Abstract
Antisense oligomer-induced manipulation of dystrophin pre-mRNA processing can remove exons carrying mutations, or exclude exons flanking frameshifting mutations, and restore dystrophin expression in dystrophinopathy models and in Duchenne muscular dystrophy (DMD) patients. Splice intervention can also be used to manipulate the normal dystrophin pre-mRNA processing and ablate dystrophin expression in wild-type mice, with signs of pathology being induced in selected muscles within 4 weeks of commencing treatment. The disruption of normal dystrophin pre-mRNA processing to alter the reading frame can be very efficient and offers an alternative mechanism to RNA silencing for gene suppression. In addition, it is possible to remove in-frame exon blocks from the DMD gene transcript and induce specific dystrophin isoforms that retain partial functionality, without having to generate transgenic animal models. Specific exon removal to yield in-frame dystrophin transcripts will facilitate mapping of functional protein domains, based upon exon boundaries, and will be particularly relevant where there is either limited, or conflicting information as to the consequences of in-frame dystrophin exon deletions on the clinical severity and progression of the dystrophinopathy.
Introduction
Elucidation of many of the molecular mechanisms that are important in disease provides opportunities for new bio-therapeutics. Antisense agents have proved to be valuable tools to inhibit gene expression by a variety of mechanisms, and to manipulate exon selection during primary transcript processing to overcome disease-causing gene lesions. We have developed antisense strategies to bypass disease-causing mutations in the DMD gene that lead to Duchenne muscular dystrophy (DMD), a relentlessly progressive muscle wasting disorder with a predictable course and fatal outcome (for review see ref. 1).
Although predominantly expressed at low levels, dystrophin serves a crucial structural role in muscle. Mutations that prevent synthesis of a functional dystrophin result in muscle fibers prone to mechanical damage, with secondary changes in signaling and metabolism, causing the severe muscle wasting and pathology associated with DMD (for review see ref. 2). Selected exon exclusion mediated by antisense oligomers can bypass premature termination codons3,4,5 or restore the reading frame around frameshifting mutations in the DMD gene,6,7,8,9 and can lead to functional protein expression and localization. Although whole-exon dystrophin deletions are clustered in hotspots,10 nondeletion mutations are spread across the entire gene.11 In-frame dystrophin deletions, particularly those in the central rod domain, generally result in the production of partially functional protein and the less severe, allelic disorder, Becker muscular dystrophy (BMD).12 Examination of the genomic organization in mildly affected BMD patients provides templates for a number of potentially functional dystrophin isoforms. However, in-frame deletions in the latter third of the gene are rare11 (Figure 1) and the optimal exon-skipping strategies to bypass many DMD gene mutations, and the functionality of the induced dystrophin isoforms13 remain to be determined. The paucity of BMD-causing mutations in the 3′ region of the DMD gene probably reflects a combination of this area not being deletion-prone and the exon arrangement being such that most exon junctions occur within codons, thereby predisposing exon deletions or duplications to disrupting the reading frame.
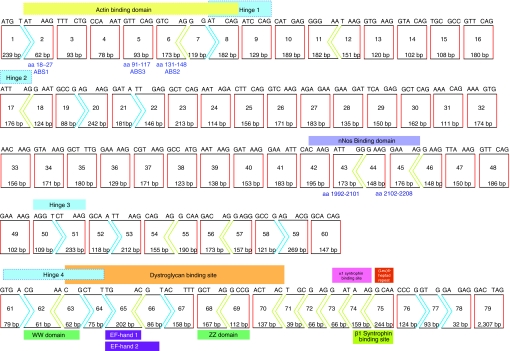
Figure 1.
Dystrophin transcript structure,43 showing the reading frame and major functional protein coding domains (actin-binding domain; exons 2–8,20,21,44,45 central rod domain; exons 8–61, which includes nNos binding sites (exons 42–45),46 cysteine-rich domain22 (dystroglycan-binding site; EF1 domain and ZZ domain23) exons 63–70, the α-syntrophin binding site24 and carboxy-terminal domain (exons 70–79). In-frame exons are indicated by rectangles (red sides), whereas codons disrupted by exon junctions are indicated by arrows.
