Abstract
Oncolytic viruses (OVs) are promising anticancer agents but like other cancer monotherapies, the genetic heterogeneity of human malignancies can lead to treatment resistance. We used a virus/cell-based assay to screen diverse chemical libraries to identify small molecules that could act in synergy with OVs to destroy tumor cells that resist viral infection. Several molecules were identified that aid in viral oncolysis, enhancing virus replication and spread as much as 1,000-fold in tumor cells. One of these molecules we named virus-sensitizers 1 (VSe1), was found to target tumor innate immune response and could enhance OV efficacy in animal tumor models and within primary human tumor explants while remaining benign to normal tissues. We believe this is the first example of a virus/cell-based “pharmacoviral” screen aimed to identify small molecules that modulate cellular response to virus infection and enhance oncolytic virotherapy.
Introduction
Oncolytic viruses (OVs) target aberrant signaling pathways unique to tumor cells and have the potential to revolutionize cancer therapy. Often OVs are selected or engineered to be restricted for growth in tumors, by elimination of virulence genes, creating virus products that replicate well in malignant cells but are unable to grow in normal tissues.1 Although this type of mutation leads to a safe therapeutic, it often creates a virus with such restricted host cell range that its usefulness is limited in genetically heterogeneous primary malignancies. Virulence gene products can antagonize signaling pathways that control cellular antiviral responses and/or usurp cellular machinery providing the invading virus with a growth advantage.2,3,4 The function of these virulence proteins is required to infect normal cells which have robust antiviral responses. In OV-sensitive tumors, antiviral pathways are defective making virulence proteins targeting these pathways redundant and permitting cancer-specific viral replication in their absence.5,6,7,8 However, cancer cells that have partially responsive antiviral signaling pathways resist OV infection when viral functions required to break down remaining cellular antiviral defenses have been deleted from the therapeutic. To facilitate OV growth in tumor cells that have some residual antiviral activity, we reasoned that it is possible to select from diverse chemical libraries, small molecule “virus-sensitizers” (or VSe) that mimic the activity of viral virulence gene products. In principle, such chemicals could be used to conditionally complement engineered mutations in OVs, transiently increasing the ability of a therapeutic virus to replicate in a wider range of tumor cell types and provide tight drug control over the extent, duration, and location of oncolytic virus replication. Here, we present the results of a compound library screen that identified a variety of molecules that enhance the replication and spread of an attenuated interferon (IFN) sensitive version of an oncolytic vesicular stomatitis virus (VSVΔ51).6 One of these compounds herein referred to as VSe1, substantially enhances virus growth and spread in a variety of murine and human cancer cell lines, in an immune-competent mouse tumor model and in primary human tumor samples.
Results
A high-throughput screen for the identification of oncolytic viro-sensitizers
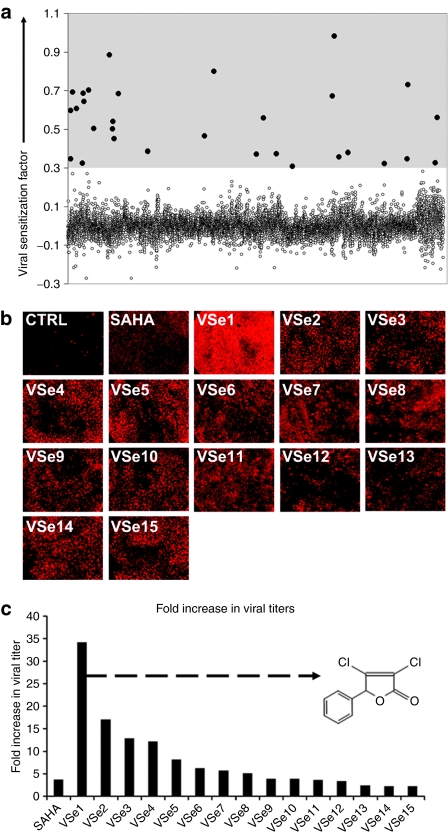
