Abstract
The bone morphogenetic protein (BMP) signaling pathway regulates multiple developmental and homeostatic processes. Mutations in the pathway can cause a variety of somatic and hereditary disorders in humans. Multiple levels of regulation, including extracellular regulation, ensure proper spatiotemporal control of BMP signaling in the right cellular context. We have identified a modulator of the BMP-like Sma/Mab pathway in C. elegans called DRAG-1. DRAG-1 is the sole member of the repulsive guidance molecule (RGM) family of proteins in C. elegans, and is crucial in regulating body size and mesoderm development. Using a combination of molecular genetic and biochemical analyses, we demonstrate that DRAG-1 is a membrane-associated protein that functions at the ligand-receptor level to modulate the Sma/Mab pathway in a cell-type-specific manner. We further show that DRAG-1 positively modulates this BMP-like pathway by using a novel Sma/Mab-responsive reporter. Our work provides a direct link between RGM proteins and BMP signaling in vivo and a simple and genetically tractable system for mechanistic studies of RGM protein regulation of BMP pathways.
Keywords: BMP, TGFβ, RGM, RGMb, DRAGON, DRAG-1, Sma/Mab signaling, Co-receptor, Body size, Mesoderm, M lineage, C. elegans
INTRODUCTION
Bone morphogenetic proteins (BMPs) belong to the transforming growth factor β (TGFβ) superfamily of ligands and the BMP signaling pathway plays roles in multiple developmental and homeostatic processes (Wu and Hill, 2009). Malfunction of the pathway causes many somatic and hereditary disorders in humans, including cardiovascular diseases and cancer (Gordon and Blobe, 2008; Massagué, 2008). All BMP family members share a common mode of signal transduction (Shi and Massagué, 2003). Upon binding of the BMP ligands, the type II receptor phosphorylates the type I receptor, which then phosphorylates the receptor-regulated SMADs (R-SMADs). These activated R-SMADs then complex with common-mediator SMADs (co-SMADs) and enter the nucleus where the complex participates in regulation of downstream gene expression. Multiple levels of regulation, including extracellular regulation, ensure proper spatiotemporal control of BMP signaling in the right cellular context (Massagué and Chen, 2000; Balemans and Van Hul, 2002; Moustakas and Heldin, 2009; Umulis et al., 2009). In particular, a family of repulsive guidance molecules (RGMs) has recently been found to bind different BMPs with high affinity and act as BMP co-receptors in tissue culture experiments (Samad et al., 2005; Babitt et al., 2005; Babitt et al., 2006).
RGM family proteins are GPI (glycosylphosphatidylinositol)-linked membrane-associated proteins (Corradini et al., 2009; Severyn et al., 2009). Vertebrates have four RGMs: RGMa, RGMb (DRAGON), RGMc (hemojuvelin or HJV) and RGMd (Camus and Lambert, 2007). RGMa, RGMb and RGMc can bind selected BMP molecules as well as type I and type II BMP receptors to enhance BMP signaling in tissue cultures (Babitt et al., 2005; Babitt et al., 2006; Samad et al., 2004; Samad et al., 2005; Xia et al., 2008; Xia et al., 2010; Andriopoulos et al., 2009). Overexpression of mouse RGMb in Xenopus embryos can augment SMAD-1-induced mesodermal and endodermal marker expression (Samad et al., 2005). Several lines of in vivo evidence suggest that RGMc acts as the co-receptor for BMP6 in regulating iron metabolism in mice (Babitt et al., 2007; Xia et al., 2008; Andriopoulos et al., 2009). However, there is, as yet, no evidence showing that RGMa and RGMb are required for modulating BMP signaling in vivo.
In this study, we describe the identification and functional characterization of the sole C. elegans RGM homolog DRAG-1 in modulating a BMP-like signaling pathway. There are two TGFβ-related pathways in C. elegans, a TGFβ-like pathway that controls dauer formation and a BMP-like Sma/Mab pathway that regulates body size, male tail formation, mesoderm development and innate immunity (Savage-Dunn, 2005; Foehr et al., 2006; Moustakas and Heldin, 2009; Partridge et al., 2010). The ligand of the Sma/Mab pathway is a BMP-like molecule DBL-1 (Morita et al., 1999; Suzuki et al., 1999). Additional members of this pathway include the type I receptor SMA-6 (Krishna et al., 1999), type II receptor DAF-4 (Estevez et al., 1993), and SMADs SMA-2, SMA-3 and SMA-4 (Savage et al., 1996). Loss-of-function mutations in any of these pathway members cause small body size and male tail sensory ray formation defects.
The Sma/Mab pathway also plays a role in patterning the C. elegans postembryonic mesoderm. The hermaphrodite postembryonic mesodermal M lineage arises from a single pluripotent precursor cell, the M mesoblast. During larval development, the M mesoblast divides to produce a dorsal lineage that gives rise to macrophage-like coelomocytes (CCs) and striated bodywall muscles (BWMs), and a ventral lineage that produces BWMs and the sex muscle precursor cells, the sex myoblasts (SMs; Fig. 1C) (Sulston and Horvitz, 1977). We have previously shown that mutations in the schnurri homolog sma-9 lead to a dorsal-to-ventral fate transformation in the M lineage (Foehr et al., 2006; Foehr and Liu, 2008). Furthermore, mutations in the core components of the Sma/Mab pathway suppress the dorsoventral patterning defects of sma-9 mutants, suggesting that SMA-9 functions by antagonizing Sma/Mab signaling to pattern the postembryonic mesoderm along the dorsoventral axis.
Fig. 1.
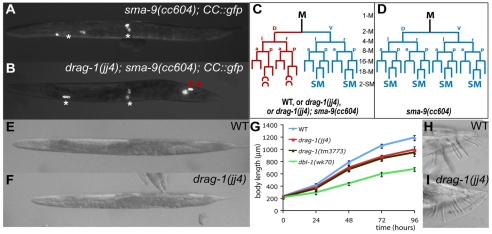
drag-1 mutants exhibit body size and mesodermal defects. All images are ventral or lateral views with anterior to the left. (A-D) Mesoderm phenotype. CC::gfp labels all the coelomocytes (CCs), including embryonically-derived CCs (*) and M-derived CCs (red arrowheads). sma-9(cc604) mutants (A,D) show a dorsal-to-ventral fate transformation in the postembryonic mesoderm lineage, the M lineage, thus lacking M-derived CCs compared with wild-type (C) or drag-1(jj4); sma-9(cc604) double mutants (B,C). (E,F) A wild-type adult (E) and drag-1(jj4) adult (F) at the same stage. (G) Growth curves of wild-type (N2), drag-1(jj4), drag-1(tm3773) and dbl-1(wk70) worms. Fifty to 85 animals were examined for each strain at each time point. By Student's t-test, the body lengths of drag-1(jj4) and drag-1(tm3773) animals are significantly different (P<0.0001) from the body lengths of wild-type and dbl-1(wk70) animals at 24, 48, 72 and 96 hours. (H,I) Tails of a wild-type male (H) and a drag-1(jj4) male (I). SM, sex myoblast. a, anterior; d, dorsal; l, left; p, posterior; r, right; v, ventral.
Among suppressors of the sma-9 phenotype, we detected a mutation in a novel locus that we have named drag-1. drag-1 mutants exhibit only a subset of phenotypes seen in the Sma/Mab pathway mutants. We show that drag-1 encodes the sole RGM homolog in C. elegans. We also show that DRAG-1 is a membrane-localized protein that functions at the ligand-receptor level in the Sma/Mab pathway to regulate body size and mesoderm patterning. Using a novel Sma/Mab-responsive reporter, we demonstrate that DRAG-1 positively modulates Sma/Mab signaling. Our work establishes a direct link between RGM proteins and BMP signaling in vivo and provides a simple and genetically tractable system for mechanistic studies on RGM protein regulation of BMP pathways in vivo.
MATERIALS AND METHODS
C. elegans strains
Strains were kept at normal conditions, as described by Brenner (Brenner, 1974). Analyses were performed at 20°C unless otherwise noted. The following mutations and integrated transgenes were used. Linkage group II (LGII): sma-6(jj6); LGIII: daf-7(m62), daf-7(e1372), daf-4(m72), sma-3(jj3), lon-1(e185), ccIs4438(intrinsic CC::gfp); LGIV: daf-1(m40), daf-1(m213); LGV: dbl-1(wk70); LGX: lon-2(e678), sma-9(cc604), sma-9(cc606), sma-9(wk55). Other strains used included VF1: unc119(ed3); gfEx[phmt-1p::gfp, unc-119(+)] (Schwartz et al., 2010); VF12.3: hmt-1(gk161) III; gfIs1[hmt-1::gfp, unc-119(+)].