Exclusion of an in-frame, multiexon block that maintains an open reading frame can result in a BMD-like protein,9,14,15,16,17,18,19 and will allow functional mapping of dystrophin domains according to exon boundaries. This will assist in devising the optimal exon-skipping strategies to overcome DMD-causing mutations, particularly in those regions of the DMD gene that are not well represented in the dystrophin mutation databases. We now extend the application of selected exon removal to induce transient in vivo models to allow specific dystrophin isoforms to be evaluated. In addition, exclusion of a frameshifting exon is another mechanism for efficient inhibition of gene expression and may be a useful technique to study the consequences of gene knockdown in vivo.
Results
Dystrophin exon arrangement
The dystrophin exon arrangement is shown in Figure 1, and the reading frame and encoded functional protein domains are indicated. The primary actin-binding domain 1 occurs within the first 90 amino acids.20 An intact actin-binding domain 1 is required for normal muscle function and to protect myofibers from necrosis.21 Genomic deletions extending downstream of exon 55 are rare,11 and it is apparent (Figure 1) that most deletions in this region will disrupt the reading frame and/or impact upon important functional domains. Deletion analysis indicates that the broad cysteine-rich domain is indispensable for dystrophin function and coincides with the in vitro identified β-dystroglycan-binding domain22 found to include EF1 and ZZ domains23 and the α-syntrophin binding site, coded for by exons 63–70,24 while the carboxy terminal is not essential for assembly of the dystrophin-associated glycoprotein complex.25 Thirty-nine of the exons are in-frame, and may be deleted individually without disrupting the reading frame.
Induction of a dystrophin isoform missing exons 19 and 20
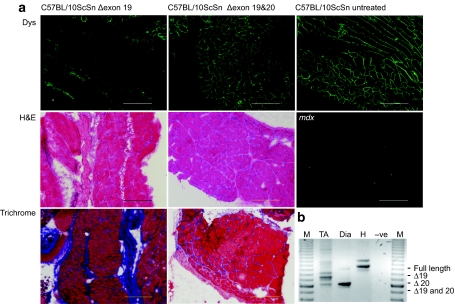
Neonatal C57BL/10ScSn mice, 3–4 days old (n = 5) were injected twice-weekly, with peptide-conjugated phosphorodiamidate morpholino oligomers (PMOs) (20 mg/kg) targeting exon 19, and exons 19 and 20 simultaneously, via the intraperitoneal route. The animals were euthanized at 10 weeks of age and dystrophin expression in selected tissues was analyzed by reverse transcription (RT)-PCR, western blotting, and immunofluorescence. The removal of exon 19 ablated dystrophin expression in diaphragm from treated C57BL/10ScSn mice, whereas the removal of exons 19 and 20 together render the diaphragm indistinguishable from wild-type, sham-treated mouse diaphragm (Figure 2a). RT-PCR analysis of dystrophin transcripts in tibialis anterior, diaphragm and heart from mice treated with oligomers targeting exon 19 and exons 19 and 20 together showed efficient splice-switching and generation of induced shortened transcripts in striated muscle, but not in heart (Figure 2b).
Figure 2.
Analysis of dystrophin expression in selected tissues from 10-week-old C57BL/10ScSn mice injected i.p. twice-weekly with peptide-conjugated PMOs (20 mg/kg) targeting dystrophin exons 19 and 20. (a) Immunofluorescent detection of dystrophin on diaphragm cryosections from mice treated with oligomers targeting exon 19 (Δ19) and exons 19 and 20 (Δ19 and 20) (upper panel), haematoxylin and eosin staining (H&E) (middle panel), and Picro Mallory trichrome (lower panel) staining revealing muscle architecture. Sections from sham-treated C57BL/10ScSn and mdx mice are included for comparison (bar = 20 µm). (b) Nested RT-PCR analysis of dystrophin transcripts in tibialis anterior (TA), diaphragm (Dia), and heart (H) from mice treated with oligomers targeting exons 19 and 20 (Δ19 and 20). Transcript product sizes in base pairs (bp) are indicated (M = 100 bp marker). i.p., intraperitoneal; PMO, phosphorodiamidate morpholino oligomer; RT, reverse transcription.