Antiviral signaling pathways involve several layers of regulation spanning from the cellular plasma membrane (e.g., toll-like receptors and IFN receptors), through the cytoplasm (e.g., IKKs, JAK, RIG-I), and into the nucleus (e.g., IRFs, STATs, NF-κB) (reviewed in refs. 9,10). Because defects within one or more of these pathways can occur within cancer cells during tumor evolution, we reasoned that a screening strategy aiming toward a single signaling molecule (e.g., IFN receptor) may isolate compounds that are useful in only a limited spectrum of resistant tumors. As a less biased alternative, we designed a virus/cell-based high-throughput screening assay that should identify compounds active at multiple levels within the cell to enhance virus replication. We screened a library of 12,280 small molecules (see Materials and Methods section for library composition) in search of compounds that enhanced the oncolytic activity of VSVΔ51 on the partially virus-resistant breast cancer cell line (4T1). Low concentrations of virus (0.03 plaque-forming units/cell) were used so that virus alone caused minimal cell death over the time of the assay, thus favoring the selection of compounds that promote virus replication and spread in cell culture. We compared the cytotoxicity of a given compound alone or in combination with a low dose of VSVΔ51 and used the ratio of these to compute a relative “viral sensitization factor” that is plotted for each compound in Figure 1a, wherein a higher value indicates compounds that enhance virus-induced killing. We used 0.3 as a threshold for selecting compounds for further validation based on the performance in this assay of suberoylanilide hydroxamic acid (SAHA, 5 µmol/l), a histone deacetylase (HDAC) inhibitor we have previously shown had some ability to enhance VSVΔ51 spread.11 Based on this threshold, 30 compounds (dots within shaded area) were tested in a second round of screening using the high-throughput screening assay format. For validation purposes, a version of VSVΔ51 encoding red fluorescent protein (RFP) was added to a monolayer of 4T1 cells in the presence of selected compounds. Twenty-four hours later, infected cultures were viewed and the extent of virus spread estimated by the expression of RFP. As expected, in vehicle-treated cultures, only small foci of RFP-expressing cells were detected whereas SAHA and 15 of the 30 compounds selected enhanced virus spread as visualized by expression of RFP in most of the cells in the monolayer (Figure 1b). At 48 hours postinfection, the supernatants from infected cultures were collected and virus titers determined. The lead compounds that showed enhanced virus spread at 24 hours also showed substantial improvements in total virus output at 48 hours when compared to vehicle-treated controls (Figure 1c).
Figure 1.
Identification of novel viral sensitizers by high-throughput screening. (a) Dot plot representation of the high-throughput screening data. The y axis represents the viral sensitization factor where positive values suggest viral sensitization. This factor is defined as the logarithm of the cytotoxicity of compounds in absence of VSV over cytotoxicity of compounds in presence of VSV (refer to Materials and Methods section). The average of assay duplicates is plotted for each compound. The x axis represents each of the 12,280 compounds. Compounds exhibiting viral sensitization factor values >0.3 were considered for further validation (dots within shaded area). (b) The potential viral sensitizers identified were retested in a 96-well plate format for VSVΔ51-enhancing activity on 4T1 cells using 10 µmol/l concentrations of drug and a VSVΔ51 MOI of 0.03. A VSVΔ51 strain expressing RFP was used to visualize virus spread after 24 hours using fluorescence microscopy. SAHA (10 µmol/l) was used as a positive control. (c) Fold change in viral titers from supernatants collected from (b) after 48-hours incubation relative to vehicle-treated control. Arrow points to inset panel showing the molecular structure of VSe1 (3,4-dichloro-5-phenyl-2,5-dihydrofuran-2-one). CTRL, control; MOI, multiplicity of infection; RFP, red fluorescent protein; SAHA, suberoylanilide hydroxamic acid; VSe, virus-sensitizers; VSV, vesicular stomatitis virus.
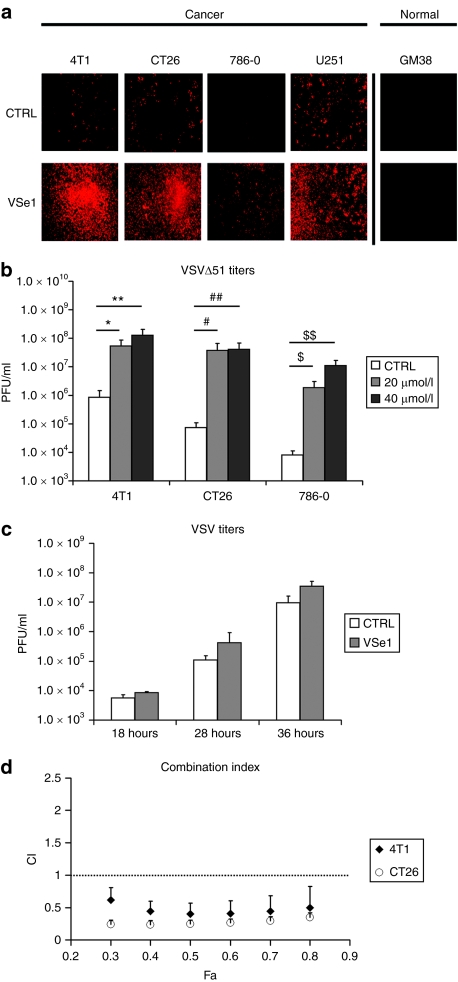
One drug herein referred to as VSe1 (3,4-dichloro-5-phenyl-2,5-dihydrofuran-2-one, Figure 1c, inset structure) had the most activity in these initial studies and was selected for more in depth characterization. We selected four cancer cell lines that were partially or highly resistant to VSVΔ51 and tested the ability of VSe1 to enhance virus replication and spread. Indeed, VSe1 was active in different types of malignancies of human and mouse origin (Figure 2a,b). Importantly, the normal fibroblast cell line GM38 remained resistant to VSVΔ51 infection, even in the presence of VSe1, suggesting that the compound is most active in transformed cell lines (Figure 2a). As expected, no virus could be recovered from VSVΔ51 or VSe1/VSVΔ51-treated GM38 fibroblasts. In contrast, VSe1 increased virus yields from resistant cancer cells (Figures 2b and 3b) in a dose-dependent fashion (starting at 2.5 µmol/l, Supplementary Figure S1). Strikingly, the most VSVΔ51-resistant cell line exhibited over 1,000-fold increases in viral titer in the presence of highest concentrations of VSe1 (786-0 cells, Figure 2b). Combination indexes calculated as described by Chou and Talalay12 revealed that the effects of VSe1 on VSVΔ51 spread also translates to truly synergistic cell killing (Figure 2d).
Figure 2.
VSe1 enhances VSVΔ51 spread and leads to synergistic cell killing in resistant cells. (a) VSVΔ51 resistant 4T1 murine breast cancer cells, CT26 murine colon cancer cells, 786-0 human renal cancer, and U251 human glioma cells were challenged with an RFP-expressing VSVΔ51 at an MOI of 0.01 following a 2–4 hours pretreatment with either VSe1 20 µmol/l or control. Normal human GM38 fibroblasts were also tested but challenged with an MOI of 0.03. Fluorescence pictures were taken 40 hours postinfection. (b) VSVΔ51 titers were determined by plaque assay on Vero cells from supernatants collected at 40 hours postinfection (VSVΔ51, MOI of 0.01) of 4T1, CT26, and 786-0 cells treated with either vehicle control, 20 or 40 µmol/l VSe1. Data represent average from three to five independent experiments *P = 0.007, **P = 0.04, #P = 0.02, ##P = 0.01, $P = 0.005, $$P = 0.009 (ANOVA). Error bars represent the SE. (c) CT26 cells were treated with VSe1 20 µmol/l or vehicle control then challenged with a wild-type VSV (MOI = 0.0003). Viral titers were assessed by plaque assay on Vero cells from supernatants collected at 18, 28, and 36 hours postinfection. (d) 4T1 and CT26 cells were treated with serial dilutions of a fixed ratio combination mixture of VSVΔ51 and VSe1 (500 PFU: 1 µmol/l VSVΔ51:VSe1). Cytotoxicity was assessed using alamar blue reagent after 48 hours. Combination indexes (CI) were calculated according to the method of Chou and Talalay12 using Calcusyn (see Materials and Methods section). Plots represent the algebraic estimate of the CI in function of the fraction of cells affected (Fa). Error bars indicate the estimate SE. ANOVA, analysis of variance; CTRL, control; MOI, multiplicity of infection; RFP, red fluorescent protein; PFU, plaque-forming units; SAHA, suberoylanilide hydroxamic acid; VSe, virus-sensitizers; VSV, vesicular stomatitis virus.
Figure 3.
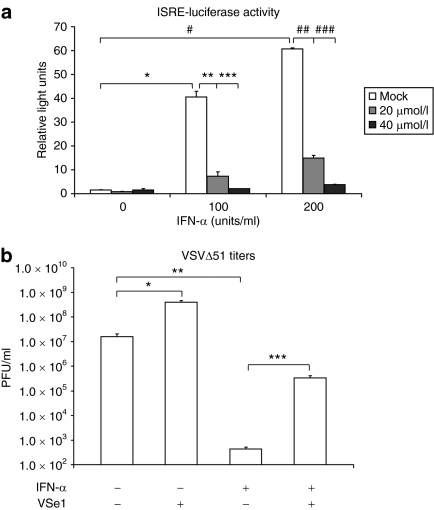
VSe1 inhibits IFN-α induced transcription and its antiviral effects. (a) 293T cells were co-transfected with an ISRE-luciferase reporter and β-galactosidase (control). Six hours post-transfection, cells were treated with indicated concentrations of VSe1 or vehicle. Twenty hours after receiving VSe1, media was replaced and cells were treated with IFN-α. The following day, cells were lysed and measured for luciferase activity. β-Galactosidase activity was also measured and used for data normalization. *P = 0.03, **P = 0.005, ***P = 0.03, #P = 0.003, ##P = 0.006, ###P = 0.002. (b) Human U251 glioma cells were cotreated with 200 U/ml Intron A and VSe1 (or vehicle) then challenged with GFP-expressing VSVΔ51 at an MOI of 0.01. Supernatants were collected 40 hours later and titered by plaque assay on Vero cells.*P = 6.4 × 10−3, **P = 1.6 × 10−4, ***P = 6.8 × 10−5 (ANOVA) error bars represent SE, n = 3. ANOVA, analysis of variance; IFN, interferon; ISRE, IFN-responsive promoter element; MOI, multiplicity of infection; PFU, plaque-forming units; RFP, red fluorescent protein; VSe, virus-sensitizers; VSV, vesicular stomatitis virus.
Our original hypothesis predicted that compounds isolated by our screen would most likely complement the engineered defect in the M-protein of VSVΔ51. Supporting this idea, we found that VSe1 had minimal ability to enhance the growth of VSV with a wild-type M gene in the CT26 cell line while it increased the titer of VSVΔ51 over 100-fold (Figure 2b,c). This suggests that VSe1 enhances VSVΔ51, likely by altering pathways that wild-type VSV already efficiently targets.
VSe1 disrupts IFN-induced antiviral response
The difference between VSVΔ51 and wild-type VSV lies on a single amino acid deletion within the M protein. An important consequence of this mutation is that the mutant M-protein can no longer block the expression of cellular antiviral genes.3,6 It is believed that VSVΔ51 is tumor selective because normal cells have multiple and redundant antiviral signaling programs whereas cancer cells harbor one or more defects within these networks making the M mutation of little consequence.6 In contrast, wild-type M expressing VSV indiscriminately blocks nuclear export and the subsequent expression of antiviral mRNAs rendering a broad range of cells, including both VSVΔ51-resistant tumor and normal cells, sensitive to virus infection and killing (Figure 2c and refs. 5, 6). Because VSe1 activity enhanced the spread of VSVΔ51 but did not augment wild-type VSV (compare Figure 2b and c), we reasoned that VSe1 may partially recapitulate M function by inhibiting the expression of one or more antiviral gene products. To test this idea, we examined the ability of VSe1 to block IFN-activated transcription programs. Human embryonic kidney 293 cells were transfected with a reporter plasmid that contains the luciferase gene under the control of an IFN-responsive promoter element. When treated with human IFN-α, the transfected cells expressed luciferase in a dose-dependent fashion; however, IFN-dependent transcription could be dampened by the addition of increasing doses of VSe1 to the cultures (Figure 3a). Supporting these findings, while IFN-α could protect the glioma cell line U251 from VSVΔ51 infection, protection could be in large part overcome by cotreatment with VSe1 (Figure 3b). In earlier studies, we had shown that several HDAC inhibitors are able to enhance oncolytic virus growth in tumor cells by interrupting IFN signaling.11 It seemed reasonable in light of these data to suggest that VSe1 could be a novel HDAC inhibitor and so we tested its ability to inhibit the activity of 11 different HDACs. We found that at the concentrations VSe1 has clear activity in our cell-based assays, it has little or no effect on the activity of any of the HDACs tested (Supplementary Figure S2). In contrast as expected, Trichostatin A used at similar concentrations was highly active against all the HDACs tested in this assay.13 Given these results and the fact that VSe1 bears no obvious structural similarity to known HDAC inhibitors, VSe1 impairment of IFN responsiveness likely occurs independently of HDAC inhibition.
VSe1 represses virus-induced cellular gene expression
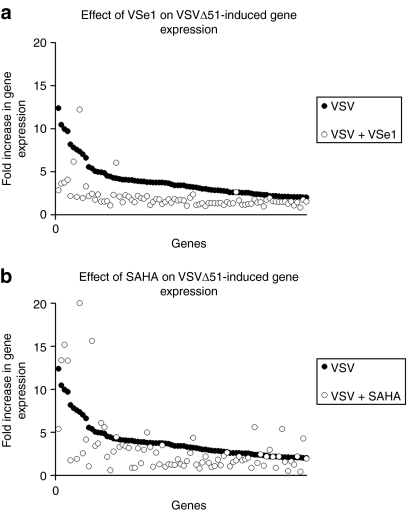
The results presented above suggest VSe1 may be active by inhibiting the expression of cellular antiviral gene transcripts. To test this idea, we used gene expression arrays and compared mRNA profiles in cells infected with VSVΔ51 in the presence or absence of VSe1. CT26 colon cancer cells were pretreated with VSe1 or vehicle and subsequently infected with VSVΔ51 (multiplicity of infection 0.03) or mock-infected with media. RNA was extracted 24 hours postinfection and mRNA expression was compared across conditions. Under these conditions VSVΔ51 infection leads to increased transcription of over 80 cellular genes (Figure 4a,b) including well-known IFN-inducible antiviral genes (e.g., OAS, Mx2,14 and see Supplementary Table S1). Consistent with its ability to enhance the replication and spread of VSVΔ51, VSe1 potently reduced the induction of ~96% of the cellular antiviral transcripts induced by virus infection alone (see Figure 4a and Supplementary Table S1). Modulation of gene expression was further confirmed for a subset of these genes by quantitative PCR analysis (Supplementary Figure S3). Similar to what has been shown previously,15 the bona fide HDAC inhibitor (SAHA) tested in the same experimental context dampened virus-induced transcription of many virus-induced genes (79%, Figure 4b). In uninfected cells, SAHA altered the transcription profile of well over 1,300 genes (Supplementary Table S2a,b) whereas VSe1 on its own affected only 111 gene transcripts (Supplementary Table S3a,b).
Figure 4.
VSe1 represses VSVΔ51-induced genes. CT26 cells were pretreated with either SAHA 5 µmol/l, VSe1 20 µmol/l, or vehicle for 4 hours then challenged with VSVΔ51 at an MOI of 0.03 (or mock treated). Twenty-four hours postinfection, cells were harvested and RNA was extracted. RNA was subsequently processed for hybridization on Affymetrix Mouse Gene 1.0 ST arrays. Expression of genes was normalized to values obtained for vehicle-treated, mock-infected control. In (a,b) points along the x axis represent each gene increased by over twofold by VSVΔ51 infection and are indicated by filled circles. (a) Fold change in gene expression of genes induced by VSVΔ51 in presence of VSe1 20 µmol/l are indicated by open circles. (b) Fold change in gene expression of genes induced by VSVΔ51 in presence of SAHA 5 µmol/l are indicated by open circles. Note that in order to maintain comparable y axes between (a) and (b), one gene (RSad2, gene rank #6, Supplementary Table S1) that was induced nearly 40-fold by SAHA/VSVΔ51 is not represented. MOI, multiplicity of infection; SAHA, suberoylanilide hydroxamic acid; VSe, virus-sensitizers; VSV, vesicular stomatitis virus.
VSe1 augments VSVΔ51 oncolytic activity in vivo and in primary human tumor samples
Since VSe1 enhanced the oncolytic activity of VSVΔ51 in cancer cells but not normal cells in vitro (Figures 1 and 2) we sought to determine whether this level of specificity would be observed in mouse models and/or in freshly explanted patient tumor material. Balb/c mice were engrafted with a CT26 colon cancer cell line and tumor growth was evaluated following treatment with vehicle control, VSe1, vehicle/VSVΔ51, or VSe1/VSVΔ51. Figure 5a shows that whereas neither VSe1 nor VSVΔ51 had a significant effect on tumor growth, the combination of VSe1 and VSVΔ51 led to a significant delay in tumor progression. Importantly, when animals were treated with VSVΔ51 harboring the GFP gene in the presence or absence of VSe1 there was no detectable virus in any of the normal tissues of treated animals (Supplementary Figure S4a) and no virus could be recovered from tissue samples following homogenization. In addition, mouse weights dipped following virus infection as is typically observed6 but did not differ significantly in presence or absence of VSe1 (Supplementary Figure S4b) further suggesting tolerability of the combination treatment.
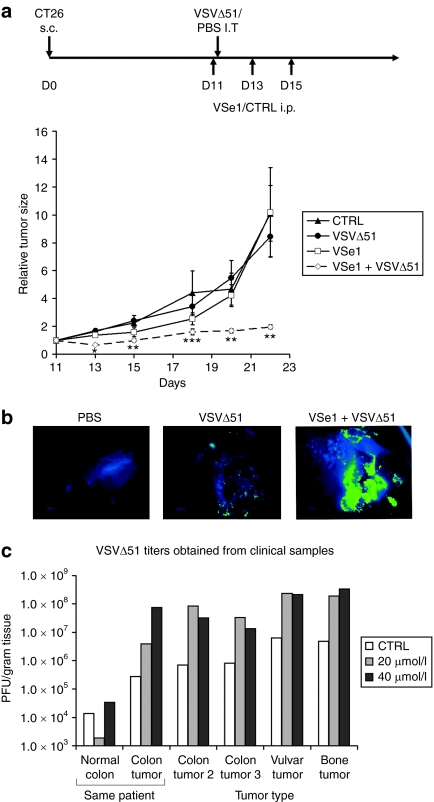
Figure 5.
VSe1 exhibits VSVΔ51-sensitizing activity in immunocompetent mice and in human clinical samples. (a) 3 × 105 VSVΔ51-resistant CT26 cells were implanted subcutaneously (s.c) in syngeneic Balb/c mice 11 days before first treatment. On day 11 VSe1 (or vehicle) was administered intraperitoneally (i.p) at 0.4 mg/mouse. Four hours later, 1 × 108 VSVΔ51 (or PBS) was administered intratumorally (i.t). Two more doses of VSe1 were administered on days 13 and 15. Mouse tumor volume was measured using caliper and average tumor volumes relative to day 11 are shown. Error bars represent SE. *P < 0.005, **P < 0.05, ***P < 0.1 (ANOVA). N = 5 mice per group. (b) False-color (LUT) fluorescence microscopy images of representative human colon tumor slices infected with 1 × 107 PFU of GFP-expressing VSVΔ51 (or PBS, left panel) 24 hours post-treatment with either vehicle (middle panel) or 40 µmol/l VSe1 (right panel). Green color is indicative of intense GFP fluorescence and virus replication whereas blue color represents low level background tissue autofluorescence. Pictures were taken after 72-hours incubation. (c) Human tumor or normal tissue slices were treated as in (b) with either 20 or 40 µmol/l VSe1. Seventy-two hours later tissue samples were collected and homogenized for subsequent tittering on Vero cells by plaque assay. ANOVA, analysis of variance; CTRL, control; GFP, green fluorescent protein; PBS, phosphate-buffered saline; PFU, plaque-forming units; VSe, virus-sensitizers; VSV, vesicular stomatitis virus.
Consistent with the in vivo results and in vitro data presented in Figure 2a, cancer-specific virus enhancement was seen when primary human tumor specimens were infected ex vivo in the presence of VSe1. An example of these experiments is shown in Figure 5b where VSVΔ51-GFP was added to a colon cancer sample in the presence or absence of VSe1. Although in this patient sample VSVΔ51-GFP replicated poorly on its own, its growth and spread (as visualized by green fluorescence) was significantly enhanced in the presence of VSe1. In Figure 5c the titers of virus produced in primary human tumor samples was determined in the presence of increasing amounts of VSe1. As was observed in our tumor cell line experiments, we found that VSe1 could increase VSVΔ51 from 10- to 100-fold in primary human tumor samples of various origins. In one colorectal cancer case, adjacent normal colon tissue was isolated and, as expected, VSVΔ51 on its own grew better in tumor versus adjacent normal tissues. Importantly, while treatment of the explants with VSe1 did not increase the replication of VSVΔ51 in normal tissues, it led to over 100-fold growth of VSVΔ51 in the tumor tissue, leading to roughly 1,000-fold differential in replication between normal and cancerous tissues.
Discussion
OVs have shown excellent therapeutic activity in a variety of inbred mouse tumor models of cancer6,7,16,17,18,19 but they have been less active in clinical trials thus far.20,21,22,23 One likely reason for this is that OVs have been engineered to be safe by deletion or mutation of virulence genes but in so doing their potency has been compromised. Several groups have attempted to improve OV efficacy by genetic manipulation (reviewed in ref. 24) and whereas this strategy shows promise, it also poses the risk that more virulent viruses may lose the excellent safety margins that have been demonstrated with the current cadre of OVs. An alternative strategy is to identify chemicals or other biologics that can complement virus mutations and transiently increase OV activity within the tumor. For the studies presented here we sought to find chemicals that could enhance the replication and spread of the mutant VSVΔ51 strain. In its wild-type form, the VSV M-protein is a multifunctional protein that blocks the nuclear export of antiviral genes, affects cell shape, sabotages normal mitochondrial function, and aids in virus assembly.3,25,26,27 VSVΔ51 harbors a mutation in its M gene that abrogates its ability to antagonize cellular antiviral responses.6 Our data suggest that VSe1 complements the M mutation by also interfering with cellular antiviral programs (Figures 2d, 3a,b, and 4a,b).
We initially suspected that VSe1 might be an HDAC inhibitor because we and others had shown that small molecules with this activity can block IFN-induced gene transcription.11 This is unlikely because to our knowledge VSe1 does not resemble any of the structures of currently known HDAC inhibitors and does not have significant activity against any of the HDACs that we tested (Supplementary Figure S2). Importantly, whereas VSe1 profoundly dampens the antiviral response similarly to HDAC inhibitors, its impact on global gene expression is significantly narrower, affecting less than a tenth of the number of genes modulated by SAHA.
Our “pharmacoviral” approach, complementing defective or deleted viral genes with small molecules, suggests a new strategy for both cancer treatment and drug discovery in general. In principle, one can foresee using a virus/cell-based strategy to find other drugs that complement defects within other virus mutants. Alternately, it may also be of interest to screen for small molecules that enhance the activity of wild-type viruses in order to find drugs that might increase oncolysis by mechanisms other than the complementation of missing genes. This screening method permits one to probe for compounds that mimic effects elicited by specific viral functions, thus favoring the discovery of molecules that affect entire pathways as opposed to single molecules. Another advantage of our high-throughput screening strategy is that because of the assay design and selection criteria, minimally cytotoxic compounds are identified.
The screening strategy presented here did not include a step to select against compounds that sensitize normal cells to virus infection. Despite this, our data demonstrate that VSe1 selectively enhances virus growth in tumor cells but not in normal tissues (Figures 2a, 5c, and Supplementary Figure S4a). Although it remains unclear why this is the case, we suspect it is related to the several layers of redundancy in cell signaling that are found in normal tissues. It has become clear from deep sequencing of cancer patient genomes that tumor cells have many accumulated mutations.28 As malignancies evolve, they appear to shed the layers of protection a normal genome maintains to protect against virus invasion.6,29,30 Although these types of mutations likely provide the tumor with a growth and immune evasion advantage they may also position the cancer cell on the precipice of catastrophy when faced with an unexpected stress. Perhaps compounds like VSe1 are the “last straw” required to break tumor cell antiviral defenses and remove the final barrier to virus growth. In contrast, the robust and redundant antiviral networks residing in normal cells may be only partially affected by VSe1-type compounds.
Materials and Methods
Drugs and chemicals. Screened compounds were a selected subset from the Maybridge HitFinder, Chembridge DIVERSet, Microsource Spectrum, Prestwick, BIOMOL, and Sigma LOPAC screening collections based on chemical diversity and nonoverlap. VSe1 (3,4-dichloro-5-phenyl-2,5-dihydrofuran-2-one) was obtained from Ryan Scientific (Mt Pleasant, SC). SAHA was obtained from Exclusive Chemistry (Obninsk, Russia). IFN-α (Intron A) was obtained from Schering-Plough (Kenilworth, NJ).
Cell lines. 4T1 (breast), CT26 (colon) mouse cancer cells; 786-0 (renal cancer), U251 (glioma), human embryonic kidney 293T (embryonic kidney), U2OS (osteosarcoma) human cells; Vero (monkey kidney cells); and GM38 normal human fibroblasts, were obtained from the American Type Culture Collection (Manassas, VA). Cells were cultured in HyQ high-glucose Dulbecco's modified Eagle's medium (Hyclone, Waltham, MA) supplemented with 10 or 20% (GM38) fetal calf serum (CanSera, Etobicoke, Ontario, Canada). All cell lines were incubated at 37 °C in a 5% CO2 humidified incubator.
Viruses. The Indiana serotype of VSV (VSVΔ51 or wild type) was used throughout this study and was propagated in Vero cells. VSVΔ51-expressing RFP or GFP are recombinant derivatives of VSVΔ51. All viruses were purified as described previously.6 For mouse studies, virus was further purified on 5–50% Optiprep (Sigma, St Louis, MO) gradient.
High-throughput screen. 4T1 cells were plated in HEPES-buffered, phenol red free Dulbecco's modified Eagle's medium in 96-well plates and allowed to adhere overnight. The next day cells were pretreated for 4 hours with a 10 µmol/l concentration of library compounds (or control dimethyl sulfoxide) added using a Biomek FX liquid handler (Beckman Coulter, Fullerton, CA), and subsequently challenged with VSVΔ51 at an multiplicity of infection of 0.0325 or a control added using a µFill liquid handler (Biotek, Winooski, VT). Duplicates were run for each condition. Forty hours later, plates were incubated with alamar blue and fluorescence emission rate was assessed using an EnVision plate reader (PerkinElmer, Waltham, MA). Cytotoxicity of each drug was determined in both presence and absence of virus and was used to calculate a viral sensitization factor (see below).
Assessment of combination index. 25 000 4T1 or CT26 cells were plated per well in 96-well plates and left to adhere over night. The following day, cells were pretreated for 4 hours with serial dilutions of Vse1 (200–1.5 µmol/l, 1:2 dilution steps) then infected with serial dilutions of VSVΔ51 (100,000–780 plaque-forming units) keeping a fixed ratio combination of VSVΔ51 and VSe1 (500 plaque-forming unit to 1 µmol/l). Cytotoxicity was assessed using alamar blue reagent after 48 hours. Combination indexes were calculated using the Calcusyn Software (Biosoft, Ferguson, MO) according to the method of Chou and Talalay.12
Reporter assays. Human embryonic kidney 293T cells were plated at 1.3 × 105 cells/well in 24-well dishes. The following day, cells were co-transfected with an IFN-responsive promoter element-driven luciferase reporter plasmid and a cytomegalovirus-driven β-galactosidase control plasmid as described previously.31 Six hours post-transfection, cells were treated with VSe1 or mock treated with vehicle. Approximately 20 hours after receiving VSe1, cells were then treated with IFN-α with a complete media change. The following day, cells were lysed and measured for luciferase using the BD Monolight kit (Becton Dickinson, Franklin Lakes, NJ). β-Galactosidase activity was measured using the Luminescent β-galactosidase kit (Clontech, Mountainview, CA).
Microarray. CT26 cells were plated at a density of 1.5 × 106 in 100-mm petris and allowed to adhere overnight. The next day, cells were treated with dimethyl sulfoxide, 20 µmol/l VSe1 or 5 µmol/l SAHA. Four hours later, VSVΔ51 (or control media) was added at an multiplicity of infection of 0.03. Twenty-four hours postinfection RNA was collected (see Supplementary Materials and Methods) and pooled RNAs form duplicate experiments were used for hybridization on Affymetrix mouse gene 1.0 ST arrays according to manufacturer instructions. Low signal genes (<50) were removed. Genes were normalized to average overall signal for each array. Fold change in gene expression was calculated for each gene in relation to uninfected control. A twofold change in gene expression relative to the control was used as a cutoff for selection of treatment-perturbed genes. Analysis was done using Microsoft Excel.
Animal tumor model. Six-week-old female Balb/c mice were given subcutaneous tumors by injecting 3 × 105 syngeneic CT26 cells suspended in 100 µl phosphate-buffered saline. Eleven days postimplantation (average tumor size = 220 mm3), mice were treated with a 0.4 mg dose of VSe1 freshly resuspended in 30% ethanol, 5% dimethyl sulfoxide, 65% phosphate-buffered saline (or vehicle control) administered intraperitoneally. VSVΔ51 (1 × 108 plaque-forming unit) was introduced intratumorally 4 hours following the first VSe1 dose. Subsequently, VSe1 (or vehicle) was readministered on days 13 and 15 postimplantation (0.4 mg/injection/mouse). Tumor sizes were measured using an electronic caliper. Tumor volume was calculated as = (length × width2)/2. Initial tumor size measured on day 11 was used to calculate relative tumor size. All experiments were performed in accordance with institutional guidelines for animal care.
Treatment and processing of primary tissue specimens. Primary tissue specimens were processed within 48 hours postsurgery from consenting patients who underwent tumor resection. Three hundred micrometer tissue slices were obtained using a Krumdieck tissue slicer (Alabama Research and Development, Munford, AL) