The sequencing of the human genome has greatly facilitated our ability to identify the gene candidates that are critical for developmental regulation and disease progression. However, investigators seeking to use such data to gain a better understanding of the developmental biology and therapeutic potential of pluripotent stem cell types may be frustrated by the limitations of current genetic engineering techniques. Targeted genomic manipulation of mammalian cells is inefficient, and unintended effects of such manipulation on the cells' developmental and proliferative potential remain uninvestigated. To this end, the recent demonstration of targeted gene correction in human induced pluripotent stem cells (hiPSCs) before and after reprogramming, as reported in this issue of Molecular Therapy, warrants special mention.1
The successful isolation of human embryonic stem cells (hESCs) has enabled the implementation of renewable generation of a wide variety of cell types in regenerative medicine approaches.2 The prospect of implementing cell replacement therapies for diverse pathologies, including neurodegeneration, cardiovascular disease, and diabetes, is now conceivable owing to the ability to generate large quantities of functional differentiated cell types. However, the use of hESCs remains controversial, and cell transplantation therapy as a field still faces major obstacles such as acute donor cell death, low engraftment rates, immune rejection, and tumorigenicity,3,4 which have impeded efforts toward clinical translation.
The recent development of hiPSCs has offered exciting solutions to allay the ethical concerns and mitigate the possibility of immune rejection by providing a mechanism for the creation of “patient-specific” pluripotent stem cells from somatic cell types.5,6 It is possible to generate hiPSCs without destroying human embryos, and autologous hiPSC grafts have the potential benefit of being less likely to provoke an immune response. Nevertheless, challenges such as low derivation efficiency and exposure to viral factors during the reprogramming process remain to be solved, but new advances have recently been made.7
Although the utility of gene targeting in hESCs has been confirmed by several reports,8,9 these techniques remain unimplemented on a large scale. By contrast, relatively facile reprogramming of somatic cell types from virtually any genetic background into hiPSCs has enabled the generation of myriad cell lines containing disease-conferring mutations.10,11,12 The use of these hiPSCs and their derivatives for disease modeling is expected to significantly enhance investigators' ability to probe the mechanics of a multitude of pathologies. As the repertoire of genetically diverse hiPSC lines expands, the ability to site-specifically modify the genome in order to alter genes of interest will become increasingly important. To realize the full potential of hESCs and hiPSCs, efficient methods for genetic modifications are needed.
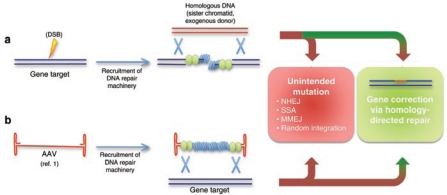
Techniques for precise targeting of defined modifications to the genome rely on activation of the cell's homologous recombination (HR) machinery (Figure 1). In the presence of a donor DNA sequence substituting for the sister chromatid, HR can yield targeted clones with an appropriate experimental system. However, the donor DNA often enters other pathways of DNA repair, most notably undergoing nonhomologous end joining at a higher frequency than HR. Therefore, targeted HR is a relatively inefficient process, occurring at a frequency between 10−5 and 10−7 in treated mammalian cells.13 This process may be enhanced by some orders of magnitude with the introduction of a DNA double-strand break at the genomic locus of interest.14 Other strategies for enhancing HR involve augmenting the activity of nucleofilament proteins RAD51 and RAD52, either by transient overexpression or via small molecules,15 although some of these studies have yielded conflicting results. Further investigation into the cellular determinants of the choice between the intended (i.e., HR) versus unintended (e.g., nonhomologous end joining, single-strand annealing, microhomology-mediated end joining, and random integration) target is required to yield satisfying insight into potential methods for improving the relative frequency of HR.
Figure 1.
Methods for gene correction in human embryonic and induced pluripotent stem cells. (a) A double-strand break (DSB) induced by ionizing radiation or a zinc-finger nuclease signals recruitment of DNA repair machinery. DNA repair then proceeds by a variety of pathways depending on the cell cycle stage and extent of 3¢ end resection. Homology-directed repair results in successful gene targeting, whereas nonhomologous end joining (NHEJ), single-strand annealing (SSA), or microhomology-mediated end joining (MMEJ) will result in nonspecific mutations. (b) A recombinant adeno-associated virus (AAV) expression vector consists of single-stranded DNA (ssDNA) flanked by palindromic inverted terminal repeats. After removal of the viral capsid, DNA repair machinery components coat the ssDNA to form a nucleofilament structure. Resolution of vector-chromosomal DNA intermediates results in introduction of a targeted modification at the homologous chromosomal locus.
