Abstract
Proton-transporting cells are located in several tissues where they acidify the extracellular environment. These cells express the vacuolar H+-ATPase (V-ATPase) B1 subunit (ATP6V1B1) in their plasma membrane. We provide here a comprehensive catalog of the proteins that are expressed in these cells, after their isolation by enzymatic digestion and fluorescence-activated cell sorting (FACS) from transgenic B1-enhanced green fluorescent protein (EGFP) mice. In these mice, type A and B intercalated cells and connecting segment cells of the kidney, and narrow and clear cells of the epididymis, which all express ATP6V1B1, also express EGFP, while all other cell types are negative. The proteome of renal and epididymal EGFP-positive (EGFP+) cells was identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) and compared with their respective EGFP-negative (EGFP−) cell populations. A total of 2,297 and 1,564 proteins were detected in EGFP+ cells from the kidney and epididymis, respectively. Out of these proteins, 202 and 178 were enriched by a factor greater than 1.5 in EGFP+ cells compared with EGFP− cells, in the kidney and epididymis respectively, and included subunits of the V-ATPase (B1, a4, and A). In addition, several proteins involved in intracellular trafficking, signaling, and cytoskeletal dynamics were identified. A novel common protein that was enriched in renal and epididymal EGFP+ cells is the progesterone receptor, which might be a potential candidate for the regulation of V-ATPase-dependent proton transport. These proteomic databases provide a framework for comprehensive future analysis of the common and distinct functions of V-ATPase-B1-expressing cells in the kidney and epididymis.
Keywords: intercalated cells, mass spectrometry, proteome, proton secretion, clear cells
specialized proton-secreting cells are located in several tissues that are involved in acid/base regulation. These cells belong to the mitochondria-rich cell family, indicating that they have a high energy requirement to achieve their characteristic functions (8). Among these cells are intercalated cells (IC) of the kidney (59) and narrow and clear cells of the epididymis (6, 9). The epididymis is a long, convoluted tubule located downstream of the testis and is the site where sperm mature and are stored (32, 43, 52). Renal intercalated cells regulate systemic acid/base balance, while narrow and clear cells establish an acidic environment in the lumen of the epididymis, a process that is essential to maintain spermatozoa quiescent before ejaculation (11, 17, 46, 55, 59). Therefore, both the epididymis and the kidney have similar luminal acidifying functions. This and many other similarities in transport properties that occur in both renal and epididymal tubules have been pointed out previously (33). Accordingly, kidney intercalated cells and epididymal narrow/clear cells have in common a high expression of the vacuolar proton pumping ATPase (V-ATPase) in their plasma membrane as well as in intracellular recycling vesicles. The V-ATPase has two main functions (11, 59). It is expressed in all cell types, where it is located in intracellular organelles (e.g., endosomes, lysosomes, Golgi) and acidifies their lumen. In intercalated cells, narrow and clear cells, as well as in osteoclasts and interdental cells of the inner ear, the V-ATPase is also located in the plasma membrane, where it is involved in proton extrusion from the cell cytoplasm (2, 6, 9, 25, 49, 50, 56, 58, 59). In these specialized cells, regulation of proton secretion is achieved at least in part by recycling mechanisms. The V-ATPase also plays a major role in proton secretion and in energizing other ion transport processes in lower vertebrates and invertebrates, and it is frequently concentrated in specialized “mitochondria-rich” cells in these nonmammalian organisms (8, 61).
The V-ATPase is composed of several subunits and is divided into two distinct domains, the V0 domain that contains integral membrane proteins, labeled a, d, e, c, c′, and c″, and the V1 domain that contains cytosolic subunits, labeled A through H (reviewed in Refs. 2, 25, 59, and 61). In mammals, the a subunit is encoded by four genes (ATP6V0A1–A4) leading to the production of four isoforms (a1–a4). Similarly subunits B, H, and d have two isoforms, and subunits C and G have three isoforms. In addition, one E isoform, originally designated as ATP6E1 and later renamed ATP6V1E2, is expressed in testis and spermatozoa while its homolog, originally designated as ATP6E2 and now labeled ATP6V1E1, is expressed ubiquitously (34, 57). Kidney IC and epididymal clear cells both have an identical combination of V-ATPase subunit isoforms that are inserted into their plasma membrane. In particular, subunits B1 and a4 are enriched in these cells and can preferentially accumulate in the plasma membrane compared with the B2 and a1, a2, and a3 isoforms, which are mainly located in intracellular structures (16, 47, 48, 50). While the V-ATPase is located in the apical membrane of type A IC and epididymal narrow and clear cells, type B IC have a more complex phenotype and can have basolateral, bipolar, and even apical V-ATPase. In addition, ICs and narrow and clear cells express distinct transporters involved in their acid/base regulating function. A-ICs express the chloride/bicarbonate exchanger, AE1, in their basolateral membrane, while B-ICs express a distinct chloride/bicarbonate exchanger, pendrin, in their apical membrane (reviewed in Refs. 11 and 59). Narrow/clear cells express the ubiquitous anion exchanger AE2, and the sodium-bicarbonate cotransporter NBCe1 in their basolateral membrane (46). All these cell types also express high levels of the cytosolic carbonic anhydrase, CAII. Therefore, while these cells share identical transporters for their common proton extrusion functions, they have also established different expression patterns for partner proteins involved in this process.
To characterize the function of V-ATPase-expressing epithelial cells, we have generated a transgenic mouse model, in which the promoter of the V-ATPase B1 subunit drives expression of enhanced green fluorescent protein (EGFP) (39). In these mice, EGFP fluorescence is detectable only in cells that strongly express the B1 subunit. These include kidney A-IC and B-IC, epididymal narrow and clear cells, and nonciliated cells of the lung. Intermediate EGFP fluorescence was also detected in nonintercalated cells of the renal connecting segments that also express B1, but no green fluorescence was detected in other cell types, including renal proximal tubule cells, thick ascending limb cells, thin limbs of Henle, and collecting duct principal cells. Similarly, no other cell types in the epididymis were found to express EGFP. Thus, these mice are extremely valuable to characterize the phenotypes of positively identified B1-expressing V-ATPase-rich cells. In this study, we characterized the proteomic signature of B1-expressing renal and epididymal cells isolated by fluorescence-activated cell sorting (FACS).
MATERIALS AND METHODS
Animals.
Generation and breeding of V-ATPase B1-EGFP (B1-EGFP) mice have been described previously (39). Adult B1-EGFP mice were housed under standard conditions and maintained on a standard rodent diet. All animal studies were approved by the Massachusetts General Hospital (MGH) Subcommittee on Research Animal Care, in accordance with National Institutes of Health, Department of Agriculture, and Accreditation of Laboratory Animal Care requirements.
Isolation of B1-expressing cells from the epididymis and kidney.
Mice were anesthetized using pentobarbital sodium (50 mg/kg body wt ip). Epididymides and kidneys were dissected and minced immediately with scissors in prewarmed RPMI 1640 medium (Gibco Invitrogen, Carlsbad, CA) containing 1.0 mg/ml collagenase type I (Gibco Invitrogen) and 1.0 mg/ml collagenase type II (Sigma Aldrich, St. Louis, MO). Tissue dissociation was performed for 45 min in a shaking (100 rpm) 37°C water bath. Cell preparations were then passed through a cell strainer with 40-μm nylon mesh to remove undigested material, and cells were washed once with RPMI 1640 medium and once with calcium-free phosphate-buffered saline (PBS). Populations of EGFP-positive (EGFP+) cells from epididymis and kidney preparations were isolated immediately by FACS based on their green fluorescence intensity. Sorting was performed at the MGH flow cytometry core facility using a modified FACSVantage cell sorter (BD Biosciences, San Jose, CA). EGFP+ and EGFP-negative (EGFP−) cell samples were collected in PBS and used without delay for protein isolation, RNA isolation, or imaging. Cells with intermediate to low fluorescence intensity were discarded to avoid potential contamination of cells that were negative for EGFP, but had a high autofluorescence, in the EGFP+ cell preparation. A fraction of each cell sample was reanalyzed by flow cytometry to estimate the purity (>95%). For liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis, EGFP+ and EGFP− cells collected in PBS were centrifuged briefly, resuspended in Laemmli sample buffer (10 mM Tris·HCl, 6% glycerol, 1.5% SDS, 60 mg/ml dithiothreitol), heated for 5 min at 99°C, and stored at −20°C. For RNA isolation, cell pellets were resuspended in RNA extraction buffer (PicoPure RNA isolation, Molecular Devices, Sunnyvale, CA) and stored at −80°C. For preparation of Cytospin smears, cells were fixed for 2 h with PBS containing 2% paraformaldehyde and stored at 4°C.
Preparation of proteins for LC-MS/MS identification.
Proteins were separated by one-dimensional SDS-PAGE using a 4–15% gradient polyacrylamide gel (Bio-Rad, Hercules, CA) to reduce sample complexity. Gels were stained with Coomassie R-250 (Pierce Imperial Protein Stain, Thermo Fisher Scientific, Rockford, IL) for 5 min, then washed in deionized water for 1 h, and scanned. Each sample lane was sliced into 19 blocks, from the top of the gel to the dye front. Each block was further minced into small pieces (up to 2 mm3). Proteins were reduced, alkylated, and trypsin-digested in gel as we have previously described (51, 54). After trypsin digestion, the peptides were extracted with 50% acetonitrile-0.1% formic acid (FA), dried in a Speed Vac concentrator, and desalted using ZipTip C18 pipette tips (Millipore, Billerica, MA). Desalted peptides were dried and reconstituted with 0.1% FA for LC-MS/MS analysis.
LC-MS/MS protein identification.
Tryptic peptides were stratified over time by reverse-phase LC (PicoFrit, BioBasic C18 column, New Objective, Woburn, MA) before delivery to an LTQ tandem mass spectrometer (MS/MS, Thermo Fisher Scientific) equipped with a nanoelectrospray ion source. The mass-to-charge (m/z) ratios of peptides and their fragmented ions were recorded as spectra by the mass spectrometer. The spectra with a total ion current >10,000 were used to search for matches to peptides in a concatenated RefSeq database, composed of forward and reverse protein sequences derived from the National Center for Biotechnology Information on April 1, 2008. The search was performed using BioWorks software (version 3.3, Thermo Fisher Scientific) based on the SEQUEST algorithm, and InsPecT (University of California San Diego, La Jolla, CA). The search parameters included 1) precursor ion mass tolerance <2 atomic mass units (amu), 2) fragment ion mass tolerance <1 amu, 3) up to three missed tryptic cleavages allowed, and 4) amino acid modifications: cysteine carboxyamidomethylation (plus 57.02 amu) and methionine oxidation (plus 15.99 amu). A target-decoy analysis was performed to set false discovery rates below 2%. To assess relative protein abundance, we applied spectral counting normalized by molecular weight to correct for the higher probability of peptide identification in larger proteins. SEQUEST search and InsPecT search results were combined in a Microsoft Excel spreadsheet. A χ2-analysis was performed to determine the proteins that were significantly enriched in the EGFP+ populations compared with EGFP− cells. Significance was set at P < 0.05.
Immunofluorescence.
Cell suspensions were fixed in paraformaldehyde before and after FACS and were collected onto microscope slides by cytocentrifugation (Cytospin Shando, Thermo Scientific, Waltham, MA) at 500 rpm for 10 min. Microscope slides were then either mounted in Vectashield medium containing DAPI (Vector Labs, Burlingame, CA), and visualized directly for their GFP fluorescence, or immunolabeled. For immunolabeling, Cytospin smears were rehydrated in PBS and pretreated with 1% (wt/vol) SDS in PBS to retrieve antigen (10). After preincubation in 1% (wt/vol) bovine serum albumin in PBS to prevent nonspecific labeling, slides were incubated for 90 min at room temperature with an affinity-purified anti-V-ATPase B1 primary antibody (7) diluted in antibody diluent (Dako, Carpinteria, CA). Slides were washed twice in high salt (2.7% NaCl) PBS and once in PBS. They were then incubated for 1 h at room temperature with a donkey anti-rabbit Cy3-conjugated secondary antibody (Jackson ImmunoResearch, West Grove, PA) and washed again. After mounting in Vectashield containing DAPI, slides were examined under a Nikon epifluorescence microscope and images were captured using a Hamamatsu Orca digital camera and IPLab software (BD Biosciences, Rockville, MD).
Electron microscopy.
Isolated cells were fixed in 2% glutaraldehyde (in 0.1 M sodium cacodylate buffer) overnight and were rinsed in sodium cacodylate buffer. They were then stained with 2% aqueous uranyl acetate, dehydrated with graded ethanol up to 100%, rinsed with propylene oxide, and embedded in 100% Epon. Ultrathin (70 to 80 nm) sections were mounted on formvar-coated nickel grids, stained with uranyl acetate and lead citrate, and inspected and photographed with a JEOL 1011 electron microscope. Images were acquired using an AMT digital imaging system.
RT-PCR.
RNA isolation was performed using the PicoPure kit following the manufacturer's instructions. The quantity and quality of RNA samples were assessed using a 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA). RNA samples were reverse-transcribed for 1 h at 42°C in a final volume of 50 μl with 1× buffer II, 5 mM MgCl2, 1.0 mM each dNTP, 1 U/μl RNase inhibitor, 2.5 μM random hexamers, and 2.5 U/μl Moloney murine leukemia virus reverse transcriptase. Reverse transcription products were used as templates for PCR. The sequences of the PCR primer sets, synthesized by Invitrogen, are listed in Table 1. Reaction mixtures consisted of a 20 μl final volume containing 2 μl template, 1.25 units AmpliTaq Gold DNA polymerase, 1× buffer II, 1.5 mM MgCl2, 1.0 mM each dNTP, and 0.5 μM forward and reverse oligonucleotide primers. All RT and PCR reagents were from Applied Biosystems (Foster City, CA). PCR was performed in a Flexigene thermal cycler (Techne, Princeton, NJ) with the following parameters: 8 min at 95°C to activate the polymerase, followed by 35 cycles of melting for 30 s at 95°C, annealing for 30 s at 60°C, extension for 30 s at 72°C, and a final extension for 10 min at 72°C. The PCR products were analyzed by electrophoresis on a 2.5% agarose gel containing GelStar stain (Lonza, Rockland, ME).