Induction of a dystrophin isoform missing exons 52 and 53
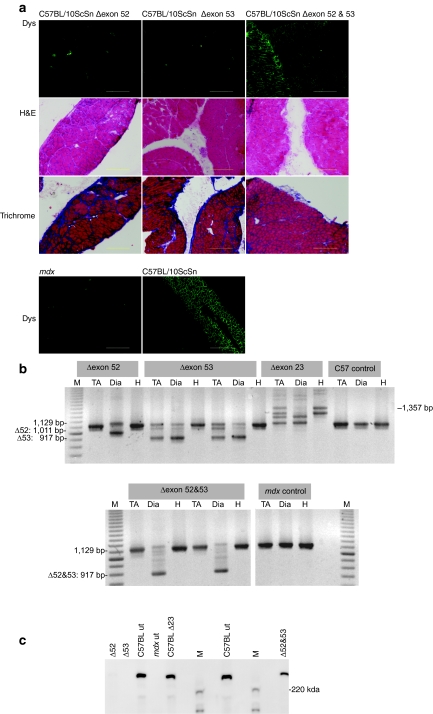
Neonatal C57BL/10ScSn mice, 2–4 days old (n = 5) were injected intraperitoneal twice-weekly, with peptide-conjugated PMOs (20 mg/kg) targeting exon 52, exon 53, and exons 52 and 53 simultaneously. Four weeks after commencing application of the oligomer targeting exon 53, severe dystrophic pathology with central nucleation, mononuclear infiltrate, and fibrosis was evident in the diaphragm. Efficient exclusion of exon 53 from the transcript was demonstrated by RT-PCR on RNA prepared from diaphragm, and to a lesser degree in tibialis anterior, but not in heart (Figure 3b) and the absence of dystrophin in diaphragm was confirmed by western blotting (Figure 3c). Dystrophin exon 52 removal was less efficient, as shown by the residual dystrophin in Figure 3c and partial dystrophin staining of some fibers in Figure 3a. However, the simultaneous removal of exons 52 and 53 maintained the dystrophin reading frame (Figure 1) and normal sarcolemmal dystrophin localization and muscle architecture (Figure 3a). RT-PCR showed the absence of a product representing the full-length transcript after dual exon skipping, consistent with the western blot that shows near-normal levels of dystrophin in diaphragm after removal of exons 52 and 53. Diaphragm muscle architecture and dystrophin expression was almost indistinguishable from that of an age-matched, untreated wild-type mouse.
Figure 3.
Analysis of dystrophin expression in C57BL/10ScSn mice treated with PMOs (20 mg/kg) targeting dystrophin 52 and 53. (a) C57BL/10ScSn mice were injected twice-weekly with peptide-conjugated PMOs (combined dosage 20 mg/kg) for 4 weeks, beginning at 4 days of age. Dystrophin expression in unfixed cryosections (6 µm) from diaphragm of C57BL/10ScSn mice treated with oligomers targeted to exon 52 (Δ52), exon 53 (Δ53), and exons 52 and 53 together (Δ52 and 53) was detected with Novocastra NCLDYS2 and Zenon Alexafluor 488 (upper panel). Sections from untreated, age-matched C57BL/10ScSn and mdx mice are included for comparison (lowest panel). Sections were also stained with haematoxylin and eosin (H&E) and Picro Mallory trichrome to reveal any pathogenic changes in muscle architecture (middle panels) (bar = 20 µm). (b) Nested RT-PCR across dystrophin exons 49–55 on RNA prepared from diaphragm (Dia), tibialis anterior (TA), and heart (H) of oligomer treated and untreated mice (M = 100 bp marker) and (c) western blot on extracts prepared from diaphragms of C57BL/10ScSn mice treated with oligomers targeted to exon 52 (Δ52), exon 53 (Δ53), and exons 52 and 53 together (Δ52 and 53). Dystrophin was visualized with NCLDYS2 (Novocastra) using Western Breeze (Invitrogen). Samples from diaphragms of C57BL/10ScSn (C57BL ut) and mdx mice (mdx ut) injected with vehicle only (saline), and a normal mouse, treated with an oligomer targeting dystrophin exon 23 (C57BL Δ23) are included for comparison (M = protein standard maker).
Discussion
Intervention by splice switching is dependant upon several parameters including appropriate oligomer design and chemistry, and efficient oligomer uptake. The use of PMOs, which have demonstrated efficient and prolonged splice-switching action in vivo,26,27 conjugated to cell-penetrating peptides for efficient systemic delivery4,28,29,30 allows exploitation of AO-mediated exon selection to study protein function in animal models. We are generating in-frame transcripts to allow mapping of functional protein domains, based upon exon boundaries. Exclusion of an in-frame, multiexon block to maintain the reading frame permits functional assessment of the dystrophin isoforms and will assist in devising exon-skipping strategies for various DMD-causing mutations.