Gene-correction efficiency may vary considerably, depending on the genomic context of the target locus, so comparing the results of various studies is challenging. Fortunately, the most frequently targeted gene in hESC studies is the HPRT1 locus encoding hypoxanthine-guanine phosphoribosyl transferase (HPRT). HPRT is a key enzyme in the purine salvage pathway, the deficiency of which is manifested as Lesch–Nyhan syndrome in patients. Also, because HPRT1 is located on the X chromosome, a single gene targeting event can lead to a complete loss of function and 6-thioguanine resistance in XY cells, providing a convenient culture system for screening correctly targeted cells from random integrants. Initial attempts at HR in hESCs at the HPRT1 and POU5F1 loci yielded successful gene targeting at frequencies on the order of 10–6. The percentage of selection marker–resistant clones that actually contained the intended gene correction as confirmed by Southern blotting, referred to as the targeting efficiency, varied between 27% and 40%, depending on the targeting construct used.8
Several studies have now validated the use of zinc-finger nucleases (ZFNs) to induce a chromosomal double-strand break at the site of the targeted gene, which greatly enhances the frequency of the homology-directed repair.14 This approach requires the engineering of zinc-finger DNA-binding domains to recognize a DNA sequence of interest. The DNA-binding domains are then fused to the nuclease domain of the FokI endonuclease to enable cleavage at a specific genomic locus. However, the potential remains for off-target cleavage events at chromosomal sequences bearing a similarity to the ZFN target site. One group has demonstrated the use of ZFNs in conjunction with integrase-defective lentiviral (IDLV) vectors to achieve high gene targeting efficiencies. Increasing concentrations of IDLVs delivering ZFNs that target the CCR5 locus resulted in 0.5–5.3% of transduced cells expressing the green fluorescent protein (GFP) transgene. However, this particular study lacked Southern blot analysis, which would have confirmed whether gene targeting had generated a single-copy GFP insertion without any random integration elsewhere. Also, the increase in viral titer concomitantly increased the frequency of mismatch mutations from 4% to 28% in the GFP+ clones.16
Subsequent studies have demonstrated the use of ZFNs to successfully target the POU5F1, PIGA, PITX3, and AAVS1 loci in hESCs and hiPSCs.17,18 Because techniques employing ZFNs require concomitant delivery of three DNA constructs (two to encode the ZFN pair and one donor DNA sequence), reported efficiencies may be lower for these studies, which employed electroporation in place of viral vectors. Using a nonviral electroporation system, these studies demonstrated successful gene targeting for a variety of loci in hESC and hiPSC lines on the order of 10−5. Targeting efficiency varied from 8% to 100%, depending on the ZFN pair used and genomic locus targeted. Notably, these studies utilized ZFNs designed by different groups using entirely different methods of validation. However, a ZFN-based gene-correction approach requires extensive design and validation of new zinc-finger binding domains for each target locus. In addition to verifying proper targeting when using ZFNs, it is of paramount importance to limit cells' exposure to ZFN-expressing plasmids to prevent off-target cleavage and even potential integration of ZFN-expressing DNA into the genome.
Viral vector delivery systems have also been implemented in order to overcome the low transient transfection efficiencies observed using electroporation in hESCs. Adenoviral and lentiviral vector gene delivery systems are well established in their ability to transduce a broad range of cell types at high efficiency, although safety concerns regarding random chromosomal integration have prevented their clinical use.19 Modified viral vectors that have been developed to allay such concerns include AAV, helper-dependent adenovirus, and IDLV. Helper-dependent adenoviruses, for example, have been modified by deletion of all viral genes from the vector genome, resulting in reduced cytotoxicity and an expanded cloning capacity to allow for insertion of larger targeting constructs. These properties have been exploited by one group to achieve successful gene correction at the HPRT1 locus in hESCs at a frequency of ~10–6, with a targeting efficiency of ~40% (ref. 20).
It is within this context that Khan et al.,1 in this issue of Molecular Therapy, make a significant step forward. Remarkably, the investigators observed successful insertion of a neomycin-resistance cassette into the HPRT1 locus in ~10–5 of hESCs and hiPSCs after transduction with AAV gene targeting vectors. Importantly, their work is also the first demonstration of successful gene targeting in differentiated somatic cells (fibroblasts) before their reprogramming into hiPSCs. A 4–base pair (bp) segment was successfully inserted into the HPRT1 locus of transduced fibroblasts, and one of these fibroblast cell lines was then successfully reprogrammed into hiPSCs using lentiviral vectors, albeit at a reduced derivation efficiency when compared with an unmodified fibroblast cell line. Once derived, these genetically modified hiPSCs were also subsequently corrected to delete the original 4-bp insertion. Despite these multiple genomic modifications, karyotypic abnormalities were observed only rarely (i.e., in one hiPSC line at later passages, as is commonly observed in hESC culture).
However, unintended mutations are a common result of gene correction attempts using any approach, including the one used in the current article. Improving the proportion of site-specific versus nonspecific integration events has remained a major challenge to the gene therapy field. Although in vitro culture allows for the careful selection and clonal expansion of the targeted cell population of interest, clinical protocols may involve transduction of hundreds of millions of cells. Currently, the approach demonstrated by Khan et al. remains limited to use in cultured cells that can subsequently be expanded to achieve the cell numbers required for downstream applications. Also, despite potential uniformity of HLA subtypes across hiPSC derivatives and their recipients, it remains to be seen whether the short exposure of transplanted cells to viral capsids is sufficient to induce an immune response in the transplanted host. It is remarkable, however, that differentiated fibroblasts, having undergone AAV transduction, retain the proliferative capacity for subsequent reprogramming into hiPSCs. Differentiated somatic cells senesce over time, and senescence is known to inhibit successful reprogramming.21 The low derivation efficiency of hiPSCs is already a well-recognized and prominent impediment in the reprogramming field. The further reduction in efficiency observed when reprogramming genetically modified fibroblasts raises further questions. Will additional increases in the degree and number of genomic modifications in fibroblasts lead to further reductions in reprogramming efficiency? Or is the reduction in reprogramming efficiency correlated only with the increased number of plating and passaging steps required for fibroblast culture prior to transduction with reprogramming factors?