Molecular analysis of drag-1
The molecular lesion of jj4 was identified by sequencing PCR fragments spanning the entire genomic region of Y71G12B.16. To determine the drag-1 cDNA sequence, the ProQuest cDNA library (Invitrogen) was used for PCR amplification using primers ForwLongpPC-86 (corresponding to vector sequences 5′-TATAACGCGTTTGGAATCACTACAGGGATGTTTAATACCAC-3′) and DS147 (complementary to sequences spanning the predicted stop codon of drag-1 5′-TCAGCATAACAATGATAAAAGAGCAAAAAAAAAG-3′). The PCR fragments were cloned into the pCR-Blunt II-TOPO vector (Invitrogen). Three of the ten resulting clones sequenced contained the entire Y71G12B.16 open reading frame (ORF) as well as additional 5′UTR sequences. One clone (pDS9.10) has 32 bp of 5′UTR sequences immediately upstream of the ATG (Y71G12B.16b; Genbank HM154524), whereas the other two clones (pDS9.5 and pDS9.6) contain 60 bp 5′UTR sequences (Y71G12B.16a; Genbank HM154523).
Plasmid constructs and transgenic lines
drag-1 reporter constructs
Four kilobases of the drag-1 upstream sequence (−3977 to −1), sequences from immediately downstream of the ATG until the end of the first intron of drag-1 (4 to 1123), the entire drag-1 protein-coding region and 1 kb of the drag-1 downstream sequences were PCR-amplified using N2 genomic DNA as a template. The PCR products were used to generate the following reporter constructs for analyzing the expression pattern of drag-1: pDS14, 4 kb drag-1p::4xNLS::gfp::unc-54 3′UTR; pDS15, 4 kb drag-1p::drag-1 1st intron::4×NLS::gfp::unc-54 3′UTR (transgenic animals carrying either pDS14 or pDS15 gave identical gfp expression patterns; data not shown); pDS27, 4 kb drag-1p::drag-1 genomic ORF::1 kb drag-1 3′UTR; pCXT15, 4 kb drag-1p::drag-1 1st intron::drag-1 cDNA::unc-54 3′UTR; pJKL849, 4 kb drag-1p::drag-1 genomic ORF::gfp::1 kb drag-1 3′UTR. GFP from the Fire Lab Vector pPD95.75 (http://www.addgene.org) was inserted between amino acid (aa) 395 and aa396 of the drag-1 coding region, just prior to the cleavage site of the putative C-terminal pro-peptide.
Constructs for tissue-specific expression of drag-1
pJKL928, myo-2p::drag-1 cDNA::unc-54 3′UTR; pJKL931, elt-2p::drag-1 cDNA::unc-54 3′UTR; pJKL933, elt-3p::drag-1 cDNA::unc-54 3′UTR; pJKL935, rol-6p::drag-1 cDNA::unc-54 3′UTR; pDS20, hlh-8p::drag-1 cDNA::unc-54 3′UTR; pCXT148, hlh-8p::drag-1 cDNA::1 kb drag-1 3′UTR.
Constructs for structure-function analysis of drag-1 (derived from pJKL849)
pCXT92 (DelC::GFP), drag-1::gfp with the putative C-terminal pro-peptide (aa387-408) deleted; pCXT115 (DelN::GFP), drag-1::gfp with the N-terminal signal peptide (aa2-23) deleted; pCXT116 (DelNC::GFP), drag-1::gfp with both the N-terminal signal peptide (aa2-23) and the C-terminal pro-peptide (aa387-408) sequences deleted; pCXT94 (LIN-12TM::GFP), drag-1::gfp with the C-terminal pro-peptide (aa387-408) replaced by the LIN-12 transmembrane domain (aa907-934 of LIN-12) (Yochem et al., 1988).
Other reporters
pJKL840, sma-6p::nls::rfp::lacZ::unc-54 3′UTR [containing 3 kb of the sma-6 promoter (−3094 to −1)].
Generating transgenic animals
Transgenic animals were generated using the plasmids pRF4 (Mello et al., 1991), pC1 (pha-1 rescuing construct) (Granato et al., 1994), pJKL449 (myo-2p::gfp::unc-54 3′UTR) (Jiang et al., 2009) or LiuFD61(mec-7p::mRFP) (Amin et al., 2009) as markers.
Reverse-transcription PCR (RT-PCR)
Total RNA was extracted from mixed-stage wild-type and drag-1(jj4) worms using an RNA extraction kit (Qiagen). Reverse transcription was performed using the Superscript III First Strand Synthesize Kit (Invitrogen) following the manufacturer's instructions. Primers used for amplifying the wild-type and jj4 cDNAs were CXT49 (annealing to exon 1, forward) and CXT50 (annealing to exon 3, reverse). The resulting PCR fragments were purified and sequenced using primer CXT67.
Body size measurement
Hermaphrodite animals at the gravid adult stage were collected and treated with hypochlorite. The resulting embryos were allowed to hatch in M9 buffer at 16°C. Synchronized L1s were plated onto nematode growth medium (NGM) plates and allowed to grow for 24, 48, 72 or 96 hours, respectively, before they were washed off the plates, treated with 0.3% sodium azide and mounted onto 2% agarose pads. Images of the worms were taken on a Leica DMRA2 compound microscope with a Hamamatsu Orca-ER camera using the Openlab software (Improvision). Body length of each animal was measured using Openlab software. Subsequent statistical analyses were performed using Microsoft Excel.
Dauer formation assays
Frequency of dauer formation was assessed under non-dauer-inducing conditions as described (Vowels and Thomas, 1992). Five to ten adult hermaphrodites were placed on a 6 cm NGM plate at the test temperature. After a short period of egg-lay (less than 12 hours at 25°C), the parents were removed from the plates and their progeny were allowed to develop at a given temperature. When the non-dauer worms became young adults, the numbers of dauers and non-dauers on each plate were counted.
Immunofluorescence staining
Animal fixation, immunostaining, microscopy and image analysis were performed as described previously (Amin et al., 2007). Guinea pig anti-FOZI-1 (1:200) and goat anti-GFP (Rockland Immunochemicals; 1:1000) were used. All secondary antibodies were from Jackson ImmunoResearch Laboratories and used at a dilution of 1:50 to 1:200.
Fractionation experiment
Transgenic worms used in the fractionation experiment were generated in the pha-1(e2123ts) mutant background and maintained at 25°C. Worms were harvested in M9 and resuspended in lysis buffer [50 mM Tris-HCI, pH 7.5; 10% glycerol; 10 mM 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride and 1 Complete Mini Protease Inhibitor Cocktail Tablet (Roche) in 50 ml lysis buffer] so that the ratio of worm volume to buffer volume was 1:1. Worms were sonicated until around 90% of the worms were lysed as assessed by microscopy. Debris was cleared by centrifugation at 4000 g for 10 minutes at 4°C. The supernatants were centrifuged at 115,000 g for 1 hour. The resulting supernatant was the soluble fraction. The pellet, which contains microsomal membrane proteins, was further cleared from soluble proteins by two more rounds of centrifugation (115,000 g) and resuspension in lysis buffer. The resulting pellet was reconstituted in the same volume of lysis buffer as the soluble fraction. Micro BCA Protein Assay Reagent Kit (Pierce) was used to measure the protein concentration. Thirty micrograms per lane of samples were loaded onto a 7% SDS-PAGE. GFP fusion proteins were detected by western blotting using goat anti-GFP antibodies (Rockland Immunochemicals; 1:1000).
Generating the RAD-SMAD and BAD-SMAD reporters
Oligonucleotides containing the wild-type (RAD-SMAD) or mutant (BAD-SMAD) SMAD binding sites and StyI restriction sites at each end were treated with T4 polynucleotide kinase, annealed and ligated. Concatenated oligonucleotides were cloned into the Fire Lab Vector L3135 (http://www.addgene.org). Upon confirmation of the plasmid sequence, their DNA was injected into N2 worms using mec-7p::mRFP as a co-injection marker. The RAD-SMAD transgene was then integrated into the genome via gamma-irradiation. Seven integrated lines were obtained, out-crossed and mapped to specific chromosomes. All lines showed similar GFP expression patterns. Two of them, LW2433: jjIs2433[pCXT51(RAD-SMAD) + LiuFD61(mec-7p::mRFP)] X and LW2436: jjIs2436[pCXT51(RAD-SMAD) + LiuFD61(mec-7p::mRFP)] (I or IV) were used for subsequent analysis. The BAD-SMAD transgenes were kept in high-efficiency extra chromosomal lines.
