Abstract
The alarming rise of hospital- and community-associated methicillin-resistant Staphylococcus aureus (HA- and CA-MRSA) infections has prompted a desperate search for novel antibiotics. We discovered the streptogramin antibiotic, etamycin, for the first time from a newly discovered marine actinomycete and characterized its activity against a panel of HA- and CA-MRSA strains. Etamycin was extracted and purified from a previously uncharacterized marine-derived actinomycete, designated strain CNS-575, as a three-rotamer species as determined by two-dimensional nuclear magnetic resonance (NMR) spectroscopy. Etamycin demonstrated potent activity against hospital- and community-associated strains of MRSA in microbroth dilution assays, with minimum inhibitory concentrations (MIC) as low as 1 – 2 mg/L against HA- and CA-MRSA strains. Furthermore, etamycin was also active against other Gram-positive and several Gram-negative pathogens and was found to be non-cytotoxic at concentrations more than 20-fold above the MIC. Etamycin displayed favorable time-kill kinetics compared to the first-line MRSA antibiotic, vancomycin, and also conferred significant protection from mortality in a murine model of systemic lethal MRSA infection. These data emphasize the utility of the marine environment as a relatively untapped source of antibiotics against major drug-resistant human pathogens. These studies will also guide future isolation and preclinical development of depsipeptide anti-MRSA compounds from marine-derived actinomycetes.
Keywords: etamycin, marine-derived actinomycete, methicillin-resistant Staphylococcus aureus, streptogramin
INTRODUCTION
The alarming rise of methicillin-resistant Staphylococcus aureus (MRSA) infections in several countries worldwide has provided urgency to the search for structurally novel antibiotics that target these organisms 1. Hospital-associated (HA)-MRSA are frequently resistant to multiple antibiotic classes, and rapidly-emergent strains of community-associated (CA)-MRSA which have demonstrated unusually high virulence and ease of transmission, now represent the most common cause of skin and soft tissue infections in the U.S. 2, 3. Furthermore, the approval of novel antibiotics for human medicine has not kept pace with the development of resistance by important bacterial pathogens such as MRSA. Just a few drugs with novel scaffolds (linezolid, daptomycin) have been introduced to the antibacterial formulary in the past eleven years, and only three new antibacterial compounds are in advanced stages of clinical development 4, 5. Both HA- and CA-MRSA strains have already demonstrated the capacity to evolve resistance to linezolid and daptomycin.
Natural products represent a vast source of potential new chemical structures possessing antimicrobial activity. At least three of every four current antibacterial agents are related in some way to natural products 6, and a majority of these discoveries have come from filamentous bacteria of the order Actinomycetales (actinomycetes). More than 9,000 biologically active molecules have been isolated from actinomycetes, yielding more than 60 pharmaceutical agents used in the medical or agricultural fields 6. While terrestrial (soil) actinomycetes were the mainstay for antibiotic discovery efforts in the previous five decades, there is evidence this resource is becoming exhausted. In contrast, the marine environment, covering more than three-fourths of the world’s surface, represents a vast and relatively untapped source of novel scaffolds with unique antimicrobial properties 7–10. Recently, structures identified from new marine actinomycete species have been shown to exhibit potent anti-MRSA activity with desirable cross activity against other bacterial pathogens 11, 12.
In the course of screening a library of extracts from previously unidentified species of marine-derived actinomycetes for anti-MRSA activity, we followed a hit and purified an active secondary metabolite that we confirmed by nuclear magnetic resonance and mass spectrometric analysis to be etamycin – a classical depsipeptide antibiotic of the streptogramin B class. Since decades have elapsed since consideration of the antibiotic properties of etamycin, we undertook an evaluation of its activity against a panel of contemporary, clinically-relevant HA- and CA-MRSA strains. We find that etamycin exhibits favorable in vitro antibacterial activity and killing kinetics and is also protective against MRSA-induced lethality in a murine systemic infection model. These data suggest etamycin could serve as the basis for in-depth study of related marine-derived molecules as MRSA-targeted therapies.
MATERIALS AND METHODS
Bacterial strains
A panel of HA- and CA-MRSA and other Gram-positive and –negative bacterial strains was used to probe the antimicrobial activity of etamycin. These included MRSA strains ATCC 33591, NRS22 (USA600), NRS71 (Sanger 252, genome strain), NRS119, USA200 (UAMS-1), NRS192, USA300 (UAMS-1182), and strain NRS386 (USA700). Isolates with the NRS designation were obtained through the Network of Antimicrobial Resistance in Staphylococcus aureus (NARSA) program supported under NIAID/NIH contract # HHSN272200700055C. The USA200 and USA300 isolates were kind gifts of Greg Somerville at the University of Nebraska, Lincoln and originally obtained from Mark Smeltzer at the University of Arkansas Medical Center. Other Gram-positive strains tested were Streptococcus pyogenes serotype M1 strain 5448 13, Streptococcus agalactiae strain COH1 14, and vancomycin-resistant Enterococcus faecalis (ATCC 51299). Gram negative strains used were Pseudomonas aeruginosa (ATCC 27853), Escherichia coli (ATCC 25922), Moraxella catarrhalis (ATCC 25238), and Haemophilus influenzae (Type b, ATCC 10211).
The actinomycete strain CNS-575 was isolated from a sediment sample collected from the Nasese shoreline, Viti Levu, Fiji at low tide from a depth of ≈ 0.5 m (coordinates 18.09′04.0 S and 178.27′11.4 E). The 16S ribosomal RNA gene sequence (accession number Bankit 1235828) places this strain within the genus Streptomyces.
Fermentation and Isolation of Etamycin
Culture Conditions
A 2 ml frozen stock of actinomycete strain CNS-575 was used to inoculate 25 ml A1 medium (10 g starch, 4 g yeast extract, 2 g peptone in 1 L seawater) in a 125 ml Erlenmeyer flask and shaken at 27°C. The 5-day-old seed culture was then used to inoculate a one-liter culture in A1bfe+C (A1 media described above, with the addition of 1 g CaCO3, 5 mL of a 2% (w/v) KBr stock solution, and 5 ml of a 0.8% (w/v) Fe2(SO4)•4H2O stock solution). Lastly, 25 ml aliquots of the 3-day-old one-liter culture were used to inoculate each of 18 one-liter cultures in 2.8 L Fernbach flasks containing medium A1bfe+C. Strain CNS-575 was allowed to grow with shaking for 7 days at 27°C prior to extraction.
Isolation
Column chromatography was carried out on silica gel (Selecto Scientific, particle size 63-200). High performance liquid chromatography (HPLC) was carried out on a Beckman System Gold liquid chromatograph (126) equipped with a Shimadzu diode array detector (SPD-M10AVP) and using an Ultracarb ODS (30) column (Phenomenex, 250 × 10.0 mm, 5 μm particle size). A 40 liter culture of strain CNS-575 was extracted using XAD-7 resin and eluted with acetone (2×). The acetone solvent was removed by evaporation, and the remaining water was extracted 3× with ethyl acetate. The obtained extract (1.83 g) was fractionated on silica gel (step gradient from 100:1 to 5:1 dichloromethane: methanol eluent). Fractions with antibacterial activity eluted with 50:1 and 20:1 dichloromethane: methanol. The active fractions (487 mg) were pooled and subjected to HPLC separation (isocratic, 60% aqueous acetonitrile eluent, 3ml/min flow rate) to give etamycin (108.9 mg) after 31 minutes.
Spectroscopic analysis of etamycin
Spectroscopic analyses of etamycin
All NMR spectra were measured using a Varian Oxford AS500 spectrometer (5 mm double-resonance inverse broadband probe) at 500 and 125 MHz for 1H and 13C NMR, respectively. Offline processing was conducted using Mestre-C NMR Software (Mestrelab Research, La Coruna, Spain; www.mestrec.com). 1H and 13C chemical shifts were referenced with the CDCl3 solvent peaks at δ 7.26 and δ 77.0, respectively (Figure 1 and Table 1). The IR spectrum was recorded in KBr on a Nicolet IR100 FTIR spectrometer (Thermo). The UV spectrum was recorded in methanol on a Beckman Coulter DU800 spectrophotometer. The optical rotation was measured in methanol on a Jasco P-2000 polarimeter. High resolution mass spectra were acquired at the UCSD Molecular Mass Spectrometry Facility.
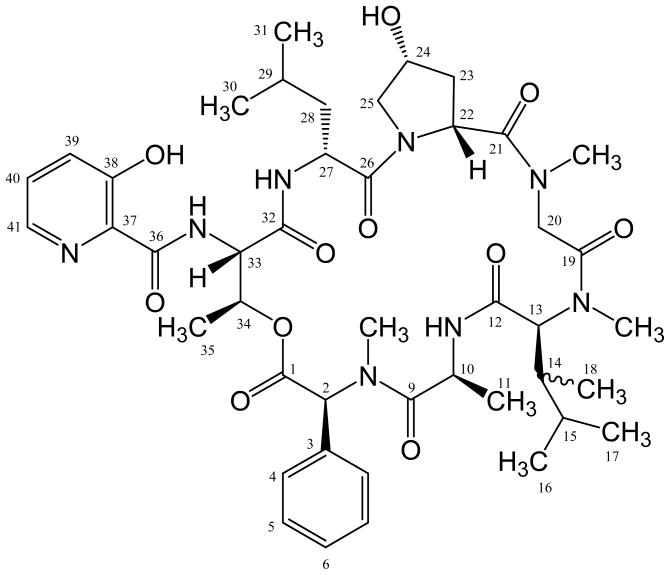
Figure 1.