Table 1.
Sequence of the primers used for PCR
| Gene | Forward Primer (5′–3′) | Reverse Primer (5′–3′) | Product Size, bp |
|---|---|---|---|
| mAnxa6 | AGGCGGTCCCTTCTGTTATC | AGTCAGGCAGGGTTATGTGG | 142 |
| mAtp6v0a4 | TGACCACCATCTTCCTGTGA | TAGAGGCAGGAGGAACCTGA | 143 |
| mAtp6v1b1 | CCCTACGATTGAGCGGATCAT | TATATCCAGGAAAGCCACGGC | 182 |
| mAqp9 | AGACGGTGTGGGCTTTGTAG | GACGGAGACAGAGAGCCACT | 177 |
| mDbnl | ACCTGAATGGGGGAGAAATC | GCACCATCATGTCTGCTCAC | 139 |
| mGapdh | TGAGCAAGAGAGGCCCTATCC | CCCTAGGCCCCTCCTGTTAT | 98 |
| mHsd17b12 | TGGGCCAAAGTGTACATCAA | GCAGTGCAGACACAACGAAT | 100 |
| mIqgap1 | GCTGCTGAATAACCCTGCTT | TCCAAAGACAAAGGATTCCAA | 142 |
| mKrt18 | AAGGTGAAGCTTGAGGCAGA | CTGCACAGTTTGCATGGAGT | 111 |
| mLrba | GCTAAGCCAGAGCCAGGTG | AAGGTGGGTGCTGCTTACAC | 100 |
| mMyo6 | CCCAGCACTAACCTGCGTAT | TGCCTGTTTGTAGTTTGACTTTG | 149 |
| mPgrmc1 | GCTGGGTGAGTTTGAAAGGA | CCAAGACCTAGACACCTGACG | 113 |
| mProm1 | GCTCGTTTTGGAGCTACCTG | GCTGAGCGACAGTTCCTTCT | 121 |
| mProm2 | CAAAGACTCCAGCAAGCACA | CCTCACCCCCAAGTTAACAA | 130 |
| mSlco4a1 | TTACTGCCTGTCCTGGAAGC | TCAGTGCAGTTTGCTTGGAC | 150 |
RESULTS
Isolation of EGFP+ cells from kidney and epididymis.
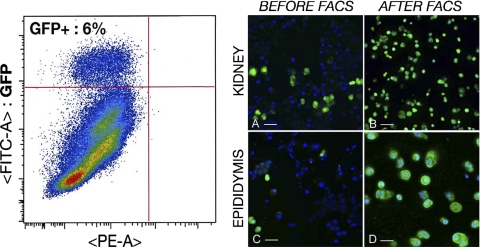
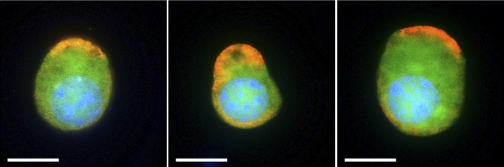
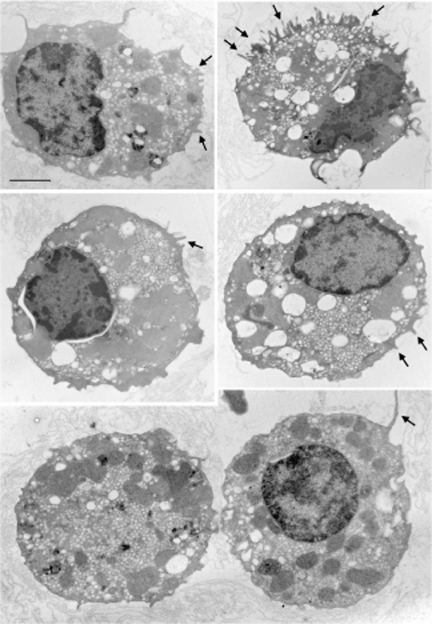
Significant numbers of EGFP+ cells were isolated after FACS sorting from kidney and epididymis cell suspensions (Fig. 1). Three different batches of EGFP+ and EGFP− cells were isolated from four B1-EGFP mice each time. On average, 330,000 and 120,000 EGFP+ cells were isolated from the kidneys and epididymides of these mice, respectively. These samples were pooled to perform proteomics analysis. Thus, a total of 1,100,000 renal EGFP+ cells and 360,000 epididymal EGFP+ cells, originating from a total of 12 mice, were analyzed. Immunofluorescence labeling of EGFP+ cells for the B1 subunit of the V-ATPase showed that they retain their polarized V-ATPase expression phenotype during the isolation procedure (Fig. 2). In addition, electron microscopy showed that EGFP+ cells retain morphological features characteristic of “mitochondria-rich” cells after FACS sorting (Fig. 3). These results confirmed that the cells isolated by FACS are indeed V-ATPase enriched and that their integrity was preserved during the isolation procedure.
Fig. 1.
Flow cytometry sorting of B1-enhanced green fluorescent protein (EGFP) cells. Left: dot plot of a representative epididymal cell sorting sample [EGFP vs. phycoerythrin channel autofluorescence (PE-A)]. Events in the top left quadrant are EGFP-positive (EGFP+) cells, representing ∼6% of the live cell population. Right: fluorescence microscopy of Cytospin smears of cells isolated from B1-EGFP mouse kidney (A and B) and epididymis (C and D) before (A and C) and after (B and D) sorting. A few vacuolar H+-ATPase (V-ATPase)-B1-expressing cells (green) are seen in the cell suspension before fluorescence-activated cell sorting (FACS). After FACS, all cells show positive EGFP fluorescence. Nuclei are visualized with DAPI (blue). Bars = 20 μm (A and C), 30 μm (B), and 15 μm (D).
Fig. 2.
Three examples of epididymal EGFP+ cells (green) labeled for the V-ATPase B1 subunit (red) visualized after FACS. Cells retained their polarized expression of the V-ATPase. Nuclei are labeled with DAPI (blue). Bars = 5 μm.
Fig. 3.
Electron microcopy showing several renal EGFP+ cells isolated by FACS. All cells retained their characteristic morphology with visible microvilli (arrows) and numerous vesicles. Bar = 3 μm.
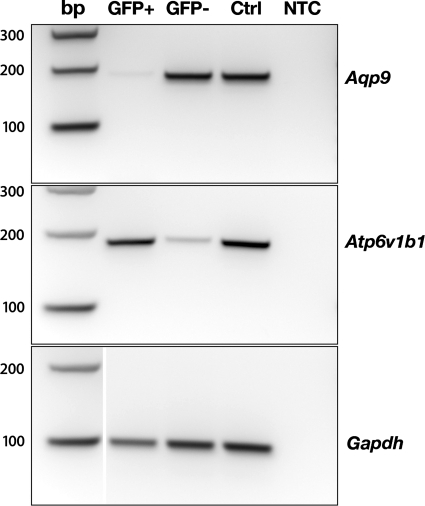
RT-PCR analysis showed a high enrichment of the V-ATPase B1 subunit as well as depletion of aquaporin 9 (a marker of epididymal principal cells) in EGFP+ cells isolated from the epididymis compared with the EGFP− population (Fig. 4), further confirming that clear cells had been positively selected by FACS. Enrichment of the V-ATPase B1 subunit was also observed in EGFP+ cells isolated from the kidney (see Fig. 6, top: Atp6v1b1).
Fig. 4.
RT-PCR analysis showing depletion of aquaporin 9 (AQP9) mRNA (top; Aqp9) and high enrichment of the V-ATPase B1 subunit (middle; Atp6v1b1) in epididymal EGFP+ cells compared with EGFP-negative (EGFP−) cells. Controls using GAPDH-specific primers (Gapdh) are shown (bottom).
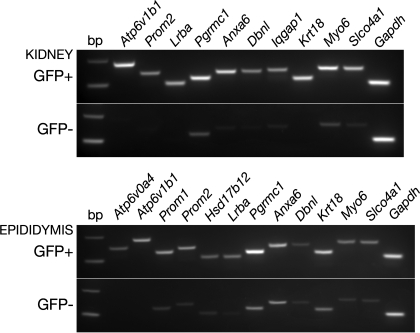
Fig. 6.
RT-PCR analysis of selected genes that were shown to be enriched in EGFP+ cells from the kidney (top; GFP+) and epididymis (bottom; GFP+) compared with their respective EGFP− samples (GFP−). Of note, the B1 subunit of the V-ATPase (Atp6v1b1) was significantly enriched in both kidney and epididymis EGFP+ cells compared with EGFP− cells. Atp6v0a4, V-ATPase a4 subunit; Prom1 and 2, Prominin 1 and 2, respectively; Lrba, LPS-responsive beige-like anchor; Pgrmc1, progesterone receptor; Anxa6, annexin A6; Dbnl, drebrin-like; Krt18, keratin 18; Myo6, myosin 6; Slco4a1, solute carrier organic anion transporter member 4a1; Hsd17b12, hydroxysteroid (17-β) dehydrogenase 12; Gapdh, GAPDH.
Mass spectrometry analysis of proteins enriched in the EGFP+ cell population compared with EGFP− cells.
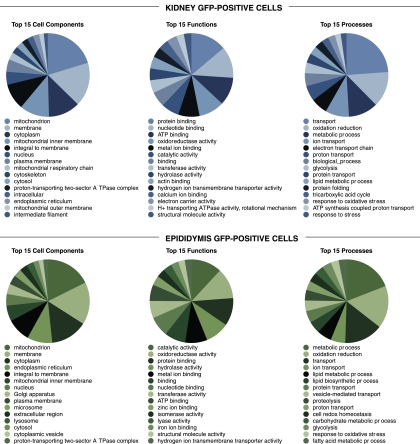
To determine the proteomic profile of B1-V-ATPase-expressing cells in the kidney and epididymis, we carried out a comparative proteomic analysis using LC-MS/MS in EGFP+ cell samples. Data were compared with proteins that were detected in the EGFP− cell samples. A total of 2,297 and 1,564 proteins were detected in EGFP+ cells from the kidney and epididymis, respectively. These proteins are listed in supplemental Tables S1 and S2 (supplemental data for this article can be found online at the American Journal of Physiology-Cell Physiology website) and are also available at the National Heart, Lung, and Blood Institute Laboratory of Kidney and Electrolyte Metabolism (NHLBI-LKEM) website (http://dir.nhlbi.nih.gov/papers/lkem/kevcpd/). We then determined the proteins that were significantly enriched in the EGFP+ populations compared with the EGFP− populations in both kidney and epididymis by performing a χ2-analysis of the spectral counts for all proteins detected. Figure 5 summarizes the gene ontology terms for these proteins, listing the 15 most frequent terms in each EGFP+ sample, grouped according to their location (cell components), their function, or their role in cellular processes. In both renal and epididymal EGFP+ cells, the most abundant group of proteins was located in mitochondria, in agreement with the fact that intercalated cells and clear cells are part of the mitochondria-rich cell family. Interestingly, in both samples the “proton-transporting two-sector ATPase complex,” the “hydrogen ion transmembrane transporter activity,” and the “proton transport” groups were listed in the top 15 cell components, functions, and processes, respectively. In addition, the “H+ transporting ATPase activity, rotational mechanism” group was identified in the top 15 functions group of kidney EGFP+ cells. These results are in agreement with the proton secretory function of renal intercalated cells and epididymal narrow and clear cells. We also grouped the proteins enriched in each EGFP+ cell population according to their established role in more specific cellular functions (Tables 2 and 3). Proteins belonging to the acid/base transport group were identified, including several V-ATPase subunits. In fact, among the proteins that were at the top of the list was the V-ATPase B1 subunit in both the kidney and epididymis EGFP+ samples, further confirming the enrichment of B1-expressing cells after FACS sorting. Pendrin, which is expressed in B-IC exclusively, and Rhbg (expressed in A-IC) were detected in renal EGFP+ cells but not in epididymal EGFP+ cells. Similarly, proteins specific for the male urogenital tract, including Crisp 1, Adam 7, and Pebp1 (14, 15, 21, 42), were identified in epididymal EGFP+ cells but not in renal EGFP+ cells. In addition, proteins known to be expressed in nonintercalated cells of the connecting segment were detected in the renal EGFP+ sample, confirming that these EGFP-expressing cells were also selected during the isolation procedure. These include calbindin-D28K (5) and aquaporin 3 (12).
Fig. 5.
Pie charts of the top 15 gene ontology terms for the proteins that were significantly enriched in the renal (top) and epididymal (bottom) EGFP+ cell populations compared with their respective EGFP− cell samples.
Table 2.