RESULTS
drag-1 mutants exhibit a subset of the phenotypes seen in mutants in the Sma/Mab pathway
In a mutagenesis screen for suppressers of the sma-9(cc604) M lineage phenotype (Foehr et al., 2006), we isolated the Sma/Mab pathway mutants as well as a single-locus recessive mutation, jj4. Unlike sma-9(cc604) animals, which lack both M lineage-derived coelomocytes (CCs; Fig. 1A,D), 98.5% (n=201) of jj4; sma-9(cc604) animals had both M-derived CCs (Fig. 1B,C). jj4 also suppressed the M lineage phenotypes of two other sma-9 alleles, cc606 and wk55 (data not shown), suggesting that this suppression is not allele-specific. Like the Sma/Mab pathway mutants, jj4 mutants by themselves did not have any M lineage defect when separated from the cc604 mutation (Fig. 1C). However, jj4 mutants were smaller than wild-type N2 worms throughout larval development, although they were not as small as the dbl-1(wk70) null mutants (Fig. 1E-G). Unlike the Sma/Mab pathway mutants, jj4 mutant males could mate and did not exhibit any male tail patterning defects (Fig. 1H,I). Thus, jj4 mutants exhibit some, but not all, phenotypes of the Sma/Mab pathway mutants.
We mapped the jj4 mutation to the left arm of chromosome I, a region not previously known to contain any genes involved in body size regulation. Thus jj4 represents a novel locus required to regulate body size. We named the locus drag-1 (see below).
drag-1 functions in the Sma/Mab pathway, possibly at the ligand-receptor level, to regulate body size
Because drag-1(jj4) mutants and the Sma/Mab pathway mutants share similar phenotypes in body size and mesoderm patterning, we tested whether drag-1 functions in the Sma/Mab pathway. We generated double mutants between jj4 (a putative null, see below) and mutations in various Sma/Mab pathway components (Fig. 2A) and measured their body sizes.
Fig. 2.
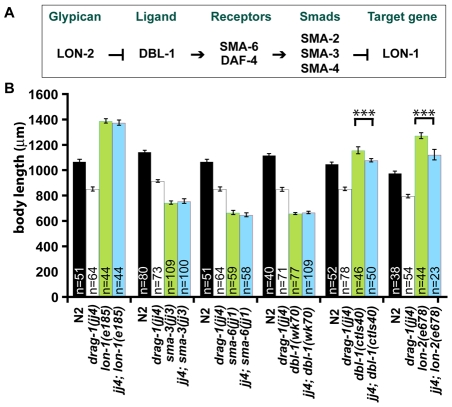
drag-1 functions at the ligand-receptor level in the Sma/Mab pathway to regulate body size. (A) A simplified schematic of the Sma/Mab pathway (see text for details). (B) Body length of wild-type (N2) and various mutant worms at 96 hours post-plating (see Materials and methods). Similar results were observed using worms at 24, 48 and 72 hours post-plating (data not shown). drag-1(jj4); lon-2(e678) and drag-1(jj4); dbl-1(ctIs40) worms show significantly different body length compared with lon-2(e678) and dbl-1(ctIs40) worms, respectively. Error bars represent 95% confidence intervals for the mean body length. The significance of difference between double mutants and the corresponding single mutants was statistically analyzed via Student's t-test. ***, P<0.0001.
As shown in Fig. 2B, double mutants between jj4 and the sma-3(jj3) null mutation were as small as sma-3(jj3) single mutants. Similarly, jj4; sma-6(jj1) and jj4; dbl-1(wk70) double mutants were as small as sma-6(jj1) and dbl-1(wk70) single mutants (both are null mutants), respectively. These observations suggest that jj4 is probably compromised for Sma/Mab pathway function, rather than affecting a pathway that functions in parallel to the Sma/Mab pathway, in regulating body size.
To further characterize the role of drag-1 in the Sma/Mab pathway, we generated the following double mutants: jj4; lon-1(e185), jj4; lon-2(e678) and jj4; dbl-1(ctIs40). e185 is a strong loss-of-function mutation in lon-1, a known downstream target of the Sma/Mab pathway (Maduzia et al., 2002; Morita et al., 2002). e678 is a null mutation in lon-2, which encodes a member of the glypican family of heparan sulfate proteoglycans and acts as a negative regulator of the Sma/Mab pathway (Gumienny et al., 2007). dbl-1(ctIs40) is a strain that overexpresses dbl-1 (Suzuki et al., 1999). As shown in Fig. 2B, jj4; lon-1(e185) mutants were as long as lon-1(e185) mutants. However, jj4; lon-2(e678) and jj4; dbl-1(ctIs40) double mutants showed intermediate body size between jj4 single mutants and lon-2(e678) or dbl-1(ctIs40) single mutants. These results suggest that drag-1 is likely to act upstream of lon-1, but in parallel to lon-2 and dbl-1. Taken together, our genetic epistasis results are consistent with drag-1 functioning at the ligand-receptor level in the Sma/Mab pathway to regulate body size.
drag-1 interacts genetically with the dauer pathway
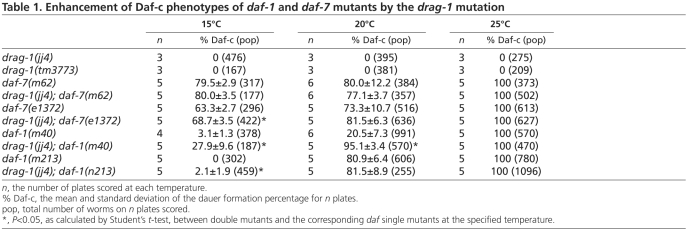
In addition to the Sma/Mab pathway, C. elegans has another TGFβ pathway that regulates dauer development (Savage-Dunn, 2005). drag-1(jj4) animals did not exhibit any constitutive or deficient dauer phenotypes on their own (Table 1). Animals carrying a deletion allele of drag-1(tm3773) (see below), did not exhibit any dauer phenotypes either (Table 1). However, drag-1(jj4) enhanced the dauer-constitutive (Daf-c) phenotype of certain daf-7 and daf-1 alleles at the permissive temperature. daf-7 and daf-1 encode the ligand and the type I receptor of the dauer pathway, respectively (Georgi et al., 1990; Ren et al., 1996). As shown in Table 1, although drag-1(jj4) did not affect the Daf-c phenotype of daf-7(m62) animals at all temperatures tested, drag-1(jj4) moderately enhanced the Daf-c phenotype of daf-7(e1372) mutants at 16°C. Similarly, drag-1(jj4) moderately enhanced the Daf-c phenotype of daf-1(m213) mutants at 16°C, but significantly enhanced the Daf-c phenotype of daf-1(m40) mutants at both 16°C and 20°C. The allele-specific and moderate enhancement of Daf-c phenotypes of daf-7 and daf-1 mutants by drag-1(jj4) are consistent with low levels of crosstalk between the Sma/Mab and dauer pathways, as suggested by previous observations on the genetic interactions between sma-6 and daf-7 and daf-1 mutations (Krishna et al., 1999).
Table 1.
Enhancement of Daf-c phenotypes of daf-1 and daf-7 mutants by the drag-1 mutation
drag-1 encodes the C. elegans homolog of the RGM family proteins
drag-1(jj4) was mapped to the left arm of chromosome I between Y92H12A.3 and Y71G12B.24 via snip-SNP mapping (Wicks et al., 2001). RNAi injection for 15 likely candidates among the 24 predicted genes located in this region showed that Y71G12B.16(RNAi) led to a small body size phenotype and the suppression of the sma-9(cc604) M lineage phenotype. For simplicity, we will refer to the suppression of the sma-9(cc604) M lineage phenotype by jj4 as the SOSMLP phenotype of jj4 in the remainder of the manuscript. Several additional lines of evidence indicated that jj4 is a loss-of-function allele of Y71G12B.16. First, DNA fragments containing the genomic DNA sequence of Y71G12B.16 rescued both the small body size and the SOSMLP phenotype of jj4 mutants (Fig. 3C,D; Fig. 5O). Second, sequencing of Y71G12B.16 in jj4 mutants revealed a T-to-A single nucleotide mutation in the splicing donor site in the first intron (Fig. 3C). RT-PCR analysis (see Materials and methods) showed that this mutation led to the production of three aberrant drag-1 transcripts, all producing mutant peptides containing the first 16 amino acids of DRAG-1 followed by 9-49 additional random amino acids (data not shown). Third, a deletion allele of drag-1, tm3773, led to the same body size and SOSMLP phenotypes as jj4 mutants (Fig. 1G; Fig. 3C,D; Fig. 5O).