Chemical structure of etamycin. (Numbered atoms correspond to those in Table 1.)
Table 1.
1H NMR, 13C NMR, and 15N NMR of etamycin (each rotamer) in CDCl3.
| rotamer 1 (major) | rotamer 2 | rotamer 3 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| δN | δCa | δH (m, J (Hz)) | δCa | δH | δCa | δH | |||
| Phg | 1 | 169.6 | C | 169.5 | 169.5 | ||||
| 2 | 62.8 | CH | 5.66 (s) | 61.6 | 5.96 | 63.2 | 5.60 | ||
| 3 | 130.3 | C | 130.9 | 130.4 | |||||
| 4, 8 | 129.7 | CH | 7.41 (m) | 129.4 | 7.46 | 129.4 | 7.36 | ||
| 5, 7 | 129b | CH | 7.4b | 129b | 7.4b | 129b | 7.4b | ||
| 6 | 129b | CH | 7.4b | 129b | 7.4b | 129b | 7.4b | ||
| 2-NCH3 | 182.5 | 31.8 | CH3 | 2.80 (s) | 31.4 | 2.87 | 32.4 | 2.78 | |
| Ala | 9 | 173.5 | C | 174.1 | 173.2 | ||||
| 10 | 45.8 | CH | 5.05 (m) | 45.3 | 5.10 | 46.2 | 4.98 | ||
| 11 | 17.9 | CH3 | 1.39 (d, 6.6) | 16.5 | 1.40 | 18.1 | 1.28 | ||
| 10-NH | 179.5 | 7.26 | 7.93 | 7.48 | |||||
| β-MeLeu | 12 | 168.9 | C | 168.4 | 167.2 | ||||
| 13 | 58.4 | CH | 5.09 (d, 10.3) | 62.8 | 3.77 | 61.9 | 3.93 | ||
| 14 | 35.4 | CH | 2.21 (m) | 37.1 | 2.26 | 36.2 | 2.35 | ||
| 15 | 28.3 | CH | 1.80 (m) | 27.3 | 1.51 | 27.2 | 1.61 | ||
| 16 | 15.4 | CH3 | 0.77 (d, 7.1) | 15.8 | 0.79 | 15.3 | 0.81 | ||
| 17 | 21.5 | CH3 | 0.94 (d, 6.2) | 21.9 | 0.90 | 21.6 | 0.95 | ||
| 18 | 8.5 | CH3 | 0.60 (d, 7.1) | 8.5 | 0.75 | 8.4 | 0.71 | ||
| 13-NCH3 | 195.5 | 30.0 | CH3 | 2.82 (s) | 29.3 | 2.93 | 28.6 | 2.91 | |
| Gly | 19 | 167.5 | C | 167.8 | 167.7 | ||||
| 20a | 52.3 | CH2 | 3.87 (d, 16.8) | 50.5 | 3.40 | 52.3 | 3.87 | ||
| 20b | 5.34 (d, 16.8) | 5.39 | 6.03 | ||||||
| 20-NCH3 | 192.5 | 35.6 | CH3 | 2.92 (s) | 33.3 | 3.21 | 36.4 | 3.02 | |
| Hyp | 21 | 173.9 | C | 173.3 | 173.7 | ||||
| 22 | 53.9 | CH | 5.17 (d, 6.1) | 55.2 | 4.82 | 56.6 | 4.53 | ||
| 23a | 37.4 | CH2 | 2.05 (d, 13.7) | 35.4 | 1.94 | 37.0 | 2.13 | ||
| 23b | 2.15 (m) | 2.26 | |||||||
| 24 | 70.8 | CH | 4.52 (m) | 71.3 | 4.44 | 70.5 | 4.38 | ||
| 25a | 58.1 | CH2 | 3.72 (dd, 5.6, 11.3) | 56.0 | 3.81 | 57.9 | 3.73 | ||
| 25b | 4.42 (dd, 5.8, 11.3) | 3.82 | |||||||
| 24-OH | 6.68 (d, 11.5) | 5.44 | 6.52 | ||||||
| 22-N | 167.5 | ||||||||
| Leu | 26 | 172.0 | C | 171.5 | 170.9 | ||||
| 27 | 49.0 | CH | 4.89 (m) | 48.9 | 5.02 | 48.8 | 5.03 | ||
| 28a | 39.4 | CH2 | 1.48 (ddd, 4.4, 9.0, 13.4) | 39.4 | 1.38 | 40.8 | 1.61 | ||
| 28b | 1.91 (m) | 2.14 | 1.80 | ||||||
| 29 | 24.2 | CH | 1.85 (m) | 24.2 | 1.87 | 24.1 | 1.69 | ||
| 30 | 21.0 | CH3 | 0.93 (d, 6.4) | 20.6 | 1.01 | 21.5 | 0.95 | ||
| 31 | 23.0 | CH3 | 0.98 (d, 7.0) | 23.3 | 0.99 | 23.2 | 0.94 | ||
| 27-NH | 174.5 | 8.33 (d, 8.5) | 8.15 | 7.84 | |||||
| Thr | 32 | 165.9 | C | 166.1 | 165.9 | ||||
| 33 | 53.0 | CH | 4.88 (m) | 53.4 | 4.84 | 53.0 | 4.93 | ||
| 34 | 70.0 | CH | 5.17 (m) | 69.7 | 5.25 | 70.0 | 5.27 | ||
| 35 | 13.4 | CH3 | 1.17 (d, 6.8) | 13.5 | 1.22 | 13.9 | 1.20 | ||
| 33-NH | 199.0 | 8.93 (d, 7.5) | 9.02 | 8.95 | |||||
| Hpca | 36 | 167.7 | C | 167.8 | 168.1 | ||||
| 37 | 130.8 | C | 130.8 | 130.6 | |||||
| 38 | 157.3 | C | 157.3 | 157.3 | |||||
| 39 | 125.6 | CH | 7.25 (m) | 125.6 | 7.25 | 125.7 | 7.27 | ||
