chromosomes. (C) Bright-field and fluorescence images of E13.5 chimaeric embryos. (D) Confocal section of genital ridge and surrounding mesonephric tissue dissected from a chimaeric embryo and immunostained for DsRed and Oct4. (E) Higher magnification confocal section of D shows cells co-stained for DsRed and Oct4, demonstrating the contribution of EG cells to germ cells. Scale bars: 100 μm in A,D; 25 μm in E.
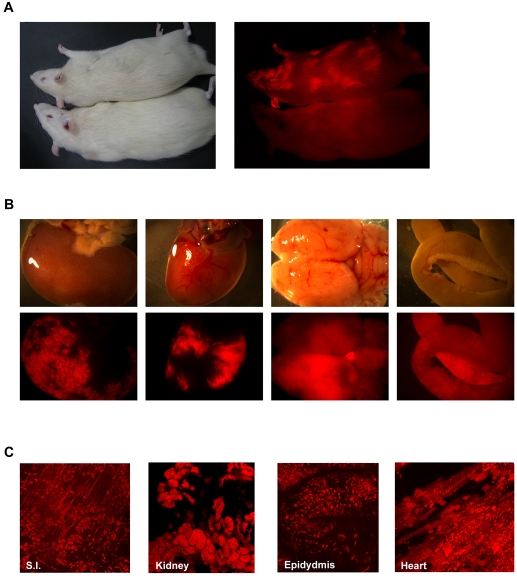
The remaining females produced 40 live-born offspring, 26 of which were overtly chimaeric by observation of DsRed using a fluorescence miner's lamp. Five of these chimaeras were graded high contribution, 9 medium and 12 low. After 8 weeks, we sacrificed 3 medium-contribution chimaeras to assess tissue chimaerism. Under an excitation lamp, significant fluorescence was emitted through the coat, as well as from the ears and paws of chimaeric animals, but not wild-type controls (Fig. 7A). Organs were dissected and chimaeric contribution assessed by fluorescence microscopy. Red fluorescence was evident in all tissues examined, including kidneys, heart, brain and intestine (Fig. 7B). Confocal sections through unfixed tissue squashes demonstrated chimaeric contribution to the tissue parenchyma as well as stromal cells (Fig. 7C). We conclude that rat EG cells are competent to give rise to healthy adult chimaeras and to differentiate into fully functional adult tissues of all three germ layers. Chimaeras have developed into healthy adults. They are fertile but their competence for germline transmission remains to be determined because they have yet to produce offspring carrying the DsRed transgene.
Fig. 7.
Contribution of rat EG cells to adult chimaeras. (A) Adult chimaera and wild-type control. Fluorescence is visible in the ears, paw and through the coat of the chimaera but not the wild-type control. (B) Bright-field and fluorescence images of kidney, heart, brain and small intestine from a medium-contribution chimaera show extensive contribution of rat EG cells (red fluorescence) to a wide range of tissues. (C) 3D projection of serial confocal sections through unfixed tissue squashes demonstrate the contribution of rat EG cells (red fluorescence) to tissue parenchyma and stromal cells.
DISCUSSION
These findings establish that mouse and rat EG cells can be efficiently derived and propagated in 2i-LIF culture conditions, indicating that they acquire an equivalent molecular and developmental ground state to ES cells (Nichols and Smith, 2009).
The barrier separating PGCs from pluripotency is considered to have a different epigenetic basis from the restriction in somatic cells (Durcova-Hills et al., 2008; McLaren, 1992). PGCs are unipotent, but they are also the cells of origin of testicular teratocarcinoma and can acquire pluripotency spontaneously when grafted to ectopic sites (Stevens, 1983). EG cell formation appears to recapitulate this conversion in vitro but without genetic transformation. PGCs express many of the core pluripotency transcription factors. They also lack somatic determination factors. Elucidating how resetting to ground state pluripotency is accomplished in PGCs complements and might inform understanding of transcription-factor-induced somatic cell reprogramming (Takahashi and Yamanaka, 2006). Attempts to delineate the stimuli and molecular transitions that mediate derestriction of PGCs to pluripotency have been frustrated, however, by the complexity and low efficiency of the culture system (Durcova-Hills and McLaren, 2004; Shamblott et al., 2004). Our findings illustrate that use of 2i-LIF opens up the possibility of a systematic description of EG cell derivation. From previous studies, it had been inferred that initial provision of SCF and 24- to 48-hour exposure to FGF are key requirements for EG cell derivation (Durcova-Hills et al., 2006). The convention was that these factors, in combination with serum factors, might initiate reprogramming (Durcova-Hills et al., 2008). However, our results demonstrate that neither SCF, bFGF or serum are necessary for formation of EG cell colonies in 2i-LIF. This suggests that 2i-LIF might be minimally sufficient, although we do not exclude a potential contribution of paracrine stimuli in the culture. Initial exposure to bFGF and serum does increase the total yield of EG cells. This might be owing to a contribution to reprogramming or might be indirect by enhancing the attachment, survival and/or initial proliferation of PGCs in vitro (Matsui et al., 1992). Whatever the effect, it should be independent of the Erk cascade, which is fully blocked in 2i. Interestingly, it has been reported that during EG cell derivation, bFGF might be substituted by hyperactivation of Akt (Kimura et al., 2008), a potent survival and proliferative stimulus that acts in part by inhibiting Gsk3.