Fig. 3.
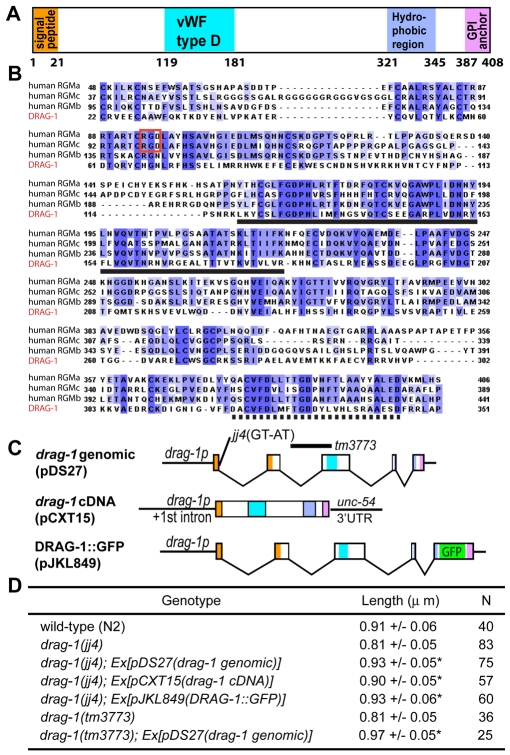
drag-1 encodes a putative GPI-anchor protein of the RGM family. (A) A schematic of the DRAG-1 protein, showing the various conserved motifs drawn in scale. (B) Alignment between DRAG-1 (the bottom line) and its human RGM homologs RGMa (GenBank AAI51133.1), RGMb (GenBank NP_001012779.2) and RGMc (GenBank NP_998818.1) in regions between the N-terminal signal peptide and the C-terminal pro-peptide. Notice the highly conserved vWF type D domain (solid underline) and hydrophobic region (dashed underline). The RGD motif (boxed in red) is present in RGMa and RGMc, but not in RGMb and DRAG-1. (C) Diagrams of the drag-1 genomic, cDNA and GFP-tagged constructs. The locations of the jj4 and tm3773 molecular lesions are shown. (D) Body size measurement of worms of various genotypes at 72 hours post-plating. Length is presented as the mean ± standard deviation. Asterisks indicate that the length of the drag-1 worms carrying a particular transgene is significantly different from that of the corresponding drag-1 single mutant. P<0.0001.
Fig. 5.
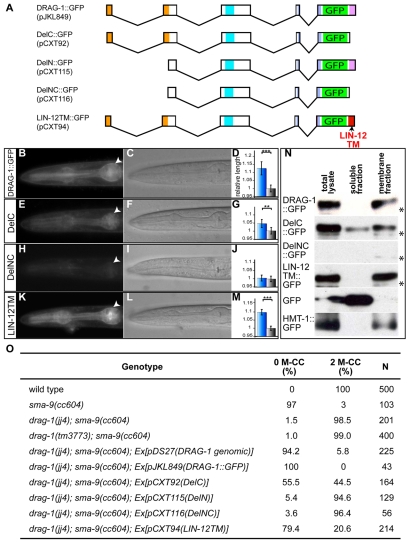
drag-1 is localized to and functions at the cell membrane. (A) Schematics of various DRAG-1 deletion constructs (see Material and methods). (B-M) GFP localization (B,E,H,K; arrowheads point to the surface of the pharynx), the corresponding DIC images (C,F,I,L) and body size measurement (D,G,J,M) of animals containing the specific transgenes. DRAG-1::GFP (B-D), DelC::GFP (E-G; abbreviated as DelC), DelNC::GFP (H-J; abbreviated as DelNC) and LIN-12TM::GFP (K-M; abbreviated as LIN-12TM). In panels D, G, J and M: blue bars, drag-1(jj4) animals carrying the transgene; grey bars, drag-1(jj4) non-transgenic animals. The y-axis shows the relative body length, with the lengths of non-transgenic worms of each group being normalized to 1. Between 30 and 70 animals were measured for each genotype. Error bars represent 95% confidence intervals for the mean relative body length. The significance of difference between transgenic and the corresponding non-transgenic animals was statistically analyzed via Student's t-test. ***, P<0.0001. **, P<0.001. (N) Western blots probed with anti-GFP antibodies showing the localization of DRAG-1::GFP fusions via fractionation experiments. Asterisks refer to non-specific bands recognized by the anti-GFP antibodies. (O) Rescue of the mesodermal phenotype of jj4 mutants by various drag-1::gfp constructs. M-CC, M-derived coelomocytes.
We isolated two cDNA variants for Y71G12B.16, Y71G12B.16a and Y71G12B.16b, which share the same coding sequences and differ only in the lengths of their 5′UTRs (see Materials and methods). The predicted Y71G12B.16 protein is 408 aa long. It contains a predicted N-terminal signal peptide, a partial von Willebrand factor D (vWF type D) domain, a hydrophobic region and a C-terminal region that meets the criteria of a pro-peptide that is cleaved off and replaced by a glycosylphosphatidylinositol (GPI) anchor during post-translational processing (Chatterjee and Mayor, 2001; Bohme and Cross, 2002). Blast search showed that Y71G12B.16 has homologs in sea urchins, tunicates, mollusks and vertebrates, but not in Drosophila. The vertebrate Y71G12B.16 homologs belong to the RGM (repulsive guidance molecule) family that includes RGMa, RGMb (DRAGON), RGMc (hemojuvelin) and the recently identified RGMd (Camus and Lambert, 2007; Corradini et al., 2009). Y71G12B.16 appears to be a distant member of the RGM family (Camus and Lambert, 2007), sharing 22%, 21% and 18% identity to human RGMb, RGMa and RGMc, respectively, in the region flanked by the N-terminal signal peptide and the C-terminal pro-peptide (Fig. 3B). In particular, Y71G12B.16 and human RGMb share 46% identity in the vWF type D domain and 40% identity in the hydrophobic region. Like RGMb, Y71G12B.16 lacks the RGD (Arg-Gly-Asp) motif found in RGMa and RGMc proteins (Fig. 3B). We therefore named Y71G12B.16 DRAG-1. Like C. elegans, other invertebrates including sea urchins, tunicates and mollusks also contain a single RGM-related protein in each genome (Camus and Lambert, 2007).
RGM proteins can function as BMP co-receptors (Babitt et al., 2005; Babitt et al., 2006; Samad et al., 2005). Our genetic evidence described above suggests that DRAG-1 might also function as a co-receptor in the Sma/Mab pathway in C. elegans.
drag-1 is expressed in the same cell types as the Sma/Mab pathway type I receptor sma-6
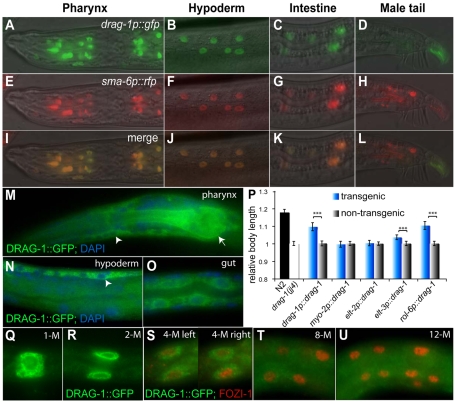
To test the hypothesis that DRAG-1 might function as a co-receptor for the Sma/Mab pathway, we first asked whether drag-1 is expressed in the same cells that express core components of the Sma/Mab pathway. A drag-1 genomic construct (pDS27), which contains the entire drag-1 genomic region including 4 kb 5′ sequences and 1 kb 3′ sequences, fully rescued the body size defects of both drag-1(jj4) and drag-1(tm3773) mutants (Fig. 3C,D). Similarly, a drag-1 cDNA construct (pCXT15; see Materials and methods), with the genomic coding region of drag-1 replaced by drag-1 cDNA and the drag-1 3′UTR replaced by unc-54 3′UTR, also rescued the body size defects of drag-1(jj4) mutants (Fig. 3C,D). These results suggest that the regulatory elements required for drag-1 function in regulating body size probably reside in the upstream sequences used in pCXT15. We then generated a transcriptional drag-1p::gfp fusion pDS15 using these upstream sequences (see Materials and methods). Transgenic lines carrying pDS15 showed GFP expression in pharyngeal, hypodermal and intestinal cells (Fig. 4A-C), the same cells that express sma-6 in hermaphrodites (Krishna et al., 1999; Yoshida et al., 2001). In fact, transgenic lines carrying both drag-1p::gfp and sma-6p::mRFP showed co-localization of the two reporters in all three tissues described above (Fig. 4A-C,E-G,I-K). Therefore, drag-1 is expressed in the same cell types as sma-6 in hermaphrodites. However, drag-1p::gfp is not expressed in the male tail cells that express sma-6p::mRFP (Fig. 4D,H,L), consistent with the lack of male tail defects in drag-1(jj4) mutant males.