| 40 | 128.5 | CH | 7.30 (m) | 128.5 | 7.29 | 128.7 | 7.3b | ||
| 41 | 139.6 | CH | 8.07 (d, 3.9) | 139.6 | 8.04 | 139.8 | 8.09 | ||
| 38-OH | 11.77 (s) | 11.74 | 11.67 | ||||||
| 41-N | 197.5 | ||||||||
13C shifts obtained from HMBC and HSQC spectra.
Overlapping peaks.
Susceptibility Testing
Minimum inhibitory concentration (MIC) assays were performed by microbroth dilution. Briefly, an overnight culture of the test strain was diluted to 1:100 in broth and grown to mid-logarithmic phase at 37°C in a shaking incubator. Todd-Hewitt broth (THB, obtained from Hardy Diagnostics, Santa Maria, CA) was used for all strains except H. influenzae, for which the susceptibility testing was done in brain-heart-infusion media supplemented with hemin (10 ug/ml) and NAD (10 ug/ml). The culture was pelleted and resuspended to OD600 = 0.4 in phosphate-buffered saline (PBS). The bacteria were then added to a 96 well test plate containing etamycin dilutions in broth; the final bacterial concentration was 5 × 105 cfu/ml in a final volume of 100 μl. Vancomycin (Hospira, Inc., Lake Forest, IL) was used as a control for all MRSA strains. Other control antibiotics included: ampicillin (Sandoz, Inc., Princeton, NJ) for E. faecalis and Streptococcus spp, cloramphenicol (Sigma Chemical Co, St. Louis, MO) for M. catarrhalis and H. influenzae, and ciprofloxacin (Bedford Laboratories, Bedford, OH) for P. aeruginosa and E. coli. The test plate containing the etamycin dilutions and target bacteria was incubated at 37°C in a shaking incubator for 18 – 20 h. The MIC was determined to be the lowest concentration of etamycin that inhibited bacterial growth as determined by turbidometric assessment at OD600.
Cytotoxicity Assay
Analysis of mammalian cell cytotoxicity was done essentially as described 15. Briefly, etamycin dilutions were added to HeLa cells (2 × 104 cells per well) in a sterile 96 well plate. Cytotoxicity was assayed by MTS at 24 hours using the CellTiter 96® AQueous non-radioactive cell proliferation assay according to the manufacturer’s instructions (Promega Madison, WI). Plates were read at A490 in a Versamax microplate reader (Molecular Devices, Mountain View, CA).
In Vitro Time-Kill Assays
Time-kill assays were done as described previously 16, 17, 18 with modifications. Briefly, MRSA strain USA300 (UAMS-1182) was cultured overnight at 37°C in THB. The bacteria were then grown to mid-logarithmic phase and prepared in PBS as for the MIC assays. Test tubes were prepared in duplicate containing vehicle control, etamycin, or vancomycin at concentrations of 1, 4 or 10 times the MIC of 4 μg/ml and 1.5 μg/ml, respectively. Bacteria were added to each tube at a starting inoculum of ~ 5 × 105 cfu/ml in a final volume of 5 ml. The cultures were incubated in a 37°C shaking incubator, and aliquots were removed from each test tube at 0, 3, 6 and 24 h. These aliquots were serially diluted in phosphate-buffered saline and plated on Todd-Hewitt agar plates in the absence of antibiotic. Colonies were enumerated after 24 hours at 37°C. The limit of detection of this assay equated to 40 cfu per ml.