EG cell generation without serum or bFGF confirms that 2i-LIF does not act merely to amplify cells that have already been reprogrammed, but directly mediates pluripotent conversion. 2i-LIF also has both inductive effects in the conversion of incompletely reprogrammed somatic cells to pluripotent status (Silva et al., 2008). It is important to note, however, that reprogramming somatic cells requires gene transfection (Takahashi and Yamanaka, 2006) and has not been effected using 2i-LIF alone. Furthermore, the frequency of PGC conversion in 2i-LIF is at least an order of magnitude higher than that of iPS cell generation after gene transfer.
Previous efforts to derive EG cells from rats have been unsuccessful. The relative ease of establishing them using 2i-LIF demonstrates that EG cells are not a mouse-specific phenomenon. Rather than any intrinsic barrier to regaining pluripotency, earlier attempts to convert rat PGCs failed owing to the use of inappropriate culture conditions. Consistent with this, when rat EG cells are cultured in serum, they either differentiate or die. There do appear to be differences in the dynamics of reprogramming between mouse and rat PGCs, with the rat EG cell colonies emerging only after passaging. This might reflect an intrinsic difference in the kinetics of EG cell induction. Alternatively, it could be due to greater sensitivity of rat cells when feeders are eliminated during initial exposure to 2i, or to sub-optimal culture conditions for clonongenic expansion of rat pluripotent stem cells (Li et al., 2008). Overall, however, our findings support the view that the sources and properties of pluripotent stem cells are broadly similar between mice and rats. The preimplantation epiblast and PGCs of both species are sources of pluripotent chimaera-forming stem cell lines. These naïve pluripotent stem cells are maintained in a self-renewing state by inhibiting Erk signalling and Gsk3, and are responsive to LIF/Stat3. Furthermore, in both species, it is possible to derive EpiSCs from the postimplantation epiblast (Brons et al., 2007; Tesar et al., 2007), which appear to represent a later, primed state of development that has acquired dependency on Erk signalling and cannot be propagated in 2i-LIF (Nichols and Smith, 2009).
Rat EG cells appear indistinguishable from rat ES cells. They show identical marker profile and differentiate similarly in culture. In the present study we obtained a higher frequency of chimaeras than in previous studies using ES cells (Buehr et al., 2008; Li et al., 2008). However, this might be because we used recipients of the same strain. Alternatively, it might be due to greater sensitivity of fluorescent reporter detection relative to coat colour pigmentation, which can be heavily skewed owing to the hooded allele in rats. The rat embryo might in fact be as amenable to pluripotent stem cell derivation as that of the mouse. Rat EG cells might offer an alternative to rat ES cells in efforts to generate gene-targeted rats and establish the platform for implementing sophisticated transgenic technologies in this important model organism.
Finally, on a practical note, it will be interesting to assess whether application of 2i-LIF might be of utility in deriving EG cell lines with ground state properties from other organisms. It has previously been noted that PGCs should be a promising source for pluripotent stem cells from livestock and other species, including human, in which epiblast development is distinct from rodents (McLaren, 1992) and facultative diapause does not occur (Nichols et al., 2001).