Khan et al. have convincingly demonstrated the feasibility of AAV-mediated gene correction in hESCs and hiPSCs. Overall, their results open many new avenues for investigators to site-specifically modify candidate genes before and after the derivation of pluripotent stem cell types. After perturbing a gene of interest, the derived iPSC lines can be differentiated into the relevant cell population to measure the gene's effect on cell function. Importantly, the ability to site-specifically modify the genome mitigates the risk of malignant transformation induced by random retroviral or lentiviral transgene insertion.22 Safe, reliable genomic modification would greatly assist the therapeutic cell transplantation field to overcome this significant obstacle to clinical realization. In any case, these advances have significantly streamlined the genetic manipulation of pluripotent stem cells, and have brought a small step closer the realization of the full potential of pluripotent stem cells for investigating disease and treating patients.
Acknowledgments
We acknowledge funding support from the National Institutes of Health (DP2OD004437 and R01AI085575), the Edward Mallinckrodt Jr. Foundation (J.C.W.), and the Howard Hughes Medical Institute (K.H.N.).
REFERENCES
- Khan IF, Hirata RK, Wang P-R, Li Y, Kho J, Nelson A.et al. (2010Engineering of human pluripotent stem cells by AAV-mediated gene targeting Mol Ther 181192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS.et al. (1998Embryonic stem cell lines derived from human blastocysts Science 2821145–1147. [DOI] [PubMed] [Google Scholar]
- Lee AS, Tang C, Cao F, Xie X, van der Bogt K, Hwang A.et al. (2009Effects of cell number on teratoma formation by human embryonic stem cells Cell Cycle 82608–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swijnenburg RJ, Schrepfer S, Govaert JA, Cao F, Ransohoff K, Sheikh AY.et al. (2008Immunosuppressive therapy mitigates immunological rejection of human embryonic stem cell xenografts Proc Natl Acad Sci USA 10512991–12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., and , Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S.et al. (2007Induced pluripotent stem cell lines derived from human somatic cells Science 3181917–1920. [DOI] [PubMed] [Google Scholar]
- Sun N, Longaker MT., and , Wu JC. Human iPS cell-based therapy: considerations before clinical applications. Cell Cycle. 2010;9:880–885. doi: 10.4161/cc.9.5.10827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaka TP., and , Thomson JA. Homologous recombination in human embryonic stem cells. Nat. Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
- Irion S, Luche H, Gadue P, Fehling HJ, Kennedy M., and , Keller G. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat Biotechnol. 2007;25:1477–1482. doi: 10.1038/nbt1362. [DOI] [PubMed] [Google Scholar]
- Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W.et al. (2008Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons Science 3211218–1221. [DOI] [PubMed] [Google Scholar]
- Lee G, Papapetrou EP, Kim H, Chambers SM, Tomishima MJ, Fasano CA.et al. (2009Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs Nature 461402–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG.et al. (2009Parkinson's disease patient–derived induced pluripotent stem cells free of viral reprogramming factors Cell 136964–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C., and , Capecchi MR. Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol Cell Biol. 1992;12:3365–3371. doi: 10.1128/mcb.12.8.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S.et al. (2005Highly efficient endogenous human gene correction using designed zinc-finger nucleases Nature 435646–651. [DOI] [PubMed] [Google Scholar]
- Jayathilaka K, Sheridan SD, Bold TD, Bochenska K, Logan HL, Weichselbaum RR.et al. (2008A chemical compound that stimulates the human homologous recombination protein RAD51 Proc Natl Acad Sci USA 10515848–15853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA.et al. (2007Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery Nat Biotechnol 251298–1306. [DOI] [PubMed] [Google Scholar]
- Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK.et al. (2009Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells Cell Stem Cell 597–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC.et al. (2009Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases Nat Biotechnol 27851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donsante A, Miller DG, Li Y, Vogler C, Brunt EM, Russell DW.et al. (2007AAV vector integration sites in mouse hepatocellular carcinoma Science 317477. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Mitsui K, Aizawa E, Hasegawa K, Kawase E, Yamagishi T.et al. (2008Highly efficient transient gene expression and gene targeting in primate embryonic stem cells with helper-dependent adenoviral vectors Proc Natl Acad Sci USA 10513781–13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I.et al. (2009Senescence impairs successful reprogramming to pluripotent stem cells Genes Dev 232134–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P.et al. (2003LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1 Science 302415–419. [DOI] [PubMed] [Google Scholar]