In Vivo Studies
Eight week old female CD1 mice (Charles River) were injected intraperitoneally (ip) with 2 × 109 cfu of HA-MRSA strain Sanger 252 in 4% mucin. At 1 and 8 h after infection, the mice were injected ip with etamycin (20 mg/kg) or an equivalent volume of vehicle control. The mice were observed for signs of sepsis, including lethargy, piloerection, and dehydration. Survival was monitored for a total of 72 h, and mice that became moribund during the study were humanely euthanized, as were any surviving mice at the end of the study. These studies were reviewed and approved by the University of California San Diego Animal Subjects Committee.
RESULTS
The actinomycete strain designated CNS-575 was cultured from a marine sediment collected along the coast of Fiji. 16S rRNA gene sequence analysis places this strain within the genus Streptomyces, however it shares < 98% sequence identity with any formally described species suggesting it may represent a new species. Solid-phase extraction and chromatographic purification yielded an 879 molecular weight peptide which appeared as three distinct rotamers in the NMR spectrum. The presence of rotamers was not affected by changes of NMR solvent or acquisition temperature, and we ultimately performed the NMR-based structure elucidation of all three rotamers simultaneously (Table 1). The resulting planar structure was consistent with that of the previously-reported terrestrially-derived streptogramin, etamycin 19, 20 (Figure 1), although a complete chemical shift assignment of etamycin has not previously been reported. Acid hydrolysis followed by analysis using advanced Marfey’s method 21 revealed the stereochemical configuration to be consistent with etamycin (see Supporting Information for a detailed discussion of the Marfey’s analysis).
Etamycin was originally isolated from terrestrial actinomycete S. griseus alongside the streptogramin-A antibiotic griseoviridin, and the two molecules together displayed synergism common for other streptogramins, including bactericidal activity primarily against some Gram-positive organisms 22. We investigated in more detail the antibiotic properties of etamycin as a single agent against a panel of clinically-relevant MRSA strains and discovered it to possess very good activity. MIC ranged from 1–2 mg/L for CA-MRSA strains and certain HA-MRSA strains, to 8–16 mg/L for the HA-MRSA strains ATCC 33591 and Sanger 252 (Table 2). Etamycin also showed a low level of activity against strains of the Gram-positive bacteria S. pyogenes and S. agalactiae (MIC = 8 mg/L) and minimal activity against vancomycin-resistant E. faecalis (MIC = 16 mg/L). The spectrum of etamycin activity extended to strains of the Gram-negative coccobacilli and respiratory tract pathogens M. catarrhalis (MIC = 1 mg/L) and to a lesser extent H. influenzae (MIC = 16 mg/L), but not to other Gram-negative pathogens tested including E. coli, S. typhimurium, and P. aeruginosa. In order to assess a potential therapeutic index for etamycin against MRSA, we analyzed its effects in a mammalian HeLa cell cytotoxicity assay. We found that etamycin exhibited no cytoxicity at concentrations up to 128 mg/L (data not shown), suggesting the potential for a favorable therapeutic index against MRSA.
Table 2.
Antibacterial activity of Etamycin
| Strain | MIC (mg/L) |
|---|---|
| HA- and CA-MRSA Strains | |
| UAMS-1 (USA200) | 1 – 2 |
| Sanger 252 | 8 – 16 (2)* |
| ATCC 33591 | 16 |
| NRS22 (USA600) | 1 – 4 |
| NRS119 (linezolid-resistant strain) | 1 – 2 |
| UAMS-1182 (USA300) | 4 |
| NRS192 | 2 |
| NRS386 (USA700)** | 2 |
| Non-MRSA Strains | |
| E. faecalis (vanB) | 16 (16) |
| S. pyogenes (M1 strain 5448) | 8 |
| S. agalactiae (serotype III strain COH1) | 8 |
| M. catarrhalis (ATCC 25238) | 1 |
| H. influenzae (type b, ATCC 10211) | 16 |
| E. coli (ATCC 25922) | >256 |
| S. typhimurium (ATCC 13311) | >256 |
| P. aeruginosa (ATCC 27853) | >256 |
Values in parentheses represent MICs for Synercid against these bacterial strains tested under identical conditions as for etamycin.
Strain NRS386 is associated with both community and healthcare infections.
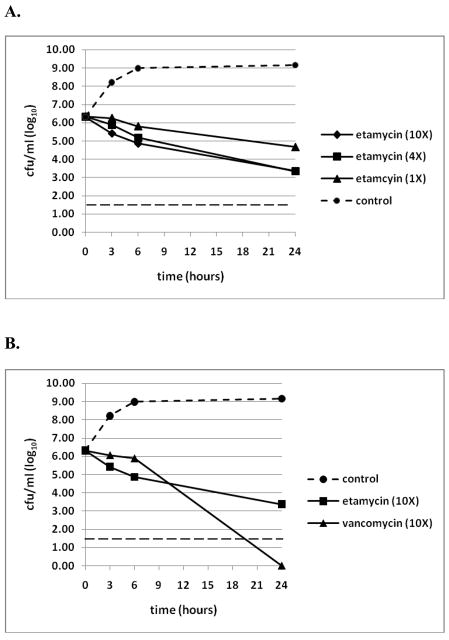
We studied the time-kill kinetics of etamycin at different concentrations for comparison against vancomycin, the most common agent used in current MRSA therapy. Evaluation was conducted at 1x, 4x and 10x the respective MIC of each drug against the CA-MRSA (USA300) strain UAMS-1182 (Figure 2A). Etamycin killed UAMS-1182 somewhat more rapidly than vancomycin at 10× MIC during the first 6 h; however, vancomycin was able reduce the number of viable bacteria to below the threshold of detection of this assay by 24 h, when etamycin killing, while substantial, remained incomplete (Figure 2B). Collectively, these data suggest etamycin acts in both time- and concentration-dependent mechanisms and is faster acting but significantly less bactericidal than vancomycin.