Supplementary Material
Acknowledgements
We thank Sam Jameson, Keith Savill and staff for excellent animal husbandry; Rachael Walker for flow cytometry support; Margaret McLeish for assistance with histology; Caroline Lee for technical assistance; and Ken Jones for assistance with tissue culture. We thank Gabriela Durcova-Hills and Petra Hajkova for helpful discussions. This study was funded by the Wellcome Trust, the Medical Research Council and the European Commission projects NeuroStemCell and EURATRANS. H.G.L. is supported by a Merck, Sharp and Dohme Award from the University of Cambridge School of Clinical Medicine MB/PhD programme and the James Baird Fund; K.B. is a Gates Cambridge Scholar; and H.A. is a Wellcome Trust Clinical Resarch Training Fellow. A.S. is a Medical Research Council Professor. We dedicate this paper to the memory of our inspirational colleague Anne McLaren. Deposited in PMC for release after 6 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.050427/-/DC1
References
- Bao S., Tang F., Li X., Hayashi K., Gillich A., Lao K., Surani M. (2009). Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature 461, 1292-1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlle-Morera L., Smith A., Nichols J. (2008). Parameters influencing derivation of embryonic stem cells from murine embryos. Genesis 46, 758-767 [DOI] [PubMed] [Google Scholar]
- Beddington R. S., Robertson E. J. (1989). An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development 105, 733-737 [DOI] [PubMed] [Google Scholar]
- Bradley A., Evans M., Kaufman M. H., Robertson E. (1984). Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature 309, 255-256 [DOI] [PubMed] [Google Scholar]
- Brons I., Smithers L., Trotter M., Rugg-Gunn P., Sun B., Chuva De Sousa Lopes S., Howlett S., Clarkson A., Ahrlund-Richter L., Pedersen R., et al. (2007). Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191-195 [DOI] [PubMed] [Google Scholar]
- Brook F. A., Gardner R. L. (1997). The origin and efficient derivation of embryonic stem cells in the mouse. Proc. Natl. Acad. Sci. USA 94, 5709-5712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehr M., Meek S., Blair K., Yang J., Ure J., Silva J., McLay R., Hall J., Ying Q., Smith A. (2008). Capture of authentic embryonic stem cells from rat blastocysts. Cell 135, 1287-1298 [DOI] [PubMed] [Google Scholar]
- Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V. B., Wong E., Orlov Y. L., Zhang W., Jiang J., et al. (2008). Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106-1117 [DOI] [PubMed] [Google Scholar]
- Dolci S., Williams D. E., Ernst M. K., Resnick J. L., Brannan C. I., Lock L. F., Lyman S. D., Boswell H. S., Donovan P. J. (1991). Requirement for mast cell growth factor for primordial germ cell survival in culture. Nature 352, 809-811 [DOI] [PubMed] [Google Scholar]
- Durcova-Hills G., McLaren A. (2004). Isolation and maintenance of murine embryonic germ cell lines. In Handbook of Stem Cells, vol. 1 (ed. Lanza R.), pp. 451-458, Burlington, San Diego, London: Elsevier Academic Press; [Google Scholar]
- Durcova-Hills G., Surani A. (2008). Reprogramming primordial germ cells (PGC) to embryonic germ (EG) cells. Curr. Protoc. Stem Cell Biol. Chapter 1: Unit 1A.3 [DOI] [PubMed] [Google Scholar]
- Durcova-Hills G., Ainscough J., McLaren A. (2001). Pluripotential stem cells derived from migrating primordial germ cells. Differentiation 68, 220-226 [DOI] [PubMed] [Google Scholar]
- Durcova-Hills G., Adams I., Barton S., Surani M., McLaren A. (2006). The role of exogenous fibroblast growth factor-2 on the reprogramming of primordial germ cells into pluripotent stem cells. Stem Cells 24, 1441-1449 [DOI] [PubMed] [Google Scholar]
- Durcova-Hills G., Tang F., Doody G., Tooze R., Surani M. (2008). Reprogramming primordial germ cells into pluripotent stem cells. PLoS ONE 3, e3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M. J., Kaufman M. H. (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154-156 [DOI] [PubMed] [Google Scholar]
- Godin I., Deed R., Cooke J., Zsebo K., Dexter M., Wylie C. C. (1991). Effects of the steel gene product on mouse primordial germ cells in culture. Nature 352, 807-809 [DOI] [PubMed] [Google Scholar]