The genotype–phenotype data available in existing dystrophinopathy databases can be compromised by inaccurate mutation and clinical information and incomplete mutation type representation. Some of the apparent discrepancies to the reading-frame rule12 in the databases are due to the limitations of diagnostic procedures available at the time of testing. Mutation confirmation at the RNA level reveals over 99.5% concordance with the reading-frame rule.31 Completion of the TREAT-NMD database32,33 will provide accurate information on defined mutations, permitting correlation of the genotype and clinical diagnosis, and assist in determining the best exon-skipping strategies for many dystrophin mutations. However, the paucity of BMD-causing mutations in some parts of the DMD gene, and in particular, the latter third, does not facilitate the design of exon-skipping strategies to bypass DMD-causing mutations in this region.
The first report of exon skipping in the dystrophin transcript described induction of abnormal splicing and exclusion of exon 19 in a patient with an intraexonic deletion.34 Subsequently, an oligodeoxynucleotide targeted to exon 19 was administered to a patient with a deletion of exon 20 in an effort to restore the dystrophin reading frame.35 There are only three records of deletions of dystrophin exons 19 and 20 in the Leiden database (www.dmd.nl/) and all are described as DMD. However, one diagnosis was carried by Southern blotting, another by multiplex PCR and follow-up investigations of the third found no record of the submitted mutation (K. Flanigan, personal communication).
The potential phenotype associated with the loss of exons 19 and 20 is not known, but would be predicted to be mild, because these exons occur within the rod domain region. Data presented here confirms that the dystrophin isoform translated from a transcript missing exons 19 and 20 is correctly localized to the sarcolemma and that the muscle architecture in diaphragm and tibialis anterior appears normal.
The human dystrophin deletion hotspot extends from exon 44 to exon 55. Antisense compounds to remove exon 51 are currently undergoing clinical trials, however, many additional compounds to exclude both single and multiple exons are required to address the majority of dystrophin mutations in this region. As in-frame deletions in this mutation prone region generally result in a BMD phenotype, we wished to demonstrate that exclusion of another dual exon block could result in functional dystrophin expression. As expected, exclusion of exons 52 and 53 induced near-normal levels of dystrophin expression and muscle architecture was indistinguishable from that of sham-treated normal mice. This technique will allow evaluation of dystrophin isoforms induced by selected exon removal, and in particular, will identify the optimal exon-skipping strategies for those mutations that may be addressed by more than one exon skipping approach. Similarly, bypassing a mutation involving exon 51 could be achieved by removal of either flanking exon, as dystrophin transcripts missing exon 50 and 51, or 51 and 52 are in-frame. Although both these dystrophin isoforms are associated with mild symptoms, it is possible one may be more functional than the other. Exons 50 and 51 encode the entire hinge 3, whereas part of hinge 3 is retained in the isoform missing exons 51 and 52.
It is also possible to efficiently disrupt the normal dystrophin mRNA reading frame by removing a frameshifting exon, and ablate dystrophin expression in the muscle of wild-type mice. Total suppression of DMD gene expression can be induced and maintained for several weeks in vivo, and a severe dystrophic pathology observed within 4 weeks of commencing treatment in neonatal normal mice. PMOs have been widely used to study gene knockdown by translational blockade,36,37,38 however, induced nonproductive splicing provides a specific and effective alternative for the transient suppression of gene expression. Grounds et al.,39 have proposed a two-tiered model of DMD, and suggest that the absence of dystrophin has a greater impact on growing muscle than it does on adult muscle. Further investigation in adult mice, using this strategy, may help to elucidate the impact of dystrophin disruption in mature muscle. Disruption of gene expression through altered splicing patterns could be applied across any part of the DMD gene transcript or to other genes, and offers the ability to induce transient animal models to study the consequences of gene suppression and splice-switching in vivo. This strategy would be particularly useful in studying the expression of genes that result in embryonic lethality when suppressed.
Materials and Methods
In vivo oligomer application. C57BL/10ScSn mice were supplied by the Animal Resources Centre (Murdoch, Australia) and housed at the Biological Research Facility, University of Western Australia according to the National Health and Medical Research Council Code of Practice. All experiments performed on animals were approved by the University of Western Australia Animal Experimentation Committee (approval number RA4/100/702). C57BL/10ScSn mice were injected twice-weekly with peptide-conjugated PMOs4 in normal saline (combined oligomer dosage of 20 mg/kg) targeting dystrophin exons as indicated, beginning at 4 days of age.
Antisense oligomers. PMOs targeting exon 19; H19A(+35+65), (GCCUGAGCUGAUCUGCUGGCAUCUUGCAGUU),40 exon 20; M20A(+23+47), (GUUCAGUUGUUCUGAAGCUUGUCUG) and M20A(+140+164), (AGUAGUUGUCAUCUGUUCCAAUUGU),41 exon 23; M23D(+7−18), (GGCCAAACCTCGGCTTACCTGAAAT),4,42 exon 52; M52A(+17+41), (UCCAAUUGGGGGCGUCUCUGUUCCA) and M52A(+42+71), (UUCAAAUUCUGGGCAGCAGUAAUGAGUUCU) and 53 M53A(+69+98) (CAGCCAUUGUGUUGAAUCCUUUAACAUUUC) and M53D(+05−25), (UUUUAAAGAUAUGCUUGACACUAACCUUGG),19 conjugated to peptide K30 were supplied by AVI Biopharma (Corvallis, OR). Oligomers were diluted in saline and combined into cocktails at the following ratios: exons 19 and 20/20, 2:1:1, exon 52, 1:1, exon 53 1:1 and exons 52 and 53, 3:1, and applied at a total dosage of 20 mg/kg per injection.
Histology and immunofluorescence. Dystrophin expression on unfixed cryosections (6 µm) from muscles of C57BL/10ScSn, mice treated with oligomers, or sham injected, was detected with NCLDYS2 (Novocastra, Newcastle Upon Tyne, UK) and Zenon Alexafluor 488 (Invitrogen, Melbourne, Australia) according to the manufacture's instructions.
Western blotting. Western blot extracts were prepared from tissue cryosections and 7.5 µg of each sample was loaded onto 4–12% NuPAGE Novex BIS/Tris gradient gels (Invitrogen) and electrophoresed. The gels were stained and densitometry of the myosin band was used to standardize loading of the samples for a second gel that was blotted as described previously.4,27 Dystrophin was visualized with NCLDYS2 (Novocastra) using the WesternBreeze Chemiluminescent anti-mouse Kit (Invitrogen). Images were captured on a Chemismart 3000 Gel documentation system (Vilber Lourmat, Marne-la-Vallee, France).
RNA extraction and RT-PCR. RNA was extracted from tissue cryosections using TRIzol Reagent (Invitrogen) and dystrophin transcripts were analyzed by nested RT-PCR across exons 18–268 and exons 49–55 for 35 and 30 cycles, as described previously.19
Acknowledgments
The authors' laboratory is supported by funding from the National Institutes of Health (2 R01 NS044146-05A1), the Muscular Dystrophy Association USA (4352), the National Health and Medical Research Council (634485) (Australia), Suneels's Light, Charley's Fund, the Killowen Fundraising Group, the Muscular Dystrophy Association WA Inc. and the Medical and Health Research Infrastructure Fund of Western Australia. The authors thank AVI Biopharma Inc. for generously providing the peptide-conjugated PMOs. This work was performed in Perth, Australia.