Figure 2.
In vitro time-kill kinetics of etamycin against CA-MRSA strain USA300. A.) Concentration-dependence of etamycin at one, four, and ten times the MIC over 24 hours. MRSA was incubated with the specified concentrations of etamycin, and surviving bacteria were enumerated at the given times by quantitative culture on agar in the absence of the compound. Data represent the mean of three independent experiments, and the dotted line at 1.6 log10 represents the limit of detection of the assay. B.) Comparison of killing kinetics of etamycin with vancomycin at ten times the MIC for each drug. Surviving bacteria were enumerated at the given times by quantitative culture on agar. The data represent the mean of three independent experiments, and the dotted line at 1.6 log10 represents the limit of detection of the assay.
As a first assessment of the in vivo efficacy of etamycin, we utilized a murine systemic infection model, conservatively choosing as the challenge strain MRSA Sanger 252, against which etamycin exhibited a higher MIC of 8 – 16 mg/L. Eight week old female CD1 mice were treated with etamycin (20 mg/kg) or vehicle control at one and eight hours after intraperitoneal MRSA infection, and general health and survival were followed for up to 72 h. Within 6 – 12 hours of MRSA infection, most mice in the control group exhibited signs of sepsis including lethargy, dehydration, and piloerection, while the severity of these symptoms was greatly reduced or absent in most of the etamycin-treated animals. As shown in Figure 3, the two doses of etamycin offered significant protection to mice infected with MRSA during the 72 h monitoring period (20% mortality vs. 75% morality in controls).
Figure 3.
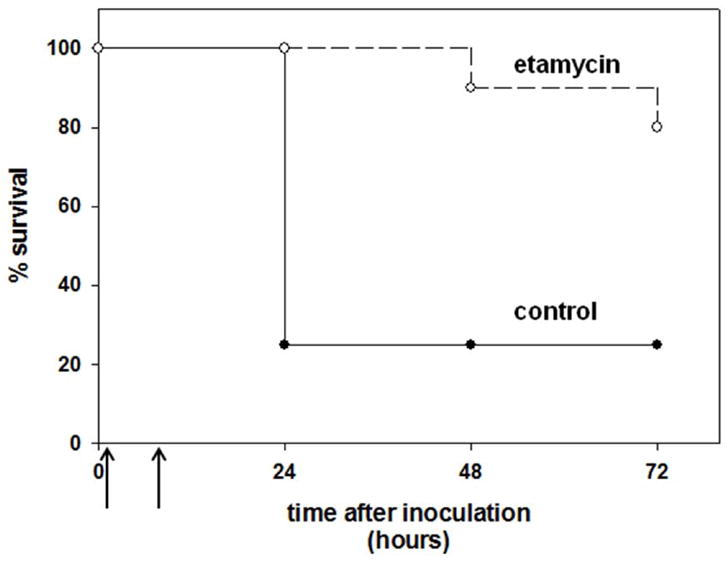
In vivo efficacy of etamycin in a murine model of MRSA sepsis. Eight week old female CD1 mice were injected intraperitoneally with 2 × 109 colony-forming units of HA-MRSA strain Sanger 252 in 1% mucin. At one and eight hours after infection (indicated by arrows), the mice were injected intraperitoneally with either 20 mg/kg etamycin (n = 10) or DMSO vehicle control (n = 8). The mice were monitored for signs of sepsis for up to 72 hours, and mice that became moribund were humanely euthanized.
DISCUSSION
For the first time from a marine-derived actinomycete we isolated the streptogramin, etamycin, and found that it had marked antibacterial activity against clinically important strains of MRSA. Although etamycin was first identified more than five decades ago, its antibiotic properties had not been characterized in detail. Those early studies also describe etamycin as a bacteriostatic compound against S. aureus strain Duncan. 22 We found that etamycin alone at high concentration may exhibit limited bactericidal activity against contemporary CA-MRSA strains, since after 24 h of exposure to etamycin at 4× MIC, < 1 % of the starting inoculum of CA-MRSA strain UAMS1182 remained viable (Figure 2), representing an approximate three-log kill by 24 hours. Furthermore, the progressive decrease in the number of surviving bacteria over time even at 1× MIC also supports a conclusion that etamycin exhibits some, though certainly incomplete, bactericidal activity over 24 hours. However, despite this result, etamycin clearly does not kill CA-MRSA as effectively as vancomycin in the 24 hour assay.