Fig. 4.
drag-1 is expressed and functions in the same cell types as the Sma/Mab pathway components. (A-L) Images shown are side views with anterior to the left and posterior to the right. drag-1p::gfp (A-D) and sma-6p::rfp (E-H) colocalize in pharyngeal (A,E,I), hypodermal (B,F,J) and intestinal (C,G,K) cells, but do not colocalize in the male tail (D,H,L). I-L are merged images of A-D and E-H, respectively. (M-O) DRAG-1::GFP (M,N) or LIN-12TM::GFP (O; see Fig. 5 for details) localization in pharyngeal (M), hypodermal (N) and intestinal (O) cells, as visualized by anti-GFP antibody staining. Note that the GFP signal (green) is located outside of the nucleus (blue, stained with DAPI), both at the cell surface and inside the cell. Arrow heads, hypodermal cells; arrow, pharynx. (P) Tissue-specific rescue of the body size phenotype of drag-1(jj4) mutants. Body size was measured in adult worms at 72 hours post-plating. Transgenic worms were distinguished from non-transgenic ones by the presence of mec-7p::rfp. The lengths of drag-1(jj4) and non-transgenic worms of each group were normalized to 1. The relative lengths of wild-type and various transgenic animals compared with their corresponding non-transgenic controls are presented. Between 30 and 80 animals were measured for each genotype. Error bars represent 95% confidence intervals for the mean of relative body length. The significance of difference between transgenic and the corresponding non-transgenic animals was statistically analyzed via Student's t-test. ***, P<0.0001. (Q-U) The M lineage expression pattern of drag-1 using LIN-12TM::GFP (green; see Fig. 5 for details). Anti-FOZI-1 antibody staining (red) was used to mark M lineage cells from the 4-M to the 12-M stage (S-U). Only one focal plane was shown for the 8-M (T) and 12-M (U) stage worms. DRAG-1::GFP is present from the 1-M to the 4-M stage (O-S), then becomes fainter at the 8-M stage (T) and undetectable after the 8-M stage (U).
The same expression pattern of drag-1 was also observed using a functional translational DRAG-1::GFP fusion (pJKL849), which has GFP inserted right before the putative cleavage site upstream of the C-terminal pro-peptide sequence (Fig. 3B). This reporter is fully functional as it rescued both the body size and the SOSMLP of jj4 mutants (Fig. 3D; Fig. 5D,O). Just like the drag-1p::gfp transcriptional fusion, DRAG-1::GFP was present in pharyngeal, hypodermal and intestinal cells (Fig. 4M-O). Robust expression in the M lineage was also detected from the 1-M to 4-M stage (Fig. 4Q-S). DRAG-1::GFP signal was reduced (40%, n=20) or undetectable (60%, n=20) at the 8-M stage (Fig. 4T), and completely undetectable beyond the 8-M stage (Fig. 4U; data not shown). The M lineage expression pattern of drag-1 is consistent with previous findings that sma-9 functions prior to the 8-M stage for proper M lineage development (Foehr et al., 2006).
drag-1 functions in the same cells as the Sma/Mab pathway receptors and SMADs to regulate body size and mesoderm patterning
The Sma/Mab pathway receptors and SMAD proteins have been shown to function in the hypodermal cells to regulate body size (Yoshida et al., 2001; Wang et al., 2002). Because drag-1 is expressed in the same tissues as sma-6 in hermaphrodites, we tested whether drag-1 also functions in hypodermal cells to regulate body size. We forced the expression of drag-1 cDNA in pharyngeal muscles [using the myo-2 promoter (Okkema et al., 1993)], intestine [using the elt-2 promoter (Fukushige et al., 1998)], and in hypodermal cells [using both the elt-3 and rol-6 promoters (Kramer and Johnson, 1993; Gilleard et al., 1999)], and tested the ability of each transgene to rescue the small body size of jj4 mutants. As a control, drag-1 cDNA under the control of its own promoter could rescue the small body size phenotype of jj4 mutants (Fig. 4P). As shown in Fig. 4P, forced expression of drag-1 in hypodermal cells rescued the small body size phenotype of jj4 mutants, whereas expression in the pharynx and intestine did not. Thus, drag-1 functions in the same cells as the Sma/Mab receptors and SMAD proteins to regulate body size.
Previous studies have shown that sma-9 and the Sma/Mab pathway function within the M lineage to regulate dorsoventral patterning of the M lineage (Foehr et al., 2006). We therefore forced the expression of drag-1 within the M lineage in drag-1(jj4); sma-9(cc604) double mutants using the hlh-8 promoter (Harfe et al., 1998). This M lineage-specific expression of drag-1 (using pCXT148) is sufficient to rescue the SOSMLP phenotype of jj4 mutants (24.5%, n=106), i.e. to reverse the M lineage phenotype of drag-1(jj4); sma-9(cc604) mutants to that of sma-9(cc604). Thus, drag-1 also functions in the same cells as sma-9 and the Sma/Mab pathway to regulate M lineage development.
DRAG-1 localizes to and functions at the cell membrane
The translational DRAG-1::GFP reporter described above allowed us to examine the sub-cellular localization pattern of DRAG-1. As shown in Fig. 4M-O and Fig. 5B-C, DRAG-1::GFP was present outside of the nucleus but localized to the cell surface as well as inside the cell. To determine if DRAG-1 protein is membrane-associated, we performed cell fractionation experiments to separate the soluble fraction and the membrane fraction using lysates from mix-staged worm populations (see Materials and methods). As controls, we used worms expressing GFP alone under the control of the hmt-1 promoter, and worms expressing the HMT-1::GFP fusion under the control of the hmt-1 promoter (Schwartz et al., 2010) (see Materials and methods). HMT-1 is a transmembrane half-molecule ATP-binding cassette transporter required for heavy metal detoxification (Vatamaniuk et al., 2005). As shown in Fig. 5N, GFP was only detected in the soluble fraction, whereas HMT-1::GFP was only detected in the membrane fraction. In the same experiment, DRAG-1::GFP was only detected in the membrane fraction. This result, in combination with results from immunostaining, demonstrates that DRAG-1 is a membrane protein, as predicted for a putative GPI-anchor protein.
To determine whether DRAG-1 functions at the cell membrane, we generated transgenic lines expressing DelC::GFP, which has the C-terminal pro-peptide deleted from DRAG-1 (Fig. 5A). DelC::GFP showed reduced signal at the cell surface (Fig. 5E,F) and became partially soluble in cell fractionation experiments (Fig. 5N). We then checked the functionality of DelC::GFP and found that it could rescue both the body size and the SOSMLP phenotypes of drag-1(jj4) mutants, but the rescuing efficiency was reduced when compared with wild-type DRAG-1::GFP (Fig. 5D,G,O). These data together suggest that the putative C-terminal pro-peptide is important, but not essential, for DRAG-1 membrane localization and function.
The membrane localization and function of DRAG-1::GFP and DelC::GFP appeared to require that the proteins enter the secretory pathway via their N-terminal signal peptide. DelN::GFP and DelNC::GFP with their N-terminal signal peptide deleted (Fig. 5A) failed to rescue the drag-1(jj4) mutant phenotypes (Fig. 5J,O) and showed a drastically reduced level of expression (Fig. 5H,I,N), possibly due to the mis-targeting and subsequent degradation of the mutant protein.