List of proteins that were identified as being significantly enriched in renal EGFP+ cells compared with renal EGFP− cells
| Protein Name | Gene Symbol | Accession No. | EGFP+ | EGFP− | Ratio |
|---|---|---|---|---|---|
| V-ATPase - acid/base | |||||
| V-ATPase subunit B1 | Atp6v1b1 | NP_598918.1 | 2007.375 | 0.00 | Unique |
| Rhesus blood group-associated B glycoprotein | Rhbg | NP_067350.2 | 645.90 | 0.00 | Unique |
| V-ATPase subunit G3 | Atp6v1 g3 | NP_796371.1 | 363.67 | 0.00 | Unique |
| V-ATPase subunit C2 | Atp6v1c2 | NP_598460.1 | 248.19 | 0.00 | Unique |
| Pendrin | Slc26a4 | NP_035997.1 | 198.40 | 0.00 | Unique |
| Carbonic anhydrase 15 | Car15 | NP_085035.1 | 140.91 | 0.00 | Unique |
| V-ATPase subunit D | Atp6v1d | NP_076210.1 | 810.74 | 70.50 | 11.50 |
| V-ATPase subunit H | Atp6v1 h | NP_598587.2 | 1,217.44 | 125.32 | 9.71 |
| V-ATPase subunit A | Atp6v1a | NP_031534.2 | 4,932.23 | 629.34 | 7.84 |
| V-ATPase subunit a4 | Atp6v0a4 | NP_536715.2 | 606.85 | 94.17 | 6.44 |
| V-ATPase subunit E1 | Atp6v1e1 | NP_031536.2 | 1,261.59 | 229.38 | 5.50 |
| Carbonic anhydrase 2 | Car2 | NP_033931.4 | 2,617.75 | 792.21 | 3.30 |
| Intracellular trafficking | |||||
| chaperonin subunit 6a (zeta) | Cct6a | NP_033968.1 | 137.92 | 17.24 | 8.00 |
| SAR1a gene homolog | Sar1a | NP_033146.1 | 535.74 | 89.29 | 6.00 |
| RAB10, member RAS oncogene family | Rab10 | NP_057885.1 | 709.82 | 177.45 | 4.00 |
| RAB1, member RAS oncogene family | Rab1 | NP_033022.1 | 1,411.07 | 529.15 | 2.67 |
| Ubiquitin B | Ubb | NP_035794.1 | 2,473.19 | 1,076.57 | 2.30 |
| Cytoskeleton | |||||
| Keratin 7 | Krt7 | NP_149064.1 | 1,222.67 | 0.00 | Unique |
| Keratin 86 | Krt86 | NP_034797.1 | 582.15 | 0.00 | Unique |
| Tubulin, β | Tubb2b | NP_076205.1 | 380.36 | 0.00 | Unique |
| WD repeat domain 1 | Wdr1 | NP_035845.1 | 346.35 | 0.00 | Unique |
| Wiskott-Aldrich syndrome homolog | Was | NP_033541.1 | 313.70 | 0.00 | Unique |
| Protein kinase C and casein kinase substrate in neurons 2 | Pacsin2 | NP_035992.1 | 250.75 | 0.00 | Unique |
| Keratin 33A | Krt33a | NP_082259.2 | 151.72 | 0.00 | Unique |
| Keratin complex 2, basic gene 18 | Krt85 | NP_058575.2 | 143.47 | 0.00 | Unique |
| Keratin 82 | Krt82 | NP_444479.1 | 122.56 | 0.00 | Unique |
| Drebrin-like | Dbnl | NP_038838.1 | 103.25 | 0.00 | Unique |
| Plastin 3 precursor | Pls3 | NP_663604.1 | 98.95 | 0.00 | Unique |
| Spectrin-β3 | Spnb3 | NP_067262.1 | 195.63 | 3.69 | 53.00 |
| CAP, adenylate cyclase-associated protein 1 | Cap1 | NP_031624.2 | 193.93 | 19.39 | 10.00 |
| Myosin, heavy polypeptide 14 | Myh14 | NP_082297.1 | 65.86 | 8.78 | 7.50 |
| Keratin 18 | Krt18 | NP_034794.1 | 863.08 | 168.41 | 5.13 |
| IQ motif containing GTPase activating protein 1 | Iqgap1 | NP_057930.1 | 651.63 | 148.34 | 4.39 |
| Gelsolin | Gsn | NP_666232.2 | 477.07 | 116.36 | 4.10 |
| Serum deprivation response | Sdpr | NP_620080.1 | 769.83 | 192.46 | 4.00 |
| Cell division cycle 42 | Cdc42 | NP_033991.1 | 940.80 | 235.20 | 4.00 |
| Spectrin-α2 | Spna2 | NP_001070022.1 | 2,358.55 | 848.10 | 2.78 |
| Myosin ID | Myo1d | NP_796364.2 | 215.37 | 77.53 | 2.78 |
| Destrin | Dstn | NP_062745.1 | 1,835.69 | 755.87 | 2.43 |
| Plectin 1 isoform 2 | Plec1 | NP_958787.1 | 121.98 | 52.28 | 2.33 |
| Myosin, heavy polypeptide 9, nonmuscle isoform 1 | Myh9 | NP_071855.2 | 1740.50 | 870.25 | 2.00 |
| Actin, α2, smooth muscle, aorta | Acta2 | NP_031418.1 | 16,448.88 | 9,140.91 | 1.80 |
| Membrane dynamics | |||||
| Myoferlin | Fer1l3 | NP_001093104.1 | 64.29 | 4.29 | 15.00 |
| Annexin A6 isoform b | Anxa6 | NP_001103681.1 | 318.77 | 26.56 | 12.00 |
| Annexin A11 | Anxa11 | NP_038497.2 | 203.40 | 18.49 | 11.00 |
| Annexin A4 | Anxa4 | NP_038499.2 | 696.07 | 83.53 | 8.33 |
| Filamin B, β | Flnb | NP_598841.1 | 867.68 | 198.84 | 4.36 |
| Ankyrin 3, epithelial isoform h | Ank3 | NP_733925.2 | 219.39 | 66.77 | 3.29 |
| Cadherin 16 | Cdh16 | NP_031689.1 | 389.49 | 166.93 | 2.33 |
| Transporters, channels | |||||
| Solute carrier family 8 (sodium/calcium exchanger) | Slc8a1 | NP_001106269.1 | 459.39 | 0.00 | Unique |
| Aquaporin 3 | Aqp3 | NP_057898.2 | 380.13 | 0.00 | Unique |
| Potassium inwardly rectifying channel J10 | Kcnj10 | NP_001034573.1 | 117.84 | 0.00 | Unique |
| Chloride channel Kb | Clcnkb | NP_062675.1 | 106.58 | 0.00 | Unique |
| Plasma membrane calcium ATPase 4 | Atp2b4 | NP_998781.1 | 120.24 | 7.51 | 16.00 |
| Chloride channel Ka | Clcnka | NP_077723.2 | 105.68 | 13.21 | 8.00 |
| Solute carrier family 12, member 3 | Slc12a3 | NP_062288.1 | 605.97 | 90.44 | 6.70 |
| Na+/K+-ATPase α1-subunit | Atp1a1 | NP_659149.1 | 10,373.32 | 4,929.98 | 2.10 |
| ATPase, Na+/K+ transporting, α2-polypeptide | Atp1a2 | NP_848492.1 | 6,371.57 | 3,092.21 | 2.06 |
| Intracellular signaling, receptors, hormones | |||||
| S100 calcium binding protein G | S100 g | NP_033919.1 | 2,564.06 | 0.00 | Unique |
| Adenylate kinase 1 | Ak1 | NP_067490.1 | 216.30 | 0.00 | Unique |
| Kallikrein 1 | Klk1 | NP_034769.4 | 556.04 | 0.00 | Unique |
| chaperonin subunit 3 (γ) | Cct3 | NP_033966.1 | 197.92 | 0.00 | Unique |
| Ataxin 2 | Atxn2 | NP_033151.2 | 73.28 | 0.00 | Unique |
| Archain 1 | Arcn1 | NP_666097.3 | 139.79 | 0.00 | Unique |
| Chapsyn-110 | Dlg2 | NP_035937.2 | 94.86 | 0.00 | Unique |
| Chaperonin subunit 8 (θ) | Cct8 | NP_033970.3 | 100.75 | 0.00 | Unique |
| Pre-B-cell colony-enhancing factor 1 | Nampt | NP_067499.1 | 126.40 | 0.00 | Unique |
| Calbindin 2 | Calb2 | NP_031612.1 | 1,625.62 | 63.75 | 25.50 |
| Calbindin-D28K | Calb1 | NP_033918.1 | 2,333.79 | 100.02 | 23.33 |
| Hydroxysteroid 11-β dehydrogenase 2 | Hsd11b2 | NP_032315.2 | 1,066.68 | 47.41 | 22.50 |
| Progesterone receptor membrane component | Pgrmc1 | NP_058063.2 | 737.52 | 92.19 | 8.00 |
| LPS-responsive beige-like anchor isoform α | Lrba | NP_109620.2 | 101.01 | 41.03 | 2.46 |
| Stress response/antioxidant | |||||
| Glutamate-cysteine ligase, catalytic subunit | Gclc | NP_034425.1 | 137.80 | 13.78 | 10.00 |
| Stress-induced phosphoprotein 1 | Stip1 | NP_058017.1 | 143.81 | 15.98 | 9.00 |
| Glutathione S-transferase, &thetas; 3 | Gstt3 | NP_598755.1 | 328.43 | 36.49 | 9.00 |
| Heat shock 70-kDa protein 1B | Hspa1b | NP_034608.2 | 384.75 | 57.00 | 6.75 |
| Peroxiredoxin 2 | Prdx2 | NP_035693.3 | 2,525.41 | 826.50 | 3.06 |
| Peroxiredoxin 3 | Prdx3 | NP_031478.1 | 1,564.33 | 568.85 | 2.75 |
| Heat shock protein 9 | Hspa9 | NP_034611.2 | 4,002.11 | 1,483.78 | 2.70 |
| Heat shock protein 2 | Hspa2 | NP_001002012.1 | 1,694.39 | 689.24 | 2.46 |
| Heat shock protein 8 | Hspa8 | NP_112442.2 | 2,370.50 | 1,100.59 | 2.15 |
| Peroxiredoxin 1 | Prdx1 | NP_035164.1 | 5,862.06 | 3,291.77 | 1.78 |
| Nuclear proteins | |||||
| Acidic (leucine-rich) nuclear phosphoprotein 32 f | Anp32a | NP_033802.2 | 175.21 | 0.00 | Unique |
| Mitochondria-ER-metabolism | |||||
| Phosphoglycerate mutase 2 | Pgam2 | NP_061358.1 | 1,318.20 | 0.00 | Unique |
| Acyl-coenzyme A dehydrogenase, short/branched cha | Acadsb | NP_080102.1 | 459.54 | 0.00 | Unique |
| NADH-ubiquinone oxidoreductase flavoprotein 3 is | 1500032D16Rik | NP_084363.2 | 297.04 | 0.00 | Unique |
| Malic enzyme 3, NADP(+)-dependent, mitochondrial | Me3 | NP_852072.1 | 267.94 | 0.00 | Unique |
| LRP16 protein | Macrod1 | NP_598908.1 | 258.11 | 0.00 | Unique |
| AAA-ATPase TOB3 | Atad3a | NP_849534.1 | 224.65 | 0.00 | Unique |
| Hydroxysteroid dehydrogenase like 2 | Hsdl2 | NP_077217.2 | 221.37 | 0.00 | Unique |
| DnaJ (Hsp40) homolog, subfamily C, member 11 | Dnajc11 | NP_766292.2 | 173.96 | 0.00 | Unique |
| Endoplasmic reticulum protein ERp29 precursor | Erp29 | NP_080405.1 | 173.47 | 0.00 | Unique |
| Acyl-CoA thioesterase 9 | Acot9 | NP_062710.2 | 158.23 | 0.00 | Unique |
| ERO1-like | Ero1l | NP_056589.1 | 147.92 | 0.00 | Unique |
| Glutaminase isoform 1 | Gls | NP_001074550.1 | 135.20 | 0.00 | Unique |
| Sulfite oxidase | Suox | NP_776094.2 | 115.22 | 0.00 | Unique |
| Ferredoxin reductase | Fdxr | NP_032023.1 | 110.70 | 0.00 | Unique |
| Malic enzyme 2, NAD(+)-dependent, mitochondrial | Me2 | NP_663469.1 | 106.38 | 0.00 | Unique |
| Aspartyl-tRNA synthetase | Dars | NP_803228.1 | 104.99 | 0.00 | Unique |
| Lectin, mannose-binding, 1 | Lman1 | NP_081676.1 | 103.83 | 0.00 | Unique |
| NADH dehydrogenase subunit 5 | ND5 | NP_904338.1 | 87.62 | 0.00 | Unique |
| Monoamine oxidase A | Maoa | NP_776101.2 | 83.91 | 0.00 | Unique |
| Leucyl/cystinyl aminopeptidase | Lnpep | NP_766415.1 | 59.67 | 0.00 | Unique |
| Hexokinase domain containing 1 | Hkdc1 | NP_663394.1 | 48.90 | 0.00 | Unique |
| Creatine kinase, mitochondrial 1, ubiquitous | Ckmt1 | NP_034027.1 | 1,531.79 | 42.55 | 36.00 |
| Glutaminase isoform 2 | Gls | NP_001106854.1 | 1,030.39 | 30.31 | 34.00 |
| Solute carrier family 25 (mitochondrial carrier, | Slc25a12 | NP_766024.1 | 1,005.77 | 67.05 | 15.00 |