- Guo G., Yang J., Nichols J., Hall J., Eyres I., Mansfield W., Smith A. (2009). Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136, 1063-1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehler J., Tolkunova E., Koschorz B., Pesce M., Gentile L., Boiani M., Lomeli H., Nagy A., McLaughlin K., Scholer H., et al. (2004). Oct4 is required for primordial germ cell survival. EMBO Rep. 5, 1078-1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., Tomooka M., Yamano N., Murayama K., Matoba S., Umehara H., Kanai Y., Nakano T. (2008). AKT signaling promotes derivation of embryonic germ cells from primordial germ cells. Development 135, 869-879 [DOI] [PubMed] [Google Scholar]
- Ko K., Tapia N., Wu G., Kim J., Bravo M. J., Sasse P., Glaser T., Ruau D., Han D., Greber B., et al. (2009). Induction of pluripotency in adult unipotent germline stem cells. Cell Stem Cell 5, 87-96 [DOI] [PubMed] [Google Scholar]
- Kurimoto K., Yabuta Y., Ohinata Y., Shigeta M., Yamanaka K., Saitou M. (2008). Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev. 22, 1617-1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labosky P. A., Barlow D. P., Hogan B. L. (1994). Mouse embryonic germ (EG) cell lines: transmission through the germline and differences in the methylation imprint of insulin-like growth factor 2 receptor (Igf2r) gene compared with embryonic stem (ES) cell lines. Development 120, 3197-3204 [DOI] [PubMed] [Google Scholar]
- Li P., Tong C., Mehrian-Shai R., Jia L., Wu N., Yan Y., Maxson R. E., Schulze E. N., Song H., Hsieh C., et al. (2008). Germline competent embryonic stem cells derived from rat blastocysts. Cell 135, 1299-1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar M. K., Feng L., Medlock E., Toksoz D., Williams D. A. (1994). Identification and mutation of primary and secondary proteolytic cleavage sites in murine stem cell factor cDNA yields biologically active, cell-associated protein. J. Biol. Chem. 269, 1237-1242 [PubMed] [Google Scholar]
- Martin G. R. (1981). Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 78, 7634-7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y. (1998). Developmental fates of the mouse germ cell line. Int. J. Dev. Biol. 42, 1037-1042 [PubMed] [Google Scholar]
- Matsui Y., Toksoz D., Nishikawa S., Williams D., Zsebo K., Hogan B. L. (1991). Effect of Steel factor and leukaemia inhibitory factor on murine primordial germ cells in culture. Nature 353, 750-752 [DOI] [PubMed] [Google Scholar]
- Matsui Y., Zsebo K., Hogan B. L. (1992). Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell 70, 841-847 [DOI] [PubMed] [Google Scholar]
- McLaren A. (1992). Embryology. The quest for immortality. Nature 359, 482-483 [DOI] [PubMed] [Google Scholar]
- McLaren A., Lawson K. A. (2005). How is the mouse germ-cell lineage established? Differentiation 73, 435-437 [DOI] [PubMed] [Google Scholar]
- Nichols J., Smith A. (2009). Naive and primed pluripotent states. Cell Stem Cell 4, 487-492 [DOI] [PubMed] [Google Scholar]
- Nichols J., Chambers I., Taga T., Smith A. (2001). Physiological rationale for responsiveness of mouse embryonic stem cells to gp130 cytokines. Development 128, 2333-2339 [DOI] [PubMed] [Google Scholar]
- Nichols J., Jones K., Phillips J. M., Newland S. A., Roode M., Mansfield W., Smith A., Cooke A. (2009a). Validated germline-competent embryonic stem cell lines from nonobese diabetic mice. Nat. Med. 15, 814-818 [DOI] [PubMed] [Google Scholar]
- Nichols J., Silva J., Roode M., Smith A. (2009b). Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development 136, 3215-3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H. (2007). How is pluripotency determined and maintained? Development 134, 635-646 [DOI] [PubMed] [Google Scholar]
- Ohinata Y., Payer B., O'Carroll D., Ancelin K., Ono Y., Sano M., Barton S. C., Obukhanych T., Nussenzweig M., Tarakhovsky A., et al. (2005). Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436, 207-213 [DOI] [PubMed] [Google Scholar]
- Okita K., Ichisaka T., Yamanaka S. (2007). Generation of germline-competent induced pluripotent stem cells. Nature 448, 313-317 [DOI] [PubMed] [Google Scholar]
- Resnick J. L., Bixler L. S., Cheng L., Donovan P. J. (1992). Long-term proliferation of mouse primordial germ cells in culture. Nature 359, 550-551 [DOI] [PubMed] [Google Scholar]
- Selwood L., Johnson M. H. (2006). Trophoblast and hypoblast in the monotreme, marsupial and eutherian mammal: evolution and origins. BioEssays 28, 128-145 [DOI] [PubMed] [Google Scholar]