The streptogramin antibiotic, quinupristin-dalfopristin (Synercid®) is currently approved for use in complicated cutaneous infections caused by multiple drug resistant Gram-positive cocci, including MRSA and vancomycin-resistant Enterococcus faecium 23. In this combination, streptogramins A and B together act to inhibit protein synthesis, with quinupristin (A) preventing polypeptide elongation by binding to the 50S ribosomal subunit and dalfopristin (B) binding to another site on the 50S ribosomal subunit and inducing a conformational change enhancing quinupristin activity 24. We found in our assays that etamycin, presumably through protein synthesis inhibition, exhibits significant activity against MRSA alone without the benefit of its streptogramin A partner. Etamycin also demonstrated a very low level of activity against E. faecalis (MIC = 16 mg/L), a frequently drug-resistant nosocomial pathogen that is associated with urinary tract and surgical wound infections 25, and this activity appeared comparable to the very low level of activity of Synercid® against this pathogen in our assay. Clearly, if etamycin were ever considered for introduction into clinical use, defined breakpoints would need to be defined.
Our isolation of etamycin from a novel marine-derived actinomycete species emphasizes the utility of the marine environment as a source of antibacterials 7, in particular marine actinomycetes that have been shown to naturally produce a wide variety of active parent molecules and analogs 6, 11, 26. Etamycin was previously isolated from the terrestrial microbe, S. griseoviridus, naturally-occurring as multiple congeners in which a hydroxyproline residue was replaced by a proline 27. Additionally, limited structure-activity relationship data point to the importance of the picolinyl moiety of etamycin for its activity 19. Future studies will investigate further the activity of etamycin analogs from marine-derived actinomycetes for characterization and optimization of activity toward further preclinical development.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health Training Program in Marine Biotechnology (T32 GM067550 to NH), by the National Institutes of Health (RO1GM084350-01 to WF and VN), by the National Institutes of Health International Cooperative Biodiversity Groups program (U01-TW007401-01 to WF and PRJ), and by the National Institutes of Health Cancer Therapeutics Training Program (T32 CA121938 to KNM). We gratefully acknowledge R. Raju and B. Aalbersberg for providing actinomycete strain CNS-575. We also acknowledge George Sakoulas, MD for many helpful discussions.
Abbreviations
- MRSA
methicillin-resistant Staphylococcus aureus
- MIC
minimum inhibitory concentration
- NMR
nuclear magnetic resonance
- cfu
colony-forming unit
Footnotes
Supplementary information is available at the Journal of Antibiotics website (http://www.nature.com/ja).
References
- 1.Cornaglia G, Rossolini GM. Forthcoming therapeutic perspectives for infections due to multidrug-resistant Gram-positive pathogens. Clin Microbiol Infect. 2009;15:218–23. doi: 10.1111/j.1469-0691.2009.02740.x. [DOI] [PubMed] [Google Scholar]
- 2.Como-Sabetti K, et al. Community-associated methicillin-resistant Staphylococcus aureus: trends in case and isolate characteristics from six years of prospective surveillance. Public Health Rep. 2009;124:427–35. doi: 10.1177/003335490912400312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel M. Community-Associated Meticillin-Resistant Staphylococcus aureus Infections: Epidemiology, Recognition and Management. Drugs. 2009;69:693–716. doi: 10.2165/00003495-200969060-00004. [DOI] [PubMed] [Google Scholar]
- 4.Boucher HW, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 5.Spellberg B, et al. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin Infect Dis. 2008;46:155–64. doi: 10.1086/524891. [DOI] [PubMed] [Google Scholar]
- 6.Demain AL. Antibiotics: Natural products essential to human health. Med Res Rev. 2009 doi: 10.1002/med.20154. [DOI] [PubMed] [Google Scholar]
- 7.Fenical W, Jensen PR. Developing a new resource for drug discovery: marine actinomycete bacteria. Nat Chem Biol. 2006;2:666–73. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]
- 8.Kwon HC, Kauffman CA, Jensen PR, Fenical W. Marinomycins A-D, antitumor-antibiotics of a new structure class from a marine actinomycete of the recently discovered genus “marinispora”. J Am Chem Soc. 2006;128:1622–32. doi: 10.1021/ja0558948. [DOI] [PubMed] [Google Scholar]
- 9.Mincer TJ, Jensen PR, Kauffman CA, Fenical W. Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl Environ Microbiol. 2002;68:5005–11. doi: 10.1128/AEM.68.10.5005-5011.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soria-Mercado IE, Prieto-Davo A, Jensen PR, Fenical W. Antibiotic terpenoid chloro-dihydroquinones from a new marine actinomycete. J Nat Prod. 2005;68:904–10. doi: 10.1021/np058011z. [DOI] [PubMed] [Google Scholar]
- 11.Hughes CC, Prieto-Davo A, Jensen PR, Fenical W. The marinopyrroles, antibiotics of an unprecedented structure class from a marine Streptomyces sp. Org Lett. 2008;10:629–31. doi: 10.1021/ol702952n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McArthur KA, et al. Lynamicins A-E, chlorinated bisindole pyrrole antibiotics from a novel marine actinomycete. J Nat Prod. 2008;71:1732–7. doi: 10.1021/np800286d. [DOI] [PubMed] [Google Scholar]
- 13.Datta V, et al. Mutational analysis of the group A streptococcal operon encoding streptolysin S and its virulence role in invasive infection. Mol Microbiol. 2005;56:681–95. doi: 10.1111/j.1365-2958.2005.04583.x. [DOI] [PubMed] [Google Scholar]
- 14.Doran KS, Chang JC, Benoit VM, Eckmann L, Nizet V. Group B streptococcal beta-hemolysin/cytolysin promotes invasion of human lung epithelial cells and the release of interleukin-8. J Infect Dis. 2002;185:196–203. doi: 10.1086/338475. [DOI] [PubMed] [Google Scholar]
- 15.Jere D, et al. Poly (amino ester) composed of poly (ethylene glycol) and aminosilane prepared by combinatorial chemistry as a gene carrier. Pharm Res. 2008;25:875–85. doi: 10.1007/s11095-007-9448-4. [DOI] [PubMed] [Google Scholar]
- 16.Ueda Y, et al. In vitro and in vivo antibacterial activities of SM-216601, a new broad-spectrum parenteral carbapenem. Antimicrob Agents Chemother. 2005;49:4185–96. doi: 10.1128/AAC.49.10.4185-4196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Credito K, Lin G, Appelbaum PC. Activity of daptomycin alone and in combination with rifampin and gentamicin against Staphylococcus aureus assessed by time-kill methodology. Antimicrob Agents Chemother. 2007;51:1504–7. doi: 10.1128/AAC.01455-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsuji BT, Yang JC, Forrest A, Kelchlin PA, Smith PF. In vitro pharmacodynamics of novel rifamycin ABI-0043 against Staphylococcus aureus. J Antimicrob Chemother. 2008;62:156–60. doi: 10.1093/jac/dkn133. [DOI] [PubMed] [Google Scholar]
- 19.Bateman KP, Thibault P, Yang K, White RL, Vining LC. Probing the substrate specificity of an enzyme catalyzing inactivation of streptogramin B antibiotics using LC-MS and LC-MS/MS. J Mass Spectrom. 1997;32:1057–63. doi: 10.1002/(SICI)1096-9888(199711)32:10<1057::AID-JMS558>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 20.Sheehan JC, Ledis SL. Total synthesis of a monocyclic peptide lactone antibiotic, etamycin. J Am Chem Soc. 1973;95:875–9. doi: 10.1021/ja00784a041. [DOI] [PubMed] [Google Scholar]
- 21.Fujii K, Ikai Y, Oka H, Suzuki M, Harada K. A Nonempirical Method Using LC/MS for Determination of the Absolute Configuration of Constituent Amino Acids in a Peptide: Combination of Marfey’s Method with Mass Spectrometry and Its Practical Application. Anal Chem. 1997;69:5146–5151. [Google Scholar]
- 22.Garcia-Mendoza C. Studies on the Mode of Action of Etamycin (Viridogrisein) Biochim Biophys Acta. 1965;97:394–6. doi: 10.1016/0304-4165(65)90121-2. [DOI] [PubMed] [Google Scholar]
- 23.Metzger R, Bonatti H, Sawyer R. Future trends in the treatment of serious Gram-positive infections. Drugs Today (Barc) 2009;45:33–45. doi: 10.1358/dot.2009.45.1.1315922. [DOI] [PubMed] [Google Scholar]
- 24.Allington DR, Rivey MP. Quinupristin/dalfopristin: a therapeutic review. Clin Ther. 2001;23:24–44. doi: 10.1016/s0149-2918(01)80028-x. [DOI] [PubMed] [Google Scholar]
- 25.Sood S, Malhotra M, Das BK, Kapil A. Enterococcal infections & antimicrobial resistance. Indian J Med Res. 2008;128:111–21. [PubMed] [Google Scholar]
- 26.Demain AL, Sanchez S. Microbial drug discovery: 80 years of progress. J Antibiot (Tokyo) 2009;62:5–16. doi: 10.1038/ja.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chopra C, et al. Congeners of etamycin produced by Streptomyces griseoviridus. J Antibiot (Tokyo) 1979;32:392–401. doi: 10.7164/antibiotics.32.392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.