To directly test whether DRAG-1 functions at the cell membrane, we replaced the putative C-terminal pro-peptide sequence of DRAG-1 with the transmembrane domain of a well-characterized transmembrane protein LIN-12, LIN-12TM::GFP (Fig. 5A). LIN-12TM::GFP was solely detected in the membrane fraction (Fig. 5N), showed cell surface localization (Fig. 5K,L) and fully rescued the body size and SOSMLP phenotypes of drag-1(jj4) mutants (Fig. 5M,O). Thus, DRAG-1 is not only present, but also functions, at the cell membrane.
DRAG-1 positively modulates Sma/Mab signaling as indicated by a Sma/Mab-responsive reporter
The phenotypes of drag-1 mutants described above suggest that DRAG-1 positively modulates Sma/Mab signaling. To test this hypothesis, we generated a Sma/Mab-responsive reporter as there are no existing phospho-SMAD antibodies or appropriate SMAD::GFP reporters or anti-SMAD antibodies that allow us to directly monitor the output of Sma/Mab signaling.
Previous work has shown that individual SMAD complexes are capable of binding to three abutting SMAD boxes (or SMAD binding site GTCT) with high affinity (Johnson et al., 1999). Furthermore, abutting SMAD boxes organized in the RLR orientation (R, rightward; L, leftward) are bound by SMAD complexes with higher affinity than those organized in the RLL or RRR orientation (Johnson et al., 1999). We decided to place multiple copies of the SMAD boxes organized in the RLR orientation upstream of the minimal pes-10 promoter and gfp (Fig. 6A) and tested whether the expression of the gfp reporter is responsive to Sma/Mab signaling. We named the reporter as RAD-SMAD (reporter acting downstream of SMAD). The expression pattern of the gfp reporter was examined using two different integrated transgenic lines carrying RAD-SMAD (see Materials and methods), which showed similar expression patterns. We detected GFP signal from late embryogenesis until adulthood. GFP signal was detected in intestinal and hypodermal cells, including those from the P-lineages (Fig. 6B) and cells in the male tail (data not shown). The expression level of the reporter appeared rather dynamic during development, with stronger intestinal expression in embryos and L1s, but fainter in adults, and stronger hypodermal expression in L2 and L3 stages (data not shown). No gfp expression was detected in dauer animals either in the wild-type background or the daf-7(e1372) background (data not shown).
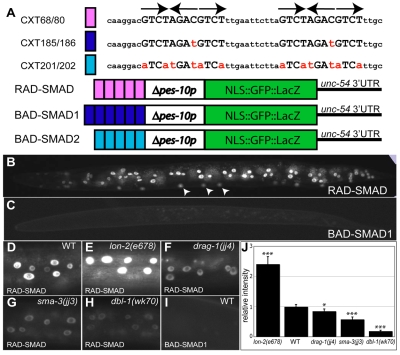
Fig. 6.
A RAD-SMAD reporter directly and positively responds to Sma/Mab signaling in vivo. (A) Schematics of the RAD-SMAD, BAD-SMAD1 and BAD-SMAD2 reporters, with the oligonucleotide sequences shown above. Mutated sequences in the BAD-SMAD1 and BAD-SMAD2 reporters are shown in red lower case. (B,D-H) Expression of the RAD-SMAD GFP reporter in wild-type (B,D), lon-2(e678) (E), drag-1(jj4) (F), sma-3(jj3) (G) and dbl-1(wk70) (H) worms at the L2 stage. Arrowheads in B point to hypodermal cells derived from the P-lineages. (C,I) The BAD-SMAD1 GFP reporter shows no expression. Hypodermal cells are shown in D-I. Pictures were taken at the same exposure. (J) Quantification of hypodermal RAD-SMAD GFP signals in various mutants. In each mutant background, the pixel intensities of GFP signals from 10 hypodermal nuclei in 10 different worms (20 worms for wild-type) at the L2 stage were measured by the Openlab software. The signal intensity in wild-type worms was set to 1 and compared with the signal intensity in each mutant background. Error bars represent 95% confidence intervals for the means of relative intensity. The significance of difference between wild-type and each mutant was statistically analyzed via Student's t-test. *, P<0.01; ***, P<0.0001.
To test the specificity of the RAD-SMAD reporter, we generated two reporters carrying different mutations in the SMAD boxes, BAD-SMAD1 and BAD-SMAD2 (bad reporter of SMAD) (Fig. 6A). Transgenic worms carrying either reporter showed no gfp expression in any cells (Fig. 6C,I; data not shown). This result indicates that the SMAD boxes are directly responsible for RAD-SMAD reporter expression.
To test if the expression level or pattern of the RAD-SMAD reporter are regulated by Sma/Mab signaling, we crossed the two integrated RAD-SMAD reporters (jjIs2433 and jjIs2436) into different Sma/Mab pathway mutants and saw similar results with both reporters. As shown in Fig. 6D,E and 6G,H, RAD-SMAD expression in all cell types was downregulated in dbl-1 and sma-3 null mutants, but upregulated in lon-2 null mutants. In fact, quantification of the GFP signal intensity in various mutants correlated well with the body sizes of each mutant (Fig. 6J, compared with the body size of each mutant in Fig. 2). These results demonstrate that RAD-SMAD positively responds to Sma/Mab signaling.
As shown in Fig. 6F,J, RAD-SMAD expression was significantly downregulated in drag-1(jj4) mutants. Furthermore, the degree of reporter downregulation in jj4 mutants was smaller than that in sma-3(jj3) and dbl-1(wk70) mutants, again correlating very well with the body sizes of these mutants. These observations indicate that DRAG-1 is a positive regulator of the Sma/Mab pathway.
DISCUSSION
DRAG-1 is a cell type-specific modulator of BMP signaling in C. elegans
Previous studies have shown that RGM proteins can enhance BMP, but not TGFβ, signaling in tissue cultures by binding to selected BMP molecules as well as type I and type II BMP receptors (Babitt et al., 2005; Babitt et al., 2006; Samad et al., 2005). However, except for RGMc, there is no evidence showing that RGMa and RGMb are essential for regulating BMP signaling in vivo (Babitt et al., 2005; Babitt et al., 2006; Babitt et al., 2007; Samad et al., 2005; Xia et al., 2008; Xia et al., 2010; Andriopoulos et al., 2009). Our data demonstrate that the sole RGM homolog DRAG-1 is an integral member of the BMP-like Sma/Mab pathway in C. elegans: (1) drag-1 mutants share similar body size and SOSMLP phenotypes as mutants in all the core members of the Sma/Mab pathway; (2) DRAG-1 is a membrane-localized protein that functions at the ligand-receptor level in the Sma/Mab pathway to regulate body size; and (3) drag-1 is expressed and functions in the same cells as the receptors and SMAD proteins of the Sma/Mab pathway to regulate body size (in hypodermal cells) and mesoderm patterning (in the M lineage). Both the mutant phenotypes and the reduced level of Sma/Mab pathway reporter (RAD-SMAD) expression in drag-1 loss-of-function mutants suggest that DRAG-1 positively regulates Sma/Mab signaling in C. elegans. DRAG-1 appears to function specifically in the BMP-like Sma/Mab pathway, but not the TGFβ-like dauer pathway, as drag-1 mutants do not have any defects in dauer formation. The genetic interaction between drag-1(jj4) and certain daf-7 and daf-1 alleles (Table 1) could be owing to the fact that both the Sma/Mab pathway and the dauer pathway share the same type II receptor DAF-4 and thus exhibit low levels of crosstalk. Consistent with this notion, previous studies have also shown genetic interactions between sma-6 and daf-7 and daf-1 mutations (Krishna et al., 1999).
Interestingly, the function of DRAG-1 in modulating Sma/Mab signaling is cell-type specific. Like other core members of the Sma/Mab pathway, drag-1 functions in both body size regulation and mesoderm patterning. However, drag-1 is not expressed in the male tail and drag-1 mutants do not have any male tail patterning defects. It has been previously suggested that body size and male tail development require different Sma/Mab signaling thresholds, with the male tail requiring a lower level of signaling activity, because a hypomorphic sma-6(e1482) allele has small body size but wild-type male tail morphology (Krishna et al., 1999). Our studies on drag-1 are consistent with this hypothesis and further demonstrate that drag-1 functions to augment the level of Sma/Mab signaling activity in the hypodermal cells to ensure proper body size regulation.