- Shamblott M. J., Axelman J., Wang S., Bugg E. M., Littlefield J. W., Donovan P. J., Blumenthal P. D., Huggins G. R., Gearhart J. D. (1998). Derivation of pluripotent stem cells from cultured human primordial germ cells. Proc. Natl. Acad. Sci. USA 95, 13726-13731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamblott M. J., Kerr C., Axelman J., Littlefield J. W., Clark G. O., Patterson E. S., Addis R. C., Kraszewski J. N., Kent K. C., Gearhart J. D. (2004). Derivation and differentiation of human embryonic germ cells. In Handbook of Stem Cells, vol. 1 (ed. Lanza R.), pp. 459-470 Burlington, San Diego, London: Elsevier Academic Press; [Google Scholar]
- Silva J., Smith A. (2008). Capturing pluripotency. Cell 132, 532-536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Barrandon O., Nichols J., Kawaguchi J., Theunissen T., Smith A., Goodell M. (2008). Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 6, e253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. (2009). Design principles of pluripotency. EMBO Mol. Med. 1, 251-254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens L. C. (1983). The origin and development of testicular, ovarian, and embryo-derived teratomas. In Cold Spring Harbor Conf. Cell Prolif. vol. 10 (ed. Silver L. M., Martin G. M., Strickland S.), pp. 23-36 New York: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Stewart C. L., Gadi I., Bhatt H. (1994). Stem cells from primordial germ cells can reenter the germ line. Dev. Biol. 161, 626-628 [DOI] [PubMed] [Google Scholar]
- Tada T., Tada M., Hilton K., Barton S. C., Sado T., Takagi N., Surani M. A. (1998). Epigenotype switching of imprintable loci in embryonic germ cells. Dev. Genes Evol. 207, 551-561 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663-676 [DOI] [PubMed] [Google Scholar]
- Tesar P. J., Chenoweth J. G., Brook F. A., Davies T. J., Evans E. P., Mack D. L., Gardner R. L., McKay R. D. (2007). New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196-199 [DOI] [PubMed] [Google Scholar]
- Tucker K. L., Wang Y., Dausman J., Jaenisch R. (1997). A transgenic mouse strain expressing four drug-selectable marker genes. Nucleic Acids Res. 25, 3745-3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnpenny L., Brickwood S., Spalluto C. M., Piper K., Cameron I. T., Wilson D. I., Hanley N. A. (2003). Derivation of human embryonic germ cells: an alternative source of pluripotent stem cells. Stem Cells 21, 598-609 [DOI] [PubMed] [Google Scholar]
- Turnpenny L., Spalluto C., Perrett R., O'Shea M., Hanley K., Cameron I., Wilson D., Hanley N. (2006). Evaluating human embryonic germ cells: concord and conflict as pluripotent stem cells. Stem Cells 24, 212-220 [DOI] [PubMed] [Google Scholar]
- Wang W., Lin C., Lu D., Ning Z., Cox T., Melvin D., Wang X., Bradley A., Liu P. (2008). Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 105, 9290-9295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M., Meissner A., Foreman R., Brambrink T., Ku M., Hochedlinger K., Bernstein B. E., Jaenisch R. (2007). In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 448, 318-324 [DOI] [PubMed] [Google Scholar]
- Wray J., Kalkan T., Smith A. (2010). The ground state of pluripotency. Biochem. Soc. Trans. (in press) [DOI] [PubMed] [Google Scholar]
- Yeom Y. I., Fuhrmann G., Ovitt C. E., Brehm A., Ohbo K., Gross M., Hubner K., Scholer H. R. (1996). Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development 122, 881-894 [DOI] [PubMed] [Google Scholar]
- Ying Q., Stavridis M., Griffiths D., Li M., Smith A. (2003a). Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 21, 183-186 [DOI] [PubMed] [Google Scholar]
- Ying Q. L., Nichols J., Chambers I., Smith A. (2003b). BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115, 281-292 [DOI] [PubMed] [Google Scholar]
- Ying Q., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. (2008). The ground state of embryonic stem cell self-renewal. Nature 453, 519-523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimizu T., Sugiyama N., De Felice M., Yeom Y. I., Ohbo K., Masuko K., Obinata M., Abe K., Scholer H. R., Matsui Y. (1999). Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Dev. Growth Differ. 41, 675-684 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.