| Cytochrome-c oxidase subunit III | COX3 | NP_904334.1 | 401.03 | 33.42 | 12.00 |
| Metaxin 1 | Mtx1 | NP_038632.1 | 308.78 | 28.07 | 11.00 |
| Phosphoglucomutase 2 | Pgm2 | NP_082408.2 | 178.81 | 16.26 | 11.00 |
| Branched chain ketoacid dehydrogenase E1, α p | Bckdha | NP_031559.2 | 178.72 | 19.86 | 9.00 |
| Mitochondrial ribosomal protein S2 | Mrps2 | NP_536700.2 | 247.37 | 30.92 | 8.00 |
| Methylcrotonoyl-coenzyme A carboxylase 2 (β) | Mccc2 | NP_084302.1 | 602.81 | 81.46 | 7.40 |
| Branched chain aminotransferase 2, mitochondrial | Bcat2 | NP_033867.1 | 657.19 | 90.65 | 7.25 |
| Isocitrate dehydrogenase 3, β-subunit | Idh3b | NP_570954.1 | 426.59 | 71.10 | 6.00 |
| Mitochondrial trifunctional protein, α-subunit | Hadha | NP_849209.1 | 3,471.64 | 737.87 | 4.70 |
| Sorting and assembly machinery component 50 homol | Samm50 | NP_848729.1 | 597.72 | 134.97 | 4.43 |
| Citrate synthase | Cs | NP_080720.1 | 1,681.59 | 386.57 | 4.35 |
| Cytochrome-c oxidase, subunit VIb polypeptide 1 | Cox6b1 | NP_079904.1 | 3,078.01 | 794.32 | 3.88 |
| Pyruvate kinase, muscle | Pkm2 | NP_035229.2 | 1,711.47 | 449.48 | 3.81 |
| Acetyl-CoA synthetase 2-like | Acss1 | NP_542142.1 | 2,572.92 | 696.83 | 3.69 |
| NADH dehydrogenase (ubiquinone) 1 α subcomple | Ndufa10 | NP_077159.1 | 1,305.31 | 369.43 | 3.53 |
| 3-Hydroxybutyrate dehydrogenase, type 1 | Bdh1 | NP_780386.2 | 548.52 | 156.72 | 3.50 |
| ATP synthase F0 subunit 8 | ATP8 | NP_904332.1 | 2,961.52 | 901.33 | 3.29 |
| Carnitine acetyltransferase | Crat | NP_031786.2 | 494.07 | 155.28 | 3.18 |
| 13-kDa differentiation-associated protein | Ndufa12 | NP_079827.2 | 2,835.57 | 907.38 | 3.13 |
| Inner membrane protein, mitochondrial | Immt | NP_083949.2 | 1,573.30 | 512.51 | 3.07 |
| NADH dehydrogenase (ubiquinone) Fe-S protein 2 | Ndufs2 | NP_694704.1 | 741.08 | 247.03 | 3.00 |
| ATP synthase, H+ transporting, mitochondrial F0 | Atp5f1 | NP_033855.2 | 3,385.28 | 1,139.94 | 2.97 |
| NADH dehydrogenase (ubiquinone) 1 α subcomple | Ndufa4 | NP_035016.1 | 3,752.63 | 1,286.62 | 2.92 |
| Voltage-dependent anion channel 3 | Vdac3 | NP_035826.1 | 845.45 | 292.66 | 2.89 |
| Choline dehydrogenase | Chdh | NP_780552.1 | 602.28 | 210.80 | 2.86 |
| Pyruvate dehydrogenase (lipoamide) β | Pdhb | NP_077183.1 | 2,414.15 | 847.52 | 2.85 |
| Isocitrate dehydrogenase 3 (NAD+) α | Idh3a | NP_083849.1 | 2,371.42 | 857.75 | 2.76 |
| Propionyl-coenzyme A carboxylase, α polypepti | Pcca | NP_659093.1 | 1,338.49 | 487.86 | 2.74 |
| ATP synthase, H+ transporting, mitochondrial F1 com | Atp5o | NP_613063.1 | 8,645.95 | 3,252.93 | 2.66 |
| Aldehyde dehydrogenase 1 family, member L1 | Aldh1l1 | NP_081682.1 | 2,360.47 | 891.51 | 2.65 |
| Cytochrome-c oxidase subunit II | COX2 | NP_904331.1 | 4,504.09 | 1,732.34 | 2.60 |
| Hexokinase 1 | Hk1 | NP_034568.1 | 1,032.45 | 397.82 | 2.60 |
| Upregulated during skeletal muscle growth 5 | Usmg5 | NP_075700.2 | 4,544.37 | 1,880.43 | 2.42 |
| NADH dehydrogenase (ubiquinone) 1 α subcomple | Ndufa8 | NP_080979.1 | 3,951.57 | 1,650.65 | 2.39 |
| Solute carrier family 25, member 5 | Slc25a5 | NP_031477.1 | 14,727.63 | 6,316.18 | 2.33 |
| Solute carrier family 25 (mitochondrial carrier) | Slc25a31 | NP_848473.1 | 6,461.13 | 2,777.15 | 2.33 |
| Isocitrate dehydrogenase 3 (NAD+), gamma | Idh3 g | NP_032349.1 | 1,028.39 | 444.08 | 2.32 |
| Superoxide dismutase 2, mitochondrial | Sod2 | NP_038699.2 | 3,983.27 | 1,747.76 | 2.28 |
| Solute carrier family 25 (mitochondrial carrier, ph | Slc25a3 | NP_598429.1 | 6,461.13 | 2,777.15 | 2.23 |
| Isocitrate dehydrogenase 2 (NADP+), mitochondrial | Idh2 | NP_766599.1 | 7,362.44 | 3,376.91 | 2.18 |
| NADH dehydrogenase (ubiquinone) Fe-S protein 1 | Ndufs1 | NP_663493.1 | 2,144.23 | 990.61 | 2.16 |
| Solute carrier family 25 (mitochondrial carrier) | Slc25a4 | NP_031476.3 | 2,704.82 | 1,276.43 | 2.12 |
| Glutamate oxaloacetate transaminase 2, mitochondrial | Got2 | NP_034455.1 | 3,648.91 | 1,729.54 | 2.11 |
| Methylcrotonoyl-coenzyme A carboxylase 1 (α) | Mccc1 | NP_076133.2 | 693.43 | 340.41 | 2.04 |
| ATP synthase, H+ transporting, mitochondrial F1F0 c | Atp5k | NP_031533.2 | 8,135.41 | 4,006.99 | 2.03 |
| NADH dehydrogenase (ubiquinone) 1 α subcomplex | Ndufa9 | NP_079634.1 | 1,223.27 | 611.63 | 2.00 |
| Aconitase 2, mitochondrial | Aco2 | NP_542364.1 | 11,677.50 | 5,873.85 | 1.99 |
| Acetyl-coenzyme A acetyltransferase 1 precursor | Acat1 | NP_659033.1 | 3,994.10 | 2,052.83 | 1.95 |
| ATP synthase, H+ transporting, mitochondrial F1 co | Atp5a1 | NP_031531.1 | 10,928.39 | 6,426.50 | 1.70 |
| Succinate dehydrogenase Fp subunit | Sdha | NP_075770.1 | 3,265.12 | 1,970.09 | 1.66 |
| ATP synthase, H+ transporting mitochondrial F1 co | Atp5b | NP_058054.2 | 14,280.52 | 9,253.92 | 1.54 |
| Oxoglutarate dehydrogenase (lipoamide) | Ogdh | NP_035086.2 | 2,370.14 | 1,545.74 | 1.53 |
| Other | |||||
| Basigin isoform 2 | Bsg | NP_001070652.1 | 539.18 | 0.00 | Unique |
| Cardiotrophin-like cytokine factor 1 | Clcf1 | NP_064336.1 | 356.28 | 0.00 | Unique |
| Ladinin | Lad1 | NP_598425.2 | 305.79 | 0.00 | Unique |
| Small glutamine-rich tetratricopeptide repeat (TP) | Sgta | NP_078775.1 | 262.22 | 0.00 | Unique |
| Tetraspanin 8 | Tspan8 | NP_666122.1 | 195.45 | 0.00 | Unique |
| Proteasome activator subunit 2 isoform 2 | Psme2 | NP_001025026.1 | 191.14 | 0.00 | Unique |
| Hypothetical protein LOC67809 | 1200015F23Rik | NP_001028308.1 | 134.54 | 0.00 | Unique |
| ROD1 regulator of differentiation 1 isoform 2 | Rod1 | NP_835458.1 | 105.82 | 0.00 | Unique |
| Protease (prosome, macropain) 26S subunit, ATPase | Psmc1 | NP_032973.1 | 101.66 | 0.00 | Unique |
| Acid sphingomyelinase-like phosphodiesterase 3a | Smpdl3a | NP_065586.3 | 100.29 | 0.00 | Unique |
| U2 small nuclear ribonucleoprotein auxiliary fac | U2af2 | NP_598432.2 | 94.13 | 0.00 | Unique |
| Asparaginyl-tRNA synthetase | Nars | NP_081626.1 | 79.28 | 0.00 | Unique |
| α-Isoform of regulatory subunit A, protein pho | Ppp2r1a | NP_058587.1 | 76.54 | 0.00 | Unique |
| Predicted: cingulin | Cgn | XP_001001375.2 | 40.13 | 0.00 | Unique |
| Predicted: similar to novel C2, FerI (NUC094) | Fer1l4 | XP_001481330.1 | 25.82 | 0.00 | Unique |
| Nascent polypeptide-associated complex α su | Naca | NP_001106670.1 | 22.68 | 0.00 | Unique |
| ATP-binding cassette, subfamily B (MDR/TAP), mem | Abcb8 | NP_083296.2 | 128.21 | 12.82 | 10.00 |
| Predicted: similar to 3-methylcrotonyl-CoA car | LOC677576 | XP_001005025.1 | 922.79 | 106.48 | 8.67 |
| Thioredoxin reductase 1 isoform 1 | Txnrd1 | NP_001035988.1 | 119.52 | 14.94 | 8.00 |
| Mannosidase 2, α1 | Man2a1 | NP_032575.1 | 60.80 | 7.60 | 8.00 |
| Glutamate oxaloacetate transaminase 1, soluble | Got1 | NP_034454.2 | 735.17 | 108.11 | 6.80 |
| Aldolase 1, A isoform | Aldoa | NP_031464.1 | 2,515.50 | 381.14 | 6.60 |
| IQ motif containing GTPase activating protein 2 | Iqgap2 | NP_081987.1 | 288.04 | 44.31 | 6.50 |
| Peptidase M20 domain containing 1 | Pm20d1 | NP_835180.1 | 197.87 | 35.98 | 5.50 |
| Aldo-keto reductase family 1, member C19 | Akr1c19 | NP_001013807.1 | 296.32 | 53.88 | 5.50 |
| Eukaryotic translation initiation factor 4B | Eif4b | NP_663600.1 | 270.29 | 49.14 | 5.50 |
| Transmembrane protein 109 | Tmem109 | NP_598903.1 | 418.15 | 76.03 | 5.50 |
| Nodal modulator 1 | Nomo1 | NP_694697.2 | 120.13 | 22.52 | 5.33 |
| Predicted: NADH dehydrogenase (ubiquinone) 1 α subcomple | Ndufa11 | XP_930081.2 | 1,389.31 | 330.79 | 4.20 |
| NADH dehydrogenase (ubiquinone) 1 β-subcomplex | Ndufb9 | NP_075661.1 | 2410.84 | 591.34 | 4.08 |
| Predicted: hypothetical protein isoform 2 | LOC100039281 | XP_001472561.1 | 8,538.62 | 2,148.08 | 3.98 |
| Phosphoglycerate kinase 1 | Pgk1 | NP_032854.2 | 1,167.22 | 314.25 | 3.71 |
| Predicted: hypothetical protein | LOC100046199 | XP_001475883.1 | 1,900.90 | 536.15 | 3.55 |
| Predicted: similar to ubiquitin A-52 residue ribos | LOC629750 | XP_899768.2 | 2,583.06 | 815.70 | 3.17 |
| Enolase 2, γ neuronal | Eno2 | NP_038537.1 | 401.72 | 126.86 | 3.17 |
| Spectrin β2-isoform 2 | Spnb2 | NP_033286.2 | 1,660.32 | 581.31 | 2.86 |
| Prothymosin-α | Ptma | NP_032998.1 | 2,692.98 | 979.26 | 2.75 |
| Nicotinamide nucleotide transhydrogenase | Nnt | NP_032736.2 | 2,028.41 | 807.85 | 2.51 |
| Histone cluster 1, H2ab | Hist1 h2ab | NP_783591.2 | 3,820.17 | 1,627.11 | 2.35 |
| Carnitine palmitoyltransferase 1a, liver | Cpt1a | NP_038523.2 | 861.18 | 373.93 | 2.30 |
| Heterogeneous nuclear ribonucleoprotein L | Hnrnpl | NP_796275.2 | 864.89 | 415.81 | 2.08 |
| Histone H3-like | OTTMUSG00000007855 | NP_001074488.1 | 6,757.74 | 3,346.07 | 2.02 |
| Histone cluster 2, H2bb | Hist2 h2bb | NP_783597.2 | 7,758.53 | 4,094.78 | 1.89 |
Values are normalized spectral counts from mass spectrometry. Final column shows ratio of normalized protein spectral count in EGFP+ cells to EGFP− cells.
Table 3.