RAD-SMAD, a Sma/Mab-responsive reporter in C. elegans
The activity of TGFβ signaling can be monitored by the phosphorylation or nuclear entry of SMADs, or the expression of direct TGFβ downstream genes (Schmierer and Hill, 2007; Wrighton et al., 2009). However, there are no existing phospho-SMAD antibodies, appropriate SMAD::GFP reporters or anti-SMAD antibodies, or direct Sma/Mab target genes that allow us to directly monitor the activity of Sma/Mab signaling in C. elegans. In this study, we generated a Sma/Mab signaling reporter RAD-SMAD that appears to directly reflect Sma/Mab signaling activity in C. elegans: (1) the reporter is active in cells that have been reported to be responsive to Sma/Mab signaling; (2) mutations in the SMAD binding sites abolished reporter expression; and (3) the level of RAD-SMAD reporter expression correlated with the level of Sma/Mab signaling in various Sma/Mab pathway mutants. The RAD-SMAD reporter appears to specifically reflect Sma/Mab signaling as no reporter expression was observed in dauer animals either in the wild-type background or the daf-7(e1372) background. We also noticed that the RAD-SMAD reporter is not expressed in the M lineage, where DRAG-1 and other Sma/Mab pathway components are expressed and function (data not shown). At present, we cannot rule out the possibility that the expression level of RAD-SMAD in the M lineage is too low to detect. An alternative explanation is that expression of Sma/Mab pathway targets in the M lineage requires additional transcriptional input. A likely candidate for the additional transcription factor is the SCHNURRI (SHN) protein SMA-9 (Liang et al., 2003; Foehr et al., 2006), as previous studies in Drosophila have identified composite SMAD-SHN binding sites that contain additional nucleotide sequences in between the two SMAD binding sites (Pyrowolakis et al., 2004; Gao et al., 2005). Despite this caveat, the RAD-SMAD reporter will be a useful tool for C. elegans researchers wishing to monitor the direct transcriptional output of the Sma/Mab signaling pathway.
In summary, our work demonstrates that DRAG-1 acts in a cell-type-specific manner to modulate the BMP-like signaling pathway in C. elegans, and establishes a direct link between RGMb proteins and BMP signaling in vivo. Because RGM proteins are not present in Drosophila, our work further provides a simple genetic system for mechanistic studies on RGM protein regulation of BMP pathways in vivo.
Acknowledgements
We thank the C. elegans Genetics Center, Dave Pryune, Shohei Mitani for strains and primers; Aalia AlBarwani, Rachel Fairbank and Amanda Lindy for help with the initial genetic analyses of drag-1(jj4); Ping Wang for help with statistical analysis; and Nirav Amin, Richard Padgett and Mariana Wolfner for helpful discussions and critical comments on the manuscript. This work was supported by NIH R01 GM066953 (to J.L.). Y.P. was a Howard Hughes Undergraduate Research Scholar and a recipient of the CALS Charitable Trust Undergraduate Research Grant at Cornell University. Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- Amin N. M., Hu K., Pruyne D., Terzic D., Bretscher A., Liu J. (2007). A Zn-finger/FH2-domain containing protein, FOZI-1, acts redundantly with CeMyoD to specify striated body wall muscle fates in the Caenorhabditis elegans postembryonic mesoderm. Development 1, 19-29 [DOI] [PubMed] [Google Scholar]
- Amin N. M., Lim S. E., Shi H., Chan T. L., Liu J. (2009). A conserved Six-Eya cassette acts downstream of Wnt signaling to direct non-myogenic versus myogenic fates in the C. elegans postembryonic mesoderm. Dev. Biol. 331, 350-360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriopoulos B., Jr, Corradini E., Xia Y., Faasse S. A., Chen S., Grgurevic L., Knutson M. D., Pietrangelo A., Vukicevic S., Lin H. Y., et al. (2009). BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat. Genet. 41, 482-487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitt J. L., Zhang Y., Samad T. A., Xia Y., Tang J., Campagna J. A., Schneyer A. L., Woolf C. J., Lin H. Y. (2005). Repulsive guidance molecule (RGMa), a DRAGON homologue, is a bone morphogenetic protein co-receptor. J. Biol. Chem. 280, 29820-29827 [DOI] [PubMed] [Google Scholar]
- Babitt J. L., Huang F. W., Wrighting D. M., Xia Y., Sidis Y., Samad T. A., Campagna J. A., Chung R. T., Schneyer A. L., Woolf C. J., et al. (2006). Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat. Genet. 38, 531-539 [DOI] [PubMed] [Google Scholar]
- Babitt J. L., Huang F. W., Xia Y., Sidis Y., Andrews N. C., Lin H. Y. (2007). Modulation of bone morphogenetic protein signaling in vivo regulates systemic iron balance. J. Clin. Invest. 117, 1933-1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balemans W., Van Hul W. (2002). Extracellular regulation of BMP signaling in vertebrates: a cocktail of modulators. Dev. Biol. 250, 231-250 [PubMed] [Google Scholar]
- Bohme U., Cross G. A. (2002). Mutational analysis of the variant surface glycoprotein GPI-Anchor signal sequence in Trypanosoma brucei. J. Cell Sci. 115, 805-816 [DOI] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus L. M., Lambert L. A. (2007). Molecular evolution of hemojuvelin and the repulsive guidance molecule family. J. Mol. Evol. 65, 68-81 [DOI] [PubMed] [Google Scholar]
- Chatterjee S., Mayor S. (2001). The GPI-anchor and protein sorting. Cell Mol. Life Sci. 58, 1969-1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradini E., Babitt J. L., Lin H. Y. (2009). The RGM/DRAGON family of BMP co-receptors. Cytokine Growth Factor Rev. 20, 389-398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez M., Attisano L., Wrana J. L., Albert P. S., Massague J., Riddle D. L. (1993). The Daf-4 gene encodes a bone morphogenetic protein receptor controlling C. elegans dauer larva development. Nature 365, 644-649 [DOI] [PubMed] [Google Scholar]
- Foehr M. L., Liu J. (2008). Dorsoventral patterning of the C. elegans postembryonic mesoderm requires both LIN-12/Notch and TGFbeta signaling. Dev. Biol. 313, 256-266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foehr M. L., Lindy A. S., Fairbank R. C., Amin N. M., Xu M., Yanowitz J., Fire A. Z., Liu J. (2006). An antagonistic role for the C. elegans schnurri homolog SMA-9 in modulating TGFbeta signaling during mesodermal patterning. Development 133, 2887-2896 [DOI] [PubMed] [Google Scholar]
- Fukushige T., Hawkins M. G., McGhee J. D. (1998). The GATA-Factor Elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev. Biol. 198, 286-302 [PubMed] [Google Scholar]
- Gao S., Steffen J., Laughon A. (2005). Dpp-responsive silencers are bound by a trimeric Mad-Medea complex. J. Biol. Chem. 280, 36158-36164 [DOI] [PubMed] [Google Scholar]
- Georgi L. L., Albert P. S., Riddle D. L. (1990). Daf-1, a C. elegans gene controlling dauer larva development, encodes a novel receptor protein kinase. Cell 61, 635-645 [DOI] [PubMed] [Google Scholar]
- Gilleard J. S., Shafi Y., Barry J. D., McGhee J. D. (1999). ELT-3: a caenorhabditis elegans GATA factor expressed in the embryonic epidermis during morphogenesis. Dev. Biol. 208, 265-280 [DOI] [PubMed] [Google Scholar]
- Gordon K. J., Blobe G. C. (2008). Role of transforming growth factor-beta superfamily signaling pathways in human disease. Biochim. Biophys. Acta 1782, 197-228 [DOI] [PubMed] [Google Scholar]
- Granato M., Schnabel H., Schnabel R. (1994). Pha-1, a selectable marker for gene transfer in C. elegans. Nucleic Acids Res. 22, 1762-1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny T. L., MacNeil L. T., Wang H., de Bono M., Wrana J. L., Padgett R. W. (2007). Glypican LON-2 is a conserved negative regulator of BMP-like signaling in Caenorhabditis elegans. Curr. Biol. 17, 159-164 [DOI] [PubMed] [Google Scholar]