REFERENCES
- Wilton SD., and , Fletcher S. Modification of pre-mRNA processing: application to dystrophin expression. Curr Opin Mol Ther. 2006;8:130–135. [PubMed] [Google Scholar]
- Deconinck N., and , Dan B. Pathophysiology of duchenne muscular dystrophy: current hypotheses. Pediatr Neurol. 2007;36:1–7. doi: 10.1016/j.pediatrneurol.2006.09.016. [DOI] [PubMed] [Google Scholar]
- Doran P, Wilton SD, Fletcher S., and , Ohlendieck K. Proteomic profiling of antisense-induced exon skipping reveals reversal of pathobiochemical abnormalities in dystrophic mdx diaphragm. Proteomics. 2009;9:671–685. doi: 10.1002/pmic.200800441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher S, Honeyman K, Fall AM, Harding PL, Johnsen RD, Steinhaus JP, et al. Morpholino oligomer-mediated exon skipping averts the onset of dystrophic pathology in the mdx mouse. Mol Ther. 2007;15:1587–1592. doi: 10.1038/sj.mt.6300245. [DOI] [PubMed] [Google Scholar]
- Goyenvalle A, Babbs A, Powell D, Kole R, Fletcher S, Wilton SD, et al. Prevention of dystrophic pathology in severely affected dystrophin/utrophin-deficient mice by morpholino-oligomer-mediated exon-skipping. Mol Ther. 2010;18:198–205. doi: 10.1038/mt.2009.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aartsma-Rus A, Bremmer-Bout M, Janson AA, den Dunnen JT, van Ommen GJ., and , van Deutekom JC. Targeted exon skipping as a potential gene correction therapy for Duchenne muscular dystrophy. Neuromuscul Disord. 2002;12 Suppl 1:S71–S77. doi: 10.1016/s0960-8966(02)00086-x. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Janson AA, Kaman WE, Bremmer-Bout M, den Dunnen JT, Baas F, et al. Therapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patients. Hum Mol Genet. 2003;12:907–914. doi: 10.1093/hmg/ddg100. [DOI] [PubMed] [Google Scholar]
- McClorey G, Fall AM, Moulton HM, Iversen PL, Rasko JE, Ryan M, et al. Induced dystrophin exon skipping in human muscle explants. Neuromuscul Disord. 2006;16:583–590. doi: 10.1016/j.nmd.2006.05.017. [DOI] [PubMed] [Google Scholar]
- McClorey G, Moulton HM, Iversen PL, Fletcher S., and , Wilton SD. Antisense oligonucleotide-induced exon skipping restores dystrophin expression in vitro in a canine model of DMD. Gene Ther. 2006;13:1373–1381. doi: 10.1038/sj.gt.3302800. [DOI] [PubMed] [Google Scholar]
- den Dunnen JT, Bakker E, Breteler EG, Pearson PL., and , van Ommen GJ. Direct detection of more than 50% of the Duchenne muscular dystrophy mutations by field inversion gels. Nature. 1987;329:640–642. doi: 10.1038/329640a0. [DOI] [PubMed] [Google Scholar]
- Prior TW, Bartolo C, Pearl DK, Papp AC, Snyder PJ, Sedra MS, et al. Spectrum of small mutations in the dystrophin coding region. Am J Hum Genet. 1995;57:22–33. [PMC free article] [PubMed] [Google Scholar]
- Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H., and , Kunkel LM. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2:90–95. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- Yokota T, Duddy W., and , Partridge T. Optimizing exon skipping therapies for DMD. Acta Myol. 2007;26:179–184. [PMC free article] [PubMed] [Google Scholar]
- Aartsma-Rus A, Janson AA, van Ommen GJ., and , van Deutekom JC. Antisense-induced exon skipping for duplications in Duchenne muscular dystrophy. BMC Med Genet. 2007;8:43. doi: 10.1186/1471-2350-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aartsma-Rus A, Kaman WE, Weij R, den Dunnen JT, van Ommen GJ., and , van Deutekom JC. Exploring the frontiers of therapeutic exon skipping for Duchenne muscular dystrophy by double targeting within one or multiple exons. Mol Ther. 2006;14:401–407. doi: 10.1016/j.ymthe.2006.02.022. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Janson AA, Kaman WE, Bremmer-Bout M, van Ommen GJ, den Dunnen JT, et al. Antisense-induced multiexon skipping for Duchenne muscular dystrophy makes more sense. Am J Hum Genet. 2004;74:83–92. doi: 10.1086/381039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T, Lu QL, Partridge T, Kobayashi M, Nakamura A, Takeda S, et al. Efficacy of systemic morpholino exon-skipping in Duchenne dystrophy dogs. Ann Neurol. 2009;65:667–676. doi: 10.1002/ana.21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fall AM, Johnsen R, Honeyman K, Iversen P, Fletcher S., and , Wilton SD. Induction of revertant fibres in the mdx mouse using antisense oligonucleotides. Genet Vaccines Ther. 2006;4:3. doi: 10.1186/1479-0556-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrpant C, Fletcher S, Iversen PL., and , Wilton SD. By-passing the nonsense mutation in the 4 CV mouse model of muscular dystrophy by induced exon skipping. J Gene Med. 2009;11:46–56. doi: 10.1002/jgm.1265. [DOI] [PubMed] [Google Scholar]
- Corrado K, Mills PL., and , Chamberlain JS. Deletion analysis of the dystrophin-actin binding domain. FEBS Lett. 1994;344:255–260. doi: 10.1016/0014-5793(94)00397-1. [DOI] [PubMed] [Google Scholar]
- Banks GB, Gregorevic P, Allen JM, Finn EE., and , Chamberlain JS. Functional capacity of dystrophins carrying deletions in the N-terminal actin-binding domain. Hum Mol Genet. 2007;16:2105–2113. doi: 10.1093/hmg/ddm158. [DOI] [PubMed] [Google Scholar]
- Chamberlain JS, Corrado K, Rafael JA, Cox GA, Hauser M., and , Lumeng C. Interactions between dystrophin and the sarcolemma membrane. Soc Gen Physiol Ser. 1997;52:19–29. [PubMed] [Google Scholar]
- Ishikawa-Sakurai M, Yoshida M, Imamura M, Davies KE., and , Ozawa E. ZZ domain is essentially required for the physiological binding of dystrophin and utrophin to beta-dystroglycan. Hum Mol Genet. 2004;13:693–702. doi: 10.1093/hmg/ddh087. [DOI] [PubMed] [Google Scholar]
- Yang B, Jung D, Rafael JA, Chamberlain JS., and , Campbell KP. Identification of α-syntrophin binding to syntrophin triplet, dystrophin, and utrophin. J Biol Chem. 1995;270:4975–4978. doi: 10.1074/jbc.270.10.4975. [DOI] [PubMed] [Google Scholar]
- Crawford GE, Faulkner JA, Crosbie RH, Campbell KP, Froehner SC., and , Chamberlain JS. Assembly of the dystrophin-associated protein complex does not require the dystrophin COOH-terminal domain. J Cell Biol. 2000;150:1399–1410. doi: 10.1083/jcb.150.6.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter J, Lou F, Rabinowitz A, Yin H, Rosenfeld J, Wilton SD, et al. Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology. Nat Med. 2006;12:175–177. doi: 10.1038/nm1345. [DOI] [PubMed] [Google Scholar]
- Fletcher S, Honeyman K, Fall AM, Harding PL, Johnsen RD., and , Wilton SD. Dystrophin expression in the mdx mouse after localised and systemic administration of a morpholino antisense oligonucleotide. J Gene Med. 2006;8:207–216. doi: 10.1002/jgm.838. [DOI] [PubMed] [Google Scholar]
- Yin H, Lu Q., and , Wood M. Effective exon skipping and restoration of dystrophin expression by peptide nucleic acid antisense oligonucleotides in mdx mice. Mol Ther. 2008;16:38–45. doi: 10.1038/sj.mt.6300329. [DOI] [PubMed] [Google Scholar]
- Yin H, Moulton HM, Betts C, Seow Y, Boutilier J, Iverson PL, et al. A fusion peptide directs enhanced systemic dystrophin exon skipping and functional restoration in dystrophin-deficient mdx mice. Hum Mol Genet. 2009;18:4405–4414. doi: 10.1093/hmg/ddp395. [DOI] [PubMed] [Google Scholar]
- Jearawiriyapaisarn N, Moulton HM, Buckley B, Roberts J, Sazani P, Fucharoen S, et al. Sustained dystrophin expression induced by peptide-conjugated morpholino oligomers in the muscles of mdx mice. Mol Ther. 2008;16:1624–1629. doi: 10.1038/mt.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aartsma-Rus A, Van Deutekom JC, Fokkema IF, Van Ommen GJ., and , Den Dunnen JT. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve. 2006;34:135–144. doi: 10.1002/mus.20586. [DOI] [PubMed] [Google Scholar]
- Bushby K, Lynn S., and , Straub T. Collaborating to bring new therapies to the patient–the TREAT-NMD model. Acta Myol. 2009;28:12–15. [PMC free article] [PubMed] [Google Scholar]
- Mercuri E, Mayhew A, Muntoni F, Messina S, Straub V, Van Ommen GJ, et al. Towards harmonisation of outcome measures for DMD and SMA within TREAT-NMD; report of three expert workshops: TREAT-NMD/ENMC workshop on outcome measures, 12th–13th May 2007, Naarden, The Netherlands; TREAT-NMD workshop on outcome measures in experimental trials for DMD, 30th June–1st July 2007, Naarden, The Netherlands; conjoint Institute of Myology TREAT-NMD meeting on physical activity monitoring in neuromuscular disorders, 11th July 2007, Paris, France. Neuromuscul Disord. 2008;18:894–903. doi: 10.1016/j.nmd.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Matsuo M, Masumura T, Nishio H, Nakajima T, Kitoh Y, Takumi T, et al. Exon skipping during splicing of dystrophin mRNA precursor due to an intraexon deletion in the dystrophin gene of Duchenne muscular dystrophy kobe. J Clin Invest. 1991;87:2127–2131. doi: 10.1172/JCI115244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima Y, Wada H, Yagi M, Ishikawa Y, Ishikawa Y, Minami R, et al. Oligonucleotides against a splicing enhancer sequence led to dystrophin production in muscle cells from a Duchenne muscular dystrophy patient. Brain Dev. 2001;23:788–790. doi: 10.1016/s0387-7604(01)00326-6. [DOI] [PubMed] [Google Scholar]
- Arora V, Devi GR., and , Iversen PL. Neutrally charged phosphorodiamidate morpholino antisense oligomers: uptake, efficacy and pharmacokinetics. Curr Pharm Biotechnol. 2004;5:431–439. doi: 10.2174/1389201043376706. [DOI] [PubMed] [Google Scholar]
- Heasman J. Morpholino oligos: making sense of antisense. Dev Biol. 2002;243:209–214. doi: 10.1006/dbio.2001.0565. [DOI] [PubMed] [Google Scholar]
- Stein DA. Inhibition of RNA virus infections with peptide-conjugated morpholino oligomers. Curr Pharm Des. 2008;14:2619–2634. doi: 10.2174/138161208786071290. [DOI] [PubMed] [Google Scholar]
- Grounds MD. Two-tiered hypotheses for Duchenne muscular dystrophy. Cell Mol Life Sci. 2008;65:1621–1625. doi: 10.1007/s00018-008-7574-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington SJ, Mann CJ, Fletcher S., and , Wilton SD. Target selection for antisense oligonucleotide induced exon skipping in the dystrophin gene. J Gene Med. 2003;5:518–527. doi: 10.1002/jgm.361. [DOI] [PubMed] [Google Scholar]
- Adams AM, Harding PL, Iversen PL, Coleman C, Fletcher S., and , Wilton SD. Antisense oligonucleotide induced exon skipping and the dystrophin gene transcript: cocktails and chemistries. BMC Mol Biol. 2007;8:57. doi: 10.1186/1471-2199-8-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebski BL, Mann CJ, Fletcher S., and , Wilton SD. Morpholino antisense oligonucleotide induced dystrophin exon 23 skipping in mdx mouse muscle. Hum Mol Genet. 2003;12:1801–1811. doi: 10.1093/hmg/ddg196. [DOI] [PubMed] [Google Scholar]
- Koenig M, Monaco AP., and , Kunkel LM. The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell. 1988;53:219–228. doi: 10.1016/0092-8674(88)90383-2. [DOI] [PubMed] [Google Scholar]
- Levine BA, Moir AJ, Patchell VB., and , Perry SV. The interaction of actin with dystrophin. FEBS Lett. 1990;263:159–162. doi: 10.1016/0014-5793(90)80728-2. [DOI] [PubMed] [Google Scholar]
- Levine BA, Moir AJ, Patchell VB., and , Perry SV. Binding sites involved in the interaction of actin with the N-terminal region of dystrophin. FEBS Lett. 1992;298:44–48. doi: 10.1016/0014-5793(92)80019-d. [DOI] [PubMed] [Google Scholar]
- Lai Y, Thomas GD, Yue Y, Yang HT, Li D, Long C, et al. Dystrophins carrying spectrin-like repeats 16 and 17 anchor nNOS to the sarcolemma and enhance exercise performance in a mouse model of muscular dystrophy. J Clin Invest. 2009;119:624–635. doi: 10.1172/JCI36612. [DOI] [PMC free article] [PubMed] [Google Scholar]