List of proteins that were identified as being significantly enriched in epididymal EGFP+ cells compared with renal EGFP− cells
| Protein Name | Gene Symbol | Accession No. | EGFP+ | EGFP− | Ratio |
|---|---|---|---|---|---|
| V-ATPase - acid/base | |||||
| V-ATPase subunit A | Atp6v1a | NP_031534.2 | 219.54 | 0.00 | Unique |
| V-ATPase subunit B1 | Atp6v1b1 | NP_598918.1 | 158.48 | 0.00 | Unique |
| V-ATPase subunit B2 | Atp6v1b2 | NP_031535.2 | 123.78 | 0.00 | Unique |
| V-ATPase subunit a4 | Atp6v0a4 | NP_536715.2 | 41.85 | 0.00 | Unique |
| Male urogenital tract | |||||
| Cysteine-rich secretory protein 1 | Crisp1 | NP_033768.3 | 216.77 | 0.00 | Unique |
| Serine (or cysteine) proteinase inhibitor, clade B, member 6b | Serpinb6b | NP_035584.1 | 117.55 | 0.00 | Unique |
| Serine proteinase inhibitor member 6C | Serpinb6c | NP_683744.2 | 93.21 | 0.00 | Unique |
| Chaperonin containing Tcp1, subunit 5 (ε) | Cct5 | NP_031663.1 | 67.09 | 0.00 | Unique |
| Lactate dehydrogenase B | Ldhb | NP_032518.1 | 492.18 | 82.03 | 6.00 |
| Dihydrolipoamide dehydrogenase | Dld | NP_031887.2 | 184.26 | 36.85 | 5.00 |
| N-acylsphingosine amidohydrolase 1 | Asah1 | NP_062708.1 | 313.41 | 67.16 | 4.67 |
| Acyl-coenzyme A oxidase 3, pristanoyl | Acox3 | NP_109646.2 | 459.16 | 102.04 | 4.50 |
| A disintegrin and metalloprotease domain 7 | Adam7 | NP_031428.2 | 168.18 | 44.85 | 3.75 |
| Phosphatidylethanolamine binding protein 1 | Pebp1 | NP_061346.2 | 1,296.18 | 528.07 | 2.45 |
| Intracellular trafficking | |||||
| RAB5A, member RAS oncogene family | Rab5a | NP_080163.1 | 211.88 | 0.00 | Unique |
| Cathepsin B preproprotein | Ctsb | NP_031824.1 | 134.12 | 0.00 | Unique |
| Adaptor protein complex AP-1, μ2-subunit isoform 2 | Ap1m2 | NP_033808.2 | 103.84 | 0.00 | Unique |
| SAC1 (supressor of actin mutations 1, homolog)-like | Sacm1l | NP_109617.1 | 89.63 | 0.00 | Unique |
| Junction plakoglobin | Jup | NP_034723.1 | 48.90 | 0.00 | Unique |
| Ubiquitin specific protease 14 isoform 2 | Usp14 | NP_001033678.1 | 95.57 | 0.00 | Unique |
| Fatty acid synthase | Fasn | NP_032014.3 | 150.50 | 7.34 | 20.50 |
| Coatomer protein complex, subunit β 2 (β prime) | Copb2 | NP_056642.1 | 107.37 | 9.76 | 11.00 |
| Solute carrier family 9 (sodium/hydrogen exchanger), isoform 3 regulator 1 | Slc9a3r1 | NP_036160.1 | 440.41 | 51.81 | 8.50 |
| ADP-ribosylation factor 5 | Arf5 | NP_031506.1 | 487.10 | 97.42 | 5.00 |
| Ubiquitin B | Ubb | NP_035794.1 | 1,338.43 | 756.51 | 1.77 |
| Clathrin, heavy polypeptide (Hc) | Cltc | NP_001003908.1 | 751.74 | 480.28 | 1.57 |
| Cytoskeleton | |||||
| Hypothetical protein LOC66302 | 2410005O16Rik | NP_079752.3 | 114.29 | 0.00 | Unique |
| Histone deacetylase 6 | Hdac6 | NP_034543.2 | 79.55 | 0.00 | Unique |
| Keratin 82 | Krt82 | NP_444479.1 | 70.04 | 0.00 | Unique |
| Keratin 7 | Krt7 | NP_149064.1 | 611.33 | 118.32 | 5.17 |
| Myosin VI | Myo6 | NP_001034635.1 | 144.10 | 34.31 | 4.20 |
| Keratin 18 | Krt18 | NP_034794.1 | 1,389.35 | 336.81 | 4.13 |
| Dynein, cytoplasmic, heavy chain 1 | Dync1 h1 | NP_084514.2 | 77.06 | 20.67 | 3.73 |
| Myosin IB | Myo1b | NP_034993.2 | 101.12 | 31.11 | 3.25 |
| Gelsolin | Gsn | NP_666232.2 | 360.71 | 174.54 | 2.07 |
| Keratin complex 2, basic, gene 8 | Krt8 | NP_112447.2 | 1,429.48 | 806.37 | 1.77 |
| Membrane dynamics | |||||
| Prominin 2 isoform 1 | Prom2 | NP_620089.1 | 171.71 | 0.00 | Unique |
| Annexin A13 | Anxa13 | NP_081487.1 | 139.19 | 0.00 | Unique |
| Dynamin 1-like isoform a | Dnm1l | NP_690029.2 | 50.29 | 0.00 | Unique |
| Prominin 1 | Prom1 | NP_032961.1 | 93.53 | 10.39 | 9.00 |
| Transporters and channels | |||||
| Solute carrier organic anion transporter family, member 4a1 | Slco4a1 | NP_683735.1 | 77.25 | 0.00 | Unique |
| Chloride intracellular channel 3 | Clic3 | NP_081361.1 | 186.25 | 0.00 | Unique |
| Solute carrier family 16, member 1 | Slc16a1 | NP_033222.1 | 262.83 | 18.77 | 14.00 |
| Intracellular signaling, receptors, hormones | |||||
| Prostaglandin E synthase | Ptges | NP_071860.1 | 347.11 | 0.00 | Unique |
| Prostaglandin H2 d-isomerase | Ptgds | NP_032989.2 | 189.88 | 0.00 | Unique |
| Hydroxysteroid (17-β) dehydrogenase 12 protein | Hsd17b12 | NP_062631.1 | 143.92 | 0.00 | Unique |
| Prostaglandin-endoperoxide synthase 2 | Ptgs2 | NP_035328.2 | 57.96 | 0.00 | Unique |
| Prostaglandin-endoperoxide synthase 1 | Ptgs1 | NP_032995.1 | 57.94 | 0.00 | Unique |
| LPS-responsive beige-like anchor isoform α | Lrba | NP_109620.2 | 22.09 | 0.00 | Unique |
| Progesterone receptor membrane component | Pgrmc1 | NP_058063.2 | 645.33 | 230.48 | 2.80 |
| Protein disulfide isomerase associated 4 | Pdia4 | NP_033917.2 | 732.35 | 428.36 | 1.71 |
| Stress response-antioxidant | |||||
| Glutathione S-transferase, μ 6 | Gstm6 | NP_032210.3 | 273.21 | 0.00 | Unique |
| Glutathione peroxidase 5 | Gpx5 | NP_034473.1 | 590.98 | 0.00 | Unique |
| Glutathione S-transferase, μ 7 | Gstm7 | NP_080948.2 | 505.65 | 77.79 | 6.50 |
| Heat shock 70-kDa protein 1B | Hspa1b | NP_034608.2 | 327.75 | 114.00 | 2.88 |
| Glutathione S-transferase, μ 4 | Gstm4 | NP_081040.1 | 1,567.43 | 548.60 | 2.86 |
| Heat shock protein 5 | Hspa5 | NP_071705.2 | 1,891.66 | 828.47 | 2.28 |
| Glutathione S-transferase, μ 5 | Gstm5 | NP_034490.1 | 1,314.06 | 713.35 | 1.84 |
| Glutathione S-transferase, μ 2 | Gstm2 | NP_032209.1 | 1,710.94 | 933.24 | 1.83 |
| Peroxiredoxin 1 | Prdx1 | NP_035164.1 | 1,758.62 | 1,037.13 | 1.70 |
| Apoptosis | |||||
| FGF receptor activating protein 1 | Frag1 | NP_663558.1 | 379.27 | 0.00 | Unique |
| Carboxylesterase 1 | Ces1 | NP_067431.2 | 143.59 | 0.00 | Unique |
| Membrane metallo endopeptidase | Mme | NP_032630.2 | 105.02 | 0.00 | Unique |
| Tripeptidyl-peptidase I | Tpp1 | NP_034036.1 | 179.32 | 32.60 | 5.50 |
| Aminolevulinate, δ-, dehydratase | Alad | NP_032551.3 | 749.51 | 166.56 | 4.50 |
| Apoptosis-inducing factor, mitochondrion-associated 1 | Aifm1 | NP_036149.1 | 134.80 | 29.96 | 4.50 |
| Voltage-dependent anion channel 1 | Vdac1 | NP_035824.1 | 747.83 | 260.12 | 2.88 |
| Prolyl 4-hydroxylase, β polypeptide | P4 hb | NP_035162.1 | 1,174.23 | 683.51 | 1.72 |
| Nuclear proteins | |||||
| Aminomethyltransferase | Amt | NP_001013836.1 | 159.06 | 0.00 | Unique |
| Synaptic nuclear envelope 2 | Syne2 | NP_001005510.2 | 16.61 | 1.28 | 13.00 |
| Nuclear mitotic apparatus protein 1 | Numa1 | NP_598708.2 | 38.19 | 4.24 | 9.00 |
| Lysosome | |||||
| Hexosaminidase B | Hexb | NP_034552.1 | 212.71 | 0.00 | Unique |
| Cathepsin D | Ctsd | NP_034113.1 | 88.98 | 0.00 | Unique |
| Mannosidase 2, α B1 | Man2b1 | NP_034894.2 | 43.61 | 0.00 | Unique |
| Epididymal secretory protein E1 | Npc2 | NP_075898.1 | 1824.59 | 121.64 | 15.00 |
| Mitochondria - ER-metabolism | |||||
| 6.8-kDa Mitochondrial proteolipid | 2010107E04Rik | NP_081636.1 | 597.19 | 0.00 | Unique |
| CDGSH iron sulfur domain 1 | Cisd1 | NP_598768.1 | 495.99 | 0.00 | Unique |
| NADPH-dependent retinol dehydrogenase/reductase isoform 1 | Dhrs4 | NP_001033027.1 | 234.01 | 0.00 | Unique |
| Alcohol dehydrogenase 5 (class III), χ-polypeptide | Adh5 | NP_031436.2 | 202.29 | 0.00 | Unique |
| Arginine-rich, mutated in early stage tumors | Armet | NP_083379.2 | 196.20 | 0.00 | Unique |
| Succinate-coenzyme A ligase, GDP-forming, β-subunit | Suclg2 | NP_035637.1 | 170.85 | 0.00 | Unique |
| Acyl-coenzyme A dehydrogenase, short/branched chain | Acadsb | NP_080102.1 | 167.11 | 0.00 | Unique |
| Creatine kinase, mitochondrial 1, ubiquitous | Ckmt1 | NP_034027.1 | 148.92 | 0.00 | Unique |
| Solute carrier family 25 (adenine nucleotide translocator), member 13 | Slc25a13 | NP_056644.1 | 147.72 | 0.00 | Unique |
| Nipsnap homolog 3A | Nipsnap3a | NP_079899.1 | 141.30 | 0.00 | Unique |
| Sideroflexin 3 | Sfxn3 | NP_444427.1 | 141.22 | 0.00 | Unique |
| Cytochrome P450, family 4, subfamily a family | Cyp4a12b | NP_758510.2 | 120.05 | 0.00 | Unique |
| Mitochondrial carrier homolog 2 | Mtch2 | NP_062732.1 | 119.41 | 0.00 | Unique |
| Sulfite oxidase | Suox | NP_776094.2 | 98.76 | 0.00 | Unique |
| Hexokinase 2 | Hk2 | NP_038848.1 | 97.53 | 0.00 | Unique |
| Methylcrotonoyl-coenzyme A carboxylase 1 (α) | Mccc1 | NP_076133.2 | 88.25 | 0.00 | Unique |
| L-specific multifunctional β-oxdiation protein | Ehhadh | NP_076226.2 | 76.63 | 0.00 | Unique |
| Hydroxysteroid dehydrogenase like 2 | Hsdl2 | NP_077217.2 | 73.79 | 0.00 | Unique |
| Proline dehydrogenase | Prodh | NP_035302.2 | 58.79 | 0.00 | Unique |
| Acetyl-CoA synthetase 2-like | Acss1 | NP_542142.1 | 53.60 | 0.00 | Unique |
| Propionyl-coenzyme A carboxylase, α polypeptide | Pcca | NP_659093.1 | 50.04 | 0.00 | Unique |
| Serine hydroxymethyltransferase 2 (mitochondrial) | Shmt2 | NP_082506.1 | 179.34 | 17.93 | 10.00 |
| Dnaj (Hsp40) homolog, subfamily C, member 10 | Dnajc10 | NP_077143.2 | 88.32 | 11.04 | 8.00 |
| Endoplasmic reticulum protein erp29 precursor | Erp29 | NP_080405.1 | 242.86 | 34.69 | 7.00 |
| Superoxide dismutase 2, mitochondrial | Sod2 | NP_038699.2 | 528.39 | 81.29 | 6.50 |
| Valyl-tRNA synthetase | Vars | NP_035820.2 | 178.28 | 28.52 | 6.25 |
| Aldehyde dehydrogenase family 3, subfamily A2 | Aldh3a2 | NP_031463.2 | 222.34 | 37.06 | 6.00 |
| Leucine-rich PPR motif-containing protein | Lrpprc | NP_082509.2 | 70.24 | 12.77 | 5.50 |
| Methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like | Mthfd1l | NP_758512.2 | 331.08 | 66.22 | 5.00 |
| Acyl-CoA thioesterase 10 | Acot10 | NP_073727.2 | 197.81 | 39.56 | 5.00 |
| Pyrroline-5-carboxylate reductase 1 | Pycr1 | NP_659044.1 | 278.01 | 61.78 | 4.50 |
| Phosphoglucomutase 2 | Pgm2 | NP_082408.2 | 146.30 | 32.51 | 4.50 |
| Acyl-CoA synthetase long-chain family member 5 | Acsl5 | NP_082252.1 | 118.10 | 26.24 | 4.50 |
| Acyl-CoA synthetase long-chain family member 1 | Acsl1 | NP_032007.2 | 166.77 | 38.49 | 4.33 |
| Solute carrier family 25, member 5 | Slc25a5 | NP_031477.1 | 1,336.12 | 334.03 | 4.00 |
| d-Lactate dehydrogenase | Ldhd | NP_081846.3 | 212.16 | 57.86 | 3.67 |
| Glutamate oxaloacetate transaminase 2, mitochondrial | Got2 | NP_034455.1 | 3,290.35 | 906.96 | 3.63 |
| Leucine aminopeptidase 3 | Lap3 | NP_077754.2 | 249.18 | 71.19 | 3.50 |
| Aldehyde dehydrogenase family 6, subfamily A1 | Aldh6a1 | NP_598803.1 | 500.73 | 155.40 | 3.22 |
| ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit | Atp5o | NP_613063.1 | 642.03 | 214.01 | 3.00 |
| ATP synthase, H+ transporting, mitochondrial F1 complex, gamma subunit isoform b | Atp5c1 | NP_001106209.1 | 760.18 | 264.41 | 2.88 |
| ER lipid raft associated 2 | Erlin2 | NP_705820.1 | 369.66 | 132.02 | 2.80 |
| Hypoxia upregulated 1 | Hyou1 | NP_067370.3 | 314.80 | 116.93 | 2.69 |
| Aconitase 2, mitochondrial | Aco2 | NP_542364.1 | 819.06 | 315.92 | 2.59 |
| Solute carrier family 25 (adenine nucleotide translocator), member 31 | Slc25a31 | NP_848473.1 | 850.15 | 340.06 | 2.50 |
| Acetyl-coenzyme A acetyltransferase 1 precursor | Acat1 | NP_659033.1 | 468.58 | 200.82 | 2.33 |
| Lactate dehydrogenase A | Ldha | NP_034829.1 | 493.17 | 219.19 | 2.25 |
| Calreticulin | Calr | NP_031617.1 | 958.44 | 458.39 | 2.09 |
| Pyruvate kinase, muscle | Pkm2 | NP_035229.2 | 708.79 | 345.75 | 2.05 |
| Aldehyde dehydrogenase family 1, subfamily A1 | Aldh1a1 | NP_038495.2 | 1,872.67 | 991.41 | 1.89 |
| ATP synthase, H+ transporting, mitochondrial F1 complex, α subunit, isoform 1 | Atp5a1 | NP_031531.1 | 1,338.85 | 719.63 | 1.86 |
| Calnexin | Canx | NP_001103970.1 | 1,040.46 | 564.82 | 1.84 |
| ATP synthase, H+ transporting mitochondrial F1 complex, β-subunit | Atp5b | NP_058054.2 | 1,705.14 | 941.38 | 1.81 |
| Aldehyde dehydrogenase 2, mitochondrial | Aldh2 | NP_033786.1 | 1,609.55 | 902.06 | 1.78 |
| Malate dehydrogenase 2, NAD (mitochondrial) | Mdh2 | NP_032643.2 | 1,516.37 | 898.59 | 1.69 |
| Other | |||||
| Predicted: similar to ubiquitin A-52 residue ribosomal protein fusion product 1 isoform 2 | LOC629750 | XP_899768.2 | 407.85 | 0.00 | Unique |
| Serine (or cysteine) proteinase inhibitor, clade B, member 9 | Serpinb9 | NP_033282.1 | 402.28 | 0.00 | Unique |
| Hypothetical protein LOC231293 | C130090K23Rik | NP_851840.1 | 396.52 | 0.00 | Unique |
| Aldo-keto reductase family 1, member C19 | Akr1c19 | NP_001013807.1 | 296.32 | 0.00 | Unique |
| Aldo-keto reductase family 1, member C13 | Akr1c13 | NP_038806.1 | 243.15 | 0.00 | Unique |
| Acetyl-coenzyme A acetyltransferase 2 | Acat2 | NP_033364.2 | 217.93 | 0.00 | Unique |
| D6Wsu176e protein | D6Wsu176e | NP_613053.3 | 202.00 | 0.00 | Unique |
| Pyrroline-5-carboxylate reductase family, member 2 | Pycr2 | Np_598466.1 | 178.26 | 0.00 | Unique |
| Predicted: similar to AFG3(ATPase family gene 3)-like 2 (yeast) | LOC100048880 | XP_001472434.1 | 149.66 | 0.00 | Unique |
| Golgi apparatus protein 1 | Glg1 | Np_033175.1 | 142.07 | 0.00 | Unique |
| Lectin, mannose-binding 2 | Lman2 | Np_080104.2 | 123.67 | 0.00 | Unique |
| Nucleobindin 2 isoform 2 | Nucb2 | Np_058053.2 | 99.30 | 0.00 | Unique |
| Farnesyl diphosphate synthetase | Fdps | Np_608219.1 | 98.57 | 0.00 | Unique |
| Hypothetical protein LOC68646 isoform 2 | 1110020G09Rik | NP_001035485.2 | 82.76 | 0.00 | Unique |
| Hydroxysteroid (17-β) dehydrogenase 4 | Hsd17b4 | Np_032318.2 | 75.49 | 0.00 | Unique |
| Eukaryotic translation initiation factor 3 subunit 6 interacting protein | Eif3eip | Np_660121.2 | 75.06 | 0.00 | Unique |
| Transmembrane 9 superfamily member 2 | Tm9 sf2 | Np_542123.2 | 66.34 | 0.00 | Unique |
| ATP citrate lyase | Acly | NP_598798.1 | 50.11 | 0.00 | Unique |
| Predicted: similar to GPI inositol-deacylase | D230012E17Rik | XP_912118.2 | 47.81 | 0.00 | Unique |
| U5 snRNP-associated 102 kDa protein | Prpf6 | NP_598462.1 | 37.48 | 0.00 | Unique |
| Predicted: Golgi autoantigen, golgin subfamily b, macrogolgin 1 | Golgb1 | XP_001481110.1 | 24.37 | 0.00 | Unique |
| Staphylococcal nuclease domain containing 1 | Snd1 | Np_062750.2 | 235.09 | 19.59 | 12.00 |
| Predicted: hypothetical protein | LOC640248 | XP_922185.2 | 556.21 | 46.35 | 12.00 |
| Carbonyl reductase 1 | Cbr1 | Np_031646.2 | 391.63 | 32.64 | 12.00 |
| Carboxylesterase 3 | Ces3 | Np_444430.2 | 291.32 | 32.37 | 9.00 |
| N-acetylneuraminic acid synthase | Nans | NP_444409.1 | 224.86 | 24.98 | 9.00 |
| Galactosidase, β 1-like 2 | Glb1l2 | Np_722498.1 | 121.68 | 13.52 | 9.00 |
| Palmitoyl-protein thioesterase 1 | Ppt1 | Np_032943.2 | 260.94 | 28.99 | 9.00 |
| Pyridoxal-dependent decarboxylase domain containing 1 isoform 3 | Pdxdc1 | Np_001034622.1 | 432.88 | 50.93 | 8.50 |
| Predicted: similar to serine proteinase inhibitor hongrES1 | Gm46 | XP_001478131.1 | 171.03 | 21.38 | 8.00 |
| Aconitase 1 | Aco1 | Np_031412.2 | 142.67 | 20.38 | 7.00 |
| Ribosomal protein S3 | Rps3 | NP_036182.1 | 262.42 | 37.49 | 7.00 |
| Endoplasmic reticulum-Golgi intermediate compartment (ERGIC) 1 | Ergic1 | NP_080446.1 | 214.97 | 30.71 | 7.00 |
| Lysophospholipase-like 1 | Lyplal1 | Np_666218.1 | 265.73 | 37.96 | 7.00 |
| Hypoxanthine guanine phosphoribosyl transferase 1 | Hprt1 | Np_038584.2 | 284.90 | 40.70 | 7.00 |
| Plasma glutamate carboxypeptidase | Pgcp | Np_061225.2 | 501.80 | 77.20 | 6.50 |
| Glycoprotein, synaptic 2 | Gpsn2 | Np_598879.1 | 1,717.91 | 332.50 | 5.17 |
| Abhydrolase domain containing 12 | Abhd12 | Np_077785.2 | 220.90 | 44.18 | 5.00 |
| 3-Phosphoglycerate dehydrogenase | Phgdh | Np_058662.2 | 247.41 | 53.02 | 4.67 |
| UDP-glucose ceramide glucosyltransferase-like 1 | Ugcgl1 | NP_942602.1 | 158.70 | 39.68 | 4.00 |
| Predicted: similar to desmoplakin | Dsp | XP_001481322.1 | 36.06 | 12.02 | 3.00 |
| Argininosuccinate lyase | Asl | Np_598529.1 | 347.90 | 135.29 | 2.57 |
| Predicted: similar to vesicle associated protein | Sec31a | XP_912694.3 | 124.94 | 48.59 | 2.57 |
| Coatomer protein complex subunit α | Copa | Np_034068.2 | 122.72 | 50.53 | 2.43 |
| UbiE-YGHL1 fusion protein | LOC554292 | NP_001019843.1 | 566.72 | 233.35 | 2.43 |
| Protein disulfide isomerase-associated 6 | Pdia6 | Np_082235.1 | 1,150.15 | 513.46 | 2.24 |
| Malate dehydrogenase 1, NAD (soluble) | Mdh1 | NP_032644.2 | 987.16 | 493.58 | 2.00 |
| Triosephosphate isomerase 1 | Tpi1 | Np_033441.1 | 1,684.60 | 973.32 | 1.73 |
Values are normalized spectral counts from mass spectrometry. Final column shows ratio of normalized protein spectral count in EGFP+ cells to EGFP− cells.
Several proteins involved in intracellular trafficking were detected in the EGFP+ populations. For example, in renal EGFP+ cells, Rab1, Rab1B, Rab2a, Rab4a, Rab5a, Rab5b, Rab5c, Rab7, Rab8a, Rab8b, Rab10, Rab11a, Rab14, Rab18, Rab21, Rab27a, Rab31, and Rab35 were detected in these cells (supplemental Table S1). However, only Rab1 and Rab10 were enriched in these cells compared with all other cell types (EGFP− cells). Similarly, out of several Rab proteins present in epididymal EGFP+ cells—Rab1, Rab2a, Rab5a, Rab5b, Rab5c, Rab6a, Rab6b, Rab7, Rab8a, Rab10, Rab11a, Rab11b, Rab12a, Rab14, and Rab35 (supplemental Table S2)—only Rab5a was shown to be enriched in these cells compared with EGFP− cells. Of note, the adaptor protein complex AP-1 mu1B, as well as Fasn, a protein that interacts with caveolin-1 in lipid rafts, β-COP, clathrin, and NHERF-1 (Slc9a3r1) were all enriched in epididymal EGFP+ cells. Several annexins (Anxa4, Anxa6, and Anxa11) were enriched in renal EGFP+ cells, while Anxa13 was enriched in epididymal EGFP+ cells.
In agreement with their dynamic endocytic function, proteins involved in cytoskeletal reorganization and vesicle recycling were shown to be enriched in the renal and epididymal EGFP+ populations. Among these proteins, Cdc42 and its downstream effector, Iqgap1 (3), as well as Pacsin 2, were identified in renal EGFP+ cells. These proteins have been implicated in actin and microtubule dynamics (3, 27). Hdac6, another protein involved in actin dynamics, was detected exclusively in epididymal EGFP+ cells.
Enriched proteins common to renal and epididymal EGFP+ cells.
A list of enriched proteins common to both renal and epididymal EGFP+ populations is provided in Table 4. As expected, among these proteins, subunits A, B1, and a4 of the V-ATPase, and gelsolin were present in both samples. Interestingly, the progesterone receptor, Pgrmc1, was enriched in both cell types, as well as the PKA-binding protein, Lrba. Members of the keratin family, keratin 7, 18, and 82, were also identified, of which keratin 82 was uniquely detected in the EGFP+ populations compared with their respective EGFP− samples.
Table 4.
List of proteins enriched in EGFP+ cells and common to both renal and epididymal samples
| Protein Name | Gene Symbol | Accession No. | Ratio Epididymis | Ratio Kidney |
|---|---|---|---|---|
| V-ATPase | ||||
| ATPase, H+ transporting, lysosomal V1 subunit B1 | Atp6v1b1 | NP_598918.1 | Unique | Unique |
| ATPase, H+ transporting, lysosomal V1 subunit A | Atp6v1a | NP_031534.2 | Unique | 7.84 |
| ATPase, H+ transporting, lysosomal V0 subunit A isoform 4 | Atp6v0a4 | NP_536715.2 | Unique | 6.44 |
| PKA binding-endosomal transport | ||||
| LPS-responsive beige-like anchor isoform α | Lrba | NP_109620.2 | Unique | 2.46 |
| Receptors | ||||
| Progesterone receptor membrane component | Pgrmc1 | NP_058063.2 | 2.80 | 8.00 |
| Cytoskeleton | ||||
| Keratin 82 | Krt82 | NP_444479.1 | Unique | Unique |
| Keratin 7 | Krt7 | NP_149064.1 | 5.17 | Unique |
| Keratin 18 | Krt18 | NP_034794.1 | 4.13 | 5.13 |
| Gelsolin | Gsn | NP_666232.2 | 2.07 | 4.10 |
| Stress response-antioxidant | ||||
| Heat shock 70-kDa protein 1B (Hspa1b) | Hspa1b | NP_034608.2 | 2.88 | 6.75 |
| Peroxiredoxin 1 | Prdx1 | NP_035164.1 | 1.70 | 1.78 |
| Mitochondria | ||||
| Hydroxysteroid dehydrogenase like 2 | Hsdl2 | NP_077217.2 | Unique | Unique |
| Acyl-coenzyme A dehydrogenase, short/branched chain | Acadsb | NP_080102.1 | Unique | Unique |
| Sulfite oxidase | Suox | NP_776094.2 | Unique | Unique |
| Creatine kinase, mitochondrial 1, ubiquitous | Ckmt1 | NP_034027.1 | Unique | 36.00 |
| Aldo-keto reductase family 1, member C19 | Akr1c19 | NP_001013807 | Unique | 5.50 |
| Acetyl-CoA synthetase 2-like [Mus musculus] | Acss1 | NP_542142.1 | Unique | 3.69 |
| Propionyl-coenzyme A carboxylase, α polypeptide | Pcca | NP_659093.1 | Unique | 2.74 |
| Superoxide dismutase 2, mitochondrial | Sod2 | NP_038699.2 | 6.50 | 2.28 |
| Solute carrier family 25, member 5 | Slc25a5 | NP_031477.1 | 4.00 | 2.33 |
| Glutamate oxaloacetate transaminase 2, mitochondrial | Got2 | NP_034455.1 | 3.63 | 2.11 |
| ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit | Atp5o | NP_613063.1 | 3.00 | 2.66 |
| Aconitase 2, mitochondrial | Aco2 | NP_542364.1 | 2.59 | 1.99 |
| Solute carrier family 25 (adenine nucleotide translocator), member 31 | Slc25a31 | NP_848473.1 | 2.50 | 2.33 |
| Acetyl-coenzyme A acetyltransferase 1 precursor | Acat1 | NP_659033.1 | 2.33 | 1.95 |
| Pyruvate kinase, muscle | Pkm2 | NP_035229.2 | 2.05 | 3.81 |
| ATP synthase, H+ transporting, mitochondrial F1 complex, α subunit, isoform 1 | Atp5a1 | NP_031531.1 | 1.86 | 1.70 |
| ATP synthase, H+ transporting mitochondrial F1 complex, β-subunit | Atp5b | NP_058054.2 | 1.81 | 1.54 |
| Glucose metabolic process | ||||
| Phosphoglucomutase 2 | Pgm2 | NP_082408.2 | 4.50 | 11.00 |
| Protein modification | ||||
| Ubiquitin B | Ubb | NP_035794.1 | 1.77 | 2.30 |
| Lumen of ER, protein secretion | ||||
| Endoplasmic reticulum protein ERp29 precursor | Erp29 | NP_080405.1 | 7.00 | Unique |
| Not classified | ||||
| Methylcrotonoyl-coenzyme A carboxylase 1 (α) | Mccc1 | NP_076133.2 | Unique | 2.04 |
| Predicted: similar to ubiquitin A-52 residue ribosomal protein fusion product 1 isoform 2 | LOC629750 | XP_899768.2 | Unique | 3.17 |
Confirmation of proteins identified by proteomics using RT-PCR.
Enrichment of some of the proteins identified by proteomics in the EGFP+ populations compared with the EGFP− populations was confirmed by RT-PCR. Figure 6 shows higher expression of genes coding for the V-ATPase B1 subunit (Atp6v1b1), prominin 2 (Prom2), the LPS-responsive beige-like anchor (Lrba), the progesterone receptor (Pgrmc1), annexin A6 (Anxa6), drebrin-like (Dbnl), keratin 18 (Krt18), myosin 6 (Myo6), and the solute carrier organic anion transporter (Slco4a1) in both the renal and epididymal EGFP+ samples, compared with their respective EGFP− samples. In addition, Iqgap1 mRNA was enriched in renal EGFP+ cells, and mRNAs coding for prominin 1 (Prom1), and the hydroxysteroid (17-β) dehydrogenase 12 protein (Hsd17b12) were enriched in epididymal EGFP+ cells.
DISCUSSION
The major aim of this study was to determine the proteomic profiles of specialized acidifying cells in the kidney and epididymis. These cells all have in common a high expression of the B1 subunit of the V-ATPase in their plasma membrane, and we used our transgenic mice that express EGFP under the control of the promoter of this subunit (39) to harvest specifically these cells by FACS. An LC-MS/MS-based proteomic analysis was carried out in EGFP+ cells, which correspond chiefly to intercalated cells in the kidney and to narrow and clear cells in the epididymis. Results were compared with the proteins detected in the respective EGFP− cell samples of both organs. To minimize false positive identification, a target-decoy analysis was performed. We also corrected for the higher probability of detecting peptides from larger proteins by normalizing spectral counts to their molecular weight. An exhaustive list of all proteins identified in renal and epididymal EGFP+ cells is provided in supplemental data and is available at the NHLBI-LKEM website (http://dir.nhlbi.nih.gov/papers/lkem/kevcpd/). These new databases provide a foundation for further analysis of the specific functions of specialized acidifying cells in the kidney and epididymis.
Out of the thousands of proteins that were identified in these cells, 202 and 178 proteins were enriched in the renal and epididymal EGFP+ populations, respectively. A large number of these proteins are located in mitochondria, in agreement with the high density of mitochondria in renal intercalated cells and epididymal clear cells (8). In addition, expression of several proteins known to be expressed in B1-expressing cells in both the kidney and epididymis was confirmed in this study. These include subunits of the V-ATPase, such as the B1 subunit itself, and other proteins involved in acid/base transport—pendrin (expressed in B-IC), Rhbg (expressed in A-IC), and Car2 (expressed in both A-IC and B-IC). Interestingly, the membrane-bound Car15 was uniquely detected in renal EGFP+ cells. This latest member of the carbonic anhydrase family has recently been described in the mouse gastrointestinal tract (44) and kidney (31). While the role of Car2 in proton secretion and bicarbonate reabsorption is well established in the kidney, it will be interesting to examine the potential participation of Car15 in these processes. It has to be noted, however, that while this isoform is expressed in several species including rodents, fish and birds, it is absent from humans and chimpanzees (30).
The presence of calbindin-28K and aquaporin 3 in renal EGFP+ cells indicates that connecting tubule cells were also positively selected during FACS sorting, in agreement with our previous report showing expression of EGFP in these cells (39). Interestingly, the closely related calcium-binding protein, calbindin 2 (also known as calretinin), was also enriched in the renal EGFP+ population. These two genes are located in the chromosomal regions where the carbonic anhydrase isozyme gene cluster (CA1, CA2, CA3) is located (45). While the role of calbindin-28K in calcium reabsorption by the kidney is well characterized (5), the role of calretinin in kidney function is not known. Calretinin is expressed in neurons and cancer cells, including colon cancer cells. It is also expressed in the chick embryonic kidney (18), but its expression in the adult kidney has not been described previously.
Proton secretion by renal intercalated cells and epididymal clear cells is dynamically regulated via recycling of the V-ATPase to and from the plasma membrane. Several proteins involved in vesicle trafficking were detected in EGFP+ cells. These include several members of the small GTPase protein family. Interestingly, only a few of these proteins, Rab5a in epididymal EGFP+ cells and Rab1 and Rab10 in renal EGFP+ cells, were actually enriched in these cells compared with all other cell types of their respective organ, indicating their potential involvement in the acidifying function of these cells. Rab5a is located in early endosomes and it will be interesting to determine its potential participation in V-ATPase recycling. Interestingly, while Rab10 is traditionally involved in Golgi export, it was recently shown to interact with myosin V proteins, indicating its participation in vesicle transport along actin filaments (53).
Several proteins involved in cytoskeletal reorganization were identified in EGFP+ cells. Of note, the actin remodeling protein, gelsolin, was concentrated in both renal and epididymal EGFP+ cells, in agreement with our previous reports showing high expression of this protein in clear cells (1) and intercalated cells (37), and its participation in the recycling of V-ATPase in clear cells (1). Pacsin 2, another actin cytoskeleton modulating protein, was enriched in renal EGFP+ cells. In addition to its role in actin dynamics, Pacsin 2 has recently been proposed to modulate the formation of microtubules (27). Similarly, Cdc42 and its downstream effector, Iqgap1, two proteins also involved in actin and microtubule remodeling, were enriched in the renal EGFP+ cells. The presence of Iqgap1 in these cells correlates with its colocalization with AE1 in the basolateral membrane of type A intercalated cells (36).
Active recycling of the V-ATPase is accompanied by marked alterations in the shape of renal type A intercalated cells and epididymal clear cells. Accumulation of the V-ATPase in their apical membrane correlates with extensive elongation of microvilli, which contain higher numbers of the pump, and increased proton secretion (11, 46, 55, 59). Interestingly, several proteins involved in membrane dynamics were detected in these cells. For example, prominin 1 and 2 were enriched in epididymal EGFP+ cells, and prominin 2 was enriched in renal EGFP+ cells. These proteins are cholesterol-binding proteins that participate in the formation of membrane protrusions (13, 23). Prominin 1 is highly expressed in the kidney and epididymis, where it is restricted to the apical membrane of epithelial cells (22, 24), while prominin 2 is present in both the apical and basolateral domains of MDCK cells (23). Interestingly, prominin 1 and 2 can be released into the extracellular space and are associated with membrane particles that are present in the urine, saliva, and seminal fluids (23, 38). The physiological role of these particles, referred to as “prominosomes,” has not been elucidated, but microvilli have been proposed to be at the origin of these prominosomes (23). In the epididymis, prominin 1 is a highly glycosylated protein that has been localized to the stereocilia of principal cells (22). In the present study, while prominin 1 was identified in the epididymal EGFP− sample, which contains principal cells, we detected an enrichment of this protein in the EGFP+ population showing its expression in clear cells also. Several prominin 1 splice variants have been identified (22), and characterization of the splice variant(s) expressed in these cells will require further studies.
Renal intercalated cells and epididymal clear cells have a very high rate of endocytosis. While this process is partially due to the active recycling of the V-ATPase in these cells, clear cells were also proposed to help in clearing the luminal content of the epididymis of proteins that are shed from spermatozoa as they transit through this organ (28, 40). In agreement with this notion, Hdac6, a protein involved in fluid-phase endocytosis (26), was detected in these cells. In addition, the sperm surface protein, Crisp1, was enriched in the epididymal EGFP+ cells, while we have shown by RT-PCR that these cells do not express Crisp1 mRNA (data not shown). These data further support the notion that clear cells might contain “foreign” proteins that have been internalized for degradation.
While thousands of proteins were detected in renal and epididymal EGFP+ cells, only a few hundreds were shown to be concentrated in these cells compared with their respective EGFP− samples. Among these proteins, only a few dozen were common to both EGFP+ preparations. This shows that while V-ATPase-rich cells of the kidney and epididymis do share a common function (transmembrane proton transport), they are nevertheless very distinct cells expressing specific sets of proteins. Interestingly, in addition to the proteins known to be expressed in these cells (V-ATPase, gelsolin, etc.), novel candidates were identified in this study that might play a role in their common acidifying function. These include the progesterone receptor, Pgrmc1, which is highly enriched in both renal and epididymal EGFP+ cells. The putative PKA-binding protein, Lrba, which participates in endosomal transport (19, 20, 60), is another potential candidate for the regulation of V-ATPase recycling in these cells. In addition, a protein involved in estradiol production, Hsd17b12 (4), was detected uniquely in the epididymal EGFP+ population. Epididymal epithelial cells express estrogen receptors and are regulated by estrogens in addition to androgens (29, 62). The present study thus points toward a potential role for clear cells in the hormonal regulation of the male reproductive tract. Interestingly, both of the ammonia-producing enzymes, glutaminase 1 and 2, were detected in renal EGFP+ cells. This might indicate the potential participation of intercalated cells or connecting tubule cells in the production of ammonia in the kidney, which is currently attributed to proximal tubules (35).
The proteins identified here will, therefore, constitute the targets for future studies on the regulation of the acidifying function, or other functions, of these cells. It has to be noted that while the LC-MS/MS approach is extremely useful to detect many proteins in a particular sample, some proteins remained undetected or their detection was below the level of confidence set during statistical analysis. This is the case for prominin 2, which was detected exclusively in the renal and epididymal EGFP+ cells and not in their respective EGFP− samples, but was marked as “nonsignificant” in renal EGFP+ cells (see Table S1). Subsequent RT-PCR analysis correlated with the high expression of this protein in both renal and epididymal EGFP+ populations. On the other hand, our proteomic analysis was very accurate in determining whether or not a given protein was enriched in the EGFP+ cells compared with EGFP− cells. For example, the drebrin-like protein, Dbnl, was detected in both renal and epididymal EGFP+ cells, but it was significantly enriched only in renal EGFP+ cells and not in epididymal EGFP+ cells. These results were confirmed by RT-PCR confirming the high accuracy of the χ2-analysis performed here.
In summary, while both renal intercalated cells and epididymal clear cells do share some common features and functions, including active proton transport, they are nevertheless very distinct cells that express individual sets of proteins. These cells have well-established roles in the maintenance of 1) systemic acid/base balance (intercalated cells) and 2) an acidic luminal environment that is essential for sperm maturation and viability in the epididymis (clear cells). This is reflected by their common expression of the V-ATPase, and proteins involved in cytoskeletal rearrangement (e.g., gelsolin). A novel common candidate for the regulation of these cells is the progesterone receptor, which was enriched in both cell populations. Further studies will be required to determine the role of progesterone in their regulation and, in particular, whether this hormone modulates V-ATPase-dependent proton secretion. In conclusion, we provide here the first comprehensive protein expression profiles of specialized V-ATPase-B1-expressing cells in the kidney and epididymis. These proteomic databases provide a unique framework for the future characterization of the common and distinct functions of these specialized acidifying cells.
GRANTS
This work was supported by National Institutes of Health Grants HD-40793 (to S. Breton), DK-38452 (to S. Breton and D. Brown), and DK-42956 (to D. Brown) as well as the intramural research budget of NHLBI Project HL-001285 (to M. A. Knepper). N. Da Silva was supported by a research career enhancement award from the American Physiological Society. The Microscopy Core facility of the MGH Program in Membrane Biology receives support from the Boston Area Diabetes and Endocrinology Research Center (DK-57521) and the Center for the Study of Inflammatory Bowel Disease (DK-43341).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Dombkowski [Massachusetts General Hospital (MGH) Flow Cytometry Core], Wells W. Wu and Guanghui Wang [National Heart, Lung, and Blood Institute (NHLBI) Proteomic Core], and Mary McKee (Program in Membrane Biology) for excellent technical assistance.
REFERENCES
- 1.Beaulieu V, Da Silva N, Pastor-Soler N, Brown CR, Smith PJ, Brown D, Breton S. Modulation of the actin cytoskeleton via gelsolin regulates vacuolar H+-ATPase (V-ATPase) recycling. J Biol Chem 280: 8452–8463, 2005 [DOI] [PubMed] [Google Scholar]
- 2.Beyenbach KW, Wieczorek H. The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. J Exp Biol 209: 577–589, 2006 [DOI] [PubMed] [Google Scholar]
- 3.Bielak-Zmijewska A, Kolano A, Szczepanska K, Maleszewski M, Borsuk E. Cdc42 protein acts upstream of IQGAP1 and regulates cytokinesis in mouse oocytes and embryos. Dev Biol 322: 21–32, 2008 [DOI] [PubMed] [Google Scholar]
- 4.Blanchard PG, Luu-The V. Differential androgen and estrogen substrates specificity in the mouse and primates type 12 17beta-hydroxysteroid dehydrogenase. J Endocrinol 194: 449–455, 2007 [DOI] [PubMed] [Google Scholar]
- 5.Boros S, Bindels RJ, Hoenderop JG. Active Ca(2+) reabsorption in the connecting tubule. Pflügers Arch 458: 99–109, 2009 [DOI] [PubMed] [Google Scholar]
- 6.Breton S, Smith PJS, Lui B, Brown D. Acidification of the male reproductive tract by a proton pumping (H+)-ATPase. Nat Med 2: 470–472, 1996 [DOI] [PubMed] [Google Scholar]
- 7.Breton S, Wiederhold T, Marshansky V, Nsumu NN, Ramesh V, Brown D. The B1 subunit of the H+-ATPase is a PDZ domain-binding protein. Colocalization with NHE-RF in renal B-intercalated cells. J Biol Chem 275: 18219–18224, 2000 [DOI] [PubMed] [Google Scholar]
- 8.Brown D, Breton S. Mitochondria-rich, proton-secreting epithelial cells. J Exp Biol 199: 2345–2358, 1996 [DOI] [PubMed] [Google Scholar]
- 9.Brown D, Lui B, Gluck S, Sabolic I. A plasma membrane proton ATPase in specialized cells of rat epididymis. Am J Physiol Cell Physiol 263: C913–C916, 1992 [DOI] [PubMed] [Google Scholar]
- 10.Brown D, Lydon J, McLaughlin M, Stuart-Tilley A, Tyzskowski R, Alper SL. Antigen retrieval in cryostat sections and cultured cells by treatment with sodium dodecyl sulfate (SDS). Histochem Cell Biol 105: 261–267, 1996 [DOI] [PubMed] [Google Scholar]
- 11.Brown D, Paunescu TG, Breton S, Marshansky V. Regulation of the V-ATPase in kidney epithelial cells: dual role in acid/base homeostasis and vesicle trafficking. J Exp Biol 212: 1762–1772, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coleman RA, Wu DC, Liu J, Wade JB. Expression of aquaporins in the renal connecting tubule. Am J Physiol Renal Physiol 279: F874–F883, 2000 [DOI] [PubMed] [Google Scholar]
- 13.Corbeil D, Roper K, Fargeas CA, Joester A, Huttner WB. Prominin: a story of cholesterol, plasma membrane protrusions and human pathology. Traffic 2: 82–91, 2001 [DOI] [PubMed] [Google Scholar]
- 14.Cornwall GA, Hsia N. ADAM7, a member of the ADAM (a disintegrin and metalloprotease) gene family is specifically expressed in the mouse anterior pituitary and epididymis. Endocrinology 138: 4262–4272, 1997 [DOI] [PubMed] [Google Scholar]
- 15.Da Ros VG, Maldera JA, Willis WD, Cohen DJ, Goulding EH, Gelman DM, Rubinstein M, Eddy EM, Cuasnicu PS. Impaired sperm fertilizing ability in mice lacking Cysteine-RIch Secretory Protein 1 (CRISP1). Dev Biol 320: 12–18, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Da Silva N, Shum WW, El-Annan J, Paunescu TG, McKee M, Smith PJ, Brown D, Breton S. Relocalization of the V-ATPase B2 subunit to the apical membrane of epididymal clear cells of mice deficient in the B1 subunit. Am J Physiol Cell Physiol 293: C199–C210, 2007 [DOI] [PubMed] [Google Scholar]
- 17.Da Silva N, Shum WW, Breton S. Regulation of vacuolar proton pumping ATPase-dependent luminal acidification in the epididymis. Asian J Androl 9: 476–482, 2007 [DOI] [PubMed] [Google Scholar]
- 18.Daneo LS, Corvetti G, Panattoni GL. Ontogeny of calbindin-D28K and calretinin in developing chick kidney. Cell Tissue Res 279: 209–213, 1995 [DOI] [PubMed] [Google Scholar]
- 19.De Souza N, Vallier LG, Fares H, Greenwald I. SEL2, the C. elegans neurobeachin/LRBA homolog, is a negative regulator of lin-12/Notch activity and affects endosomal traffic in polarized epithelial cells. Development 134: 691–702, 2007 [DOI] [PubMed] [Google Scholar]
- 20.Dyomin VG, Chaganti SR, Dyomina K, Palanisamy N, Murty VV, Dalla-Favera R, Chaganti RS. BCL8 is a novel, evolutionarily conserved human gene family encoding proteins with presumptive protein kinase A anchoring function. Genomics 80: 158–165, 2002 [DOI] [PubMed] [Google Scholar]
- 21.Eberspaecher U, Roosterman D, Kratzschmar J, Haendler B, Habenicht UF, Becker A, Quensel C, Petri T, Schleuning WD, Donner P. Mouse androgen-dependent epididymal glycoprotein CRISP-1 (DE/AEG): isolation, biochemical characterization, and expression in recombinant form. Mol Reprod Dev 42: 157–172, 1995 [DOI] [PubMed] [Google Scholar]
- 22.Fargeas CA, Joester A, Missol-Kolka E, Hellwig A, Huttner WB, Corbeil D. Identification of novel Prominin-1/CD133 splice variants with alternative C-termini and their expression in epididymis and testis. J Cell Sci 117: 4301–4311, 2004 [DOI] [PubMed] [Google Scholar]
- 23.Florek M, Bauer N, Janich P, Wilsch-Braeuninger M, Fargeas CA, Marzesco AM, Ehninger G, Thiele C, Huttner WB, Corbeil D. Prominin-2 is a cholesterol-binding protein associated with apical and basolateral plasmalemmal protrusions in polarized epithelial cells and released into urine. Cell Tissue Res 328: 31–47, 2007 [DOI] [PubMed] [Google Scholar]
- 24.Florek M, Haase M, Marzesco AM, Freund D, Ehninger G, Huttner WB, Corbeil D. Prominin-1/CD133, a neural and hematopoietic stem cell marker, is expressed in adult human differentiated cells and certain types of kidney cancer. Cell Tissue Res 319: 15–26, 2005 [DOI] [PubMed] [Google Scholar]
- 25.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol 8: 917–929, 2007 [DOI] [PubMed] [Google Scholar]
- 26.Gao YS, Hubbert CC, Lu J, Lee YS, Lee JY, Yao TP. Histone deacetylase 6 regulates growth factor-induced actin remodeling and endocytosis. Mol Cell Biol 27: 8637–8647, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grimm-Gunter EM, Milbrandt M, Merkl B, Paulsson M, Plomann M. PACSIN proteins bind tubulin and promote microtubule assembly. Exp Cell Res 314: 1991–2003, 2008 [DOI] [PubMed] [Google Scholar]
- 28.Hermo L, Dworkin J, Oko R. Role of epithelial clear cells of the rat epididymis in the disposal of the contents of cytoplasmic droplets detached from spermatozoa. Am J Anat 183: 107–124, 1988 [DOI] [PubMed] [Google Scholar]
- 29.Hess RA. Estrogen in the adult male reproductive tract: a review. Reprod Biol Endocrinol 1: 52, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hilvo M, Innocenti A, Monti SM, De Simone G, Supuran CT, Parkkila S. Recent advances in research on the most novel carbonic anhydrases, CA XIII and XV. Curr Pharm Des 14: 672–678, 2008 [DOI] [PubMed] [Google Scholar]
- 31.Hilvo M, Salzano AM, Innocenti A, Kulomaa MS, Scozzafava A, Scaloni A, Parkkila S, Supuran CT. Cloning, expression, post-translational modifications and inhibition studies on the latest mammalian carbonic anhydrase isoform, CA XV. J Med Chem 52: 646–654, 2009 [DOI] [PubMed] [Google Scholar]
- 32.Hinton BT, Palladino MA, Rudolph D, Lan ZJ, Labus JC. The role of the epididymis in the protection of spermatozoa. Curr Top Dev Biol 33: 61–102, 1996 [DOI] [PubMed] [Google Scholar]
- 33.Hinton BT, Turner TT. Is the epididymis a kidney analogue? News Physiol Sci 3: 28–31, 1988 [Google Scholar]
- 34.Imai-Senga Y, Sun-Wada GH, Wada Y, Futai M. A human gene, ATP6E1, encoding a testis-specific isoform of H(+)-ATPase subunit E. Gene 289: 7–12, 2002 [DOI] [PubMed] [Google Scholar]
- 35.Knepper MA, Packer R, Good DW. Ammonium transport in the kidney. Physiol Rev 69: 179–249, 1989 [DOI] [PubMed] [Google Scholar]
- 36.Lai LW, Yong KC, Lien YH. Site-specific expression of IQGAP1, a key mediator of cytoskeleton, in mouse renal tubules. J Histochem Cytochem 56: 659–666, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lueck A, Brown D, Kwiatkowski DJ. The actin-binding proteins adseverin and gelsolin are both highly expressed but differentially localized in kidney and intestine. J Cell Sci 111: 3633–3643, 1998 [DOI] [PubMed] [Google Scholar]
- 38.Marzesco AM, Janich P, Wilsch-Brauninger M, Dubreuil V, Langenfeld K, Corbeil D, Huttner WB. Release of extracellular membrane particles carrying the stem cell marker prominin-1 (CD133) from neural progenitors and other epithelial cells. J Cell Sci 118: 2849–2858, 2005 [DOI] [PubMed] [Google Scholar]
- 39.Miller RL, Zhang P, Smith M, Beaulieu V, Paunescu TG, Brown D, Breton S, Nelson RD. V-ATPase B1 subunit promoter drives expression of EGFP in intercalated cells of kidney, clear cells of epididymis and airway cells of lung in transgenic mice. Am J Physiol Cell Physiol 288: C1134–C1144, 2005 [DOI] [PubMed] [Google Scholar]
- 40.Moore HD, Bedford JM. The differential absorptive activity of epithelial cells of the rat epididymus before and after castration. Anat Rec 193: 313–327, 1979. [DOI] [PubMed] [Google Scholar]
- 42.Nixon B, MacIntyre DA, Mitchell LA, Gibbs GM, O'Bryan M, Aitken RJ. The identification of mouse sperm-surface-associated proteins and characterization of their ability to act as decapacitation factors. Biol Reprod 74: 275–287, 2006. [DOI] [PubMed] [Google Scholar]
- 43.Orgebin-Crist MCDB, Davies J. The epididymis in the post-genome era. In: The Third International Conference on the Epididymis, edited by Hinton BT, Turner TT. Charlottesville, VA: Van Doren, 2003, p. 2–22 [Google Scholar]
- 44.Pan PW, Rodriguez A, Parkkila S. A systematic quantification of carbonic anhydrase transcripts in the mouse digestive system. BMC Mol Biol 8: 22, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parmentier M, Passage E, Vassart G, Mattei MG. The human calbindin D28k (CALB1) and calretinin (CALB2) genes are located at 8q21.3—-q221 and 16q22—-q23, respectively, suggesting a common duplication with the carbonic anhydrase isozyme loci. Cytogenet Cell Genet 57: 41–43, 1991 [DOI] [PubMed] [Google Scholar]
- 46.Pastor-Soler N, Pietrement C, Breton S. Role of acid/base transporters in the male reproductive tract and potential consequences of their malfunction. Physiology (Bethesda) 20: 417–428, 2005 [DOI] [PubMed] [Google Scholar]
- 47.Paunescu TG, Da Silva N, Marshansky V, McKee M, Breton S, Brown D. Expression of the 56-kDa B2 subunit isoform of the vacuolar H+-ATPase in proton-secreting cells of the kidney and epididymis. Am J Physiol Cell Physiol 287: C149–C162, 2004 [DOI] [PubMed] [Google Scholar]
- 48.Paunescu TG, Russo LM, Da Silva N, Kovacikova J, Mohebbi N, Van Hoek AN, McKee M, Wagner CA, Breton S, Brown D. Compensatory membrane expression of the V-ATPase B2 subunit isoform in renal medullary intercalated cells of B1-deficient mice. Am J Physiol Renal Physiol 293: F1915–F1926, 2007 [DOI] [PubMed] [Google Scholar]
- 49.Paunescu TG, Jones AC, Tyszkowski R, Brown D. V-ATPase expression in the mouse olfactory epithelium. Am J Physiol Cell Physiol 295: C923–C930, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pietrement C, Sun-Wada GH, Silva ND, McKee M, Marshansky V, Brown D, Futai M, Breton S. Distinct expression patterns of different subunit isoforms of the V-ATPase in the rat epididymis. Biol Reprod 74: 185–194, 2006 [DOI] [PubMed] [Google Scholar]
- 51.Pisitkun T, Bieniek J, Tchapyjnikov D, Wang G, Wu WW, Shen RF, Knepper MA. High-throughput identification of IMCD proteins using LC-MS/MS. Physiol Genomics 25: 263–276, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robaire B, Viger RS. Regulation of epididymal epithelial cell functions. Biol Reprod 52: 226–236, 1995 [DOI] [PubMed] [Google Scholar]
- 53.Roland JT, Lapierre LA, Goldenring JR. Alternative splicing in class V myosins determines association with Rab10. J Biol Chem 284: 1213–1223, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sachs AN, Pisitkun T, Hoffert JD, Yu MJ, Knepper MA. LC-MS/MS analysis of differential centrifugation fractions from native inner medullary collecting duct of rat. Am J Physiol Renal Physiol 295: F1799–F1806, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shum WCW, Da Silva N, Brown D, Breton S. Regulation of luminal acidification in the male reproductive tract via cell-cell crosstalk. J Exp Biol 212: 1753–1761, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stankovic KM, Brown D, Alper SL, Adams JC. Localization of pH regulating proteins H+-ATPase and Cl−/HCO3− exchanger in the guinea pig inner ear. Hear Res 114: 21–34, 1997 [DOI] [PubMed] [Google Scholar]
- 57.Sun-Wada GH, Imai-Senga Y, Yamamoto A, Murata Y, Hirata T, Wada Y, Futai M. A proton pump ATPase with testis-specific E1-subunit isoform required for acrosome acidification. J Biol Chem 277: 18098–18105, 2002 [DOI] [PubMed] [Google Scholar]
- 58.Sun-Wada GH, Wada Y, Futai M. Diverse and essential roles of mammalian vacuolar-type proton pump ATPase: toward the physiological understanding of inside acidic compartments. Biochim Biophys Acta 1658: 106–114, 2004 [DOI] [PubMed] [Google Scholar]
- 59.Wagner CA, Finberg KE, Breton S, Marshansky V, Brown D, Geibel JP. Renal vacuolar-ATPase. Physiol Rev 84: 1263–1314, 2004 [DOI] [PubMed] [Google Scholar]
- 60.Wang JW, Howson J, Haller E, Kerr WG. Identification of a novel lipopolysaccharide-inducible gene with key features of both A kinase anchor proteins and chs1/beige proteins. J Immunol 166: 4586–4595, 2001 [DOI] [PubMed] [Google Scholar]
- 61.Wieczorek H, Beyenbach KW, Huss M, Vitavska O. Vacuolar-type proton pumps in insect epithelia. J Exp Biol 212: 1611–1619, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou Q, Nie R, Prins GS, Saunders PT, Katzenellenbogen BS, Hess RA. Localization of androgen and estrogen receptors in adult male mouse reproductive tract. J Androl 23: 870–881, 2002. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.