- Harfe B. D., Vaz Gomes A., Kenyon C., Liu J., Krause M., Fire A. (1998). Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes Dev. 12, 2623-2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Shi H., Liu J. (2009). Two hox cofactors, the Meis/Hth homolog UNC-62 and the Pbx/Exd homolog CEH-20, function together during C. elegans postembryonic mesodermal development. Dev. Biol. 334, 535-546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K., Kirkpatrick H., Comer A., Hoffmann F. M., Laughon A. (1999). Interaction of SMAD complexes with tripartite DNA-binding sites. J. Biol. Chem. 274, 20709-20716 [DOI] [PubMed] [Google Scholar]
- Kramer J. M., Johnson J. J. (1993). Analysis of mutations in the Sqt-1 and Rol-6 collagen genes of Caenorhabditis elegans. Genetics 135, 1035-1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna S., Maduzia L. L., Padgett R. W. (1999). Specificity of TGFbeta signaling is conferred by distinct type i receptors and their associated SMAD proteins in Caenorhabditis elegans. Development 126, 251-260 [DOI] [PubMed] [Google Scholar]
- Liang J., Lints R., Foehr M. L., Tokarz R., Yu L., Emmons S. W., Liu J., Savage-Dunn C. (2003). The Caenorhabditis elegans schnurri homolog Sma-9 mediates stage- and cell type-specific responses to DBL-1 BMP-related signaling. Development 130, 6453-6464 [DOI] [PubMed] [Google Scholar]
- Maduzia L. L., Gumienny T. L., Zimmerman C. M., Wang H., Shetgiri P., Krishna S., Roberts A. F., Padgett R. W. (2002). Lon-1 regulates Caenorhabditis elegans body size downstream of the Dbl-1 TGF beta signaling pathway. Dev. Biol. 246, 418-428 [DOI] [PubMed] [Google Scholar]
- Massagué J. (2008). TGFβ in cancer. Cell 134, 215-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J., Chen Y. G. (2000). Controlling TGF-β signaling. Genes Dev. 14, 627-644 [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V. (1991). Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959-3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K., Chow K. L., Ueno N. (1999). Regulation of body length and male tail ray pattern formation of Caenorhabditis elegans by a member of TGF-beta family. Development 126, 1337-1347 [DOI] [PubMed] [Google Scholar]
- Morita K., Flemming A. J., Sugihara Y., Mochii M., Suzuki Y., Yoshida S., Wood W. B., Kohara Y., Leroi A. M., Ueno N. (2002). A Caenorhabditis elegans TGF-beta, DBL-1, controls the expression of LON-1, a PR-related protein, that regulates polyploidization and body length. EMBO J. 21, 1063-1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustakas A., Heldin C. H. (2009). The regulation of TGFβ signal transduction. Development 136, 3699-3714 [DOI] [PubMed] [Google Scholar]
- Okkema P. G., Harrison S. W., Plunger V., Aryana A., Fire A. (1993). Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics 135, 385-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge F. A., Gravato-Nobre M. J., Hodgkin J. (2010). Signal transduction pathways that function in both development and innate immunity. Dev. Dyn. 239, 1330-1336 [DOI] [PubMed] [Google Scholar]
- Pyrowolakis G., Hartmann B., Müller B., Basler K., Affolter M. (2004). A simple molecular complex mediates widespread BMP-induced repression during Drosophila development. Dev. Cell 7, 229-240 [DOI] [PubMed] [Google Scholar]
- Ren P., Lim C. S., Johnsen R., Albert P. S., Pilgrim D., Riddle D. L. (1996). Control of C. elegans larval development by neuronal expression of a TGF-Beta homolog. Science 274, 1389-1391 [DOI] [PubMed] [Google Scholar]
- Samad T. A., Srinivasan A., Karchewski L. A., Jeong S. J., Campagna J. A., Ji R. R., Fabrizio D. A., Zhang Y., Lin H. Y., Bell E., et al. (2004). DRAGON: A member of the repulsive guidance molecule-related family of neuronal- and muscle-expressed membrane proteins is regulated by DRG11 and has neuronal adhesive properties. J. Neurosci. 24, 2027-2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samad T. A., Rebbapragada A., Bell E., Zhang Y., Sidis Y., Jeong S. J., Campagna J. A., Perusini S., Fabrizio D. A., Schneyer A. L., et al. (2005). DRAGON, a bone morphogenetic protein Co-receptor. J. Biol. Chem. 280, 14122-14129 [DOI] [PubMed] [Google Scholar]
- Savage C., Das P., Finelli A. L., Townsend S. R., Sun C. Y., Baird S. E., Padgett R. W. (1996). Caenorhabditis elegans genes Sma-2, Sma-3, and Sma-4 define a conserved family of transforming growth factor beta pathway components. Proc. Natl. Acad. Sci. USA 93, 790-794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage-Dunn C. (2005). TGF-β signaling. WormBook (ed. The C. elegans Research Community ). doi/10.1895/wormbook.1.22.1, http://www.wormbook.org [DOI] [PMC free article] [PubMed]
- Schmierer B., Hill C. S. (2007). TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat. Rev. Mol. Cell Biol. 8, 970-982 [DOI] [PubMed] [Google Scholar]
- Schwartz M. S., Benci J. L., Selote D. S., Sharma A. K., Chen A. G. Y., Dang H., Fares H., Vatamaniuk O. K. (2010). Detoxification of multiple heavy metals by a half-molecule ABC transporter, HMT-1, and coelomocytes of Caenorhabditis elegans. PLoS ONE 5, e9564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severyn C. J., Shinde U., Rotwein P. (2009). Molecular biology, genetics and biochemistry of the repulsive guidance molecule family. Biochem. J. 422, 393-403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Massagué J. (2003). Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113, 685-700 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Horvitz H. R. (1977). Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110-156 [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Yandell M. D., Roy P. J., Krishna S., Savage-Dunn C., Ross R. M., Padgett R. W., Wood W. B. (1999). A BMP homolog acts as a dose-dependent regulator of body size and male tail patterning in Caenorhabditis elegans. Development 126, 241-250 [DOI] [PubMed] [Google Scholar]
- Umulis D., O'Connor M. B., Blair S. S. (2009). The extracellular regulation of bone morphogenetic protein signaling. Development 136, 3715-3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatamaniuk O. K., Bucher E. A., Sundaram M. V., Rea P. A. (2005). CeHMT-1, a putative phytochelatin transporter, is required for cadmium tolerance in Caenorhabditis elegans. J. Biol. Chem. 280, 23684-23690 [DOI] [PubMed] [Google Scholar]
- Vowels J. J., Thomas J. H. (1992). Genetic analysis of chemosensory control of dauer formation in Caenorhabditis elegans. Genetics 130, 105-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tokarz R., Savage-Dunn C. (2002). The expression of TGFbeta signal transducers in the hypodermis regulates body size in C. elegans. Development 129, 4989-4998 [DOI] [PubMed] [Google Scholar]
- Wicks S. R., Yeh R. T., Gish W. R., Waterston R. H., Plasterk R. H. (2001). Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28, 160-164 [DOI] [PubMed] [Google Scholar]
- Wrighton K. H., Lin X., Feng X. H. (2009). Phospho-control of TGF-beta superfamily signaling. Cell Res. 19, 8-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M. Y., Hill C. S. (2009). Tgf-beta superfamily signaling in embryonic development and homeostasis. Dev. Cell 16, 329-343 [DOI] [PubMed] [Google Scholar]
- Xia Y., Babitt J. L., Sidis Y., Chung R. T., Lin H. Y. (2008). Hemojuvelin regulates hepcidin expression via a selective subset of BMP ligands and receptors independently of neogenin. Blood 111, 5195-5204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Babitt J. L., Bouley R., Zhang Y., Da Silva N., Chen S., Zhuang Z., Samad T. A., Brenner G. J., Anderson J. L., et al. (2010). Dragon enhances BMP signaling and increases transepithelial resistance in kidney epithelial cells. J. Am. Soc. Nephrol. 21, 666-677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem J., Weston K., Greenwald I. (1988). The Caenorhabditis elegans lin-12 gene encodes a transmembrane protein with overall similarity to Drosophila Notch. Nature 335, 547-550 [DOI] [PubMed] [Google Scholar]
- Yoshida S., Morita K., Mochii M., Ueno N. (2001). Hypodermal expression of Caenorhabditis elegans TGF-Beta Type I receptor SMA-6 is essential for the growth and maintenance of body length. Dev. Biol. 240, 32-45 [DOI] [PubMed] [Google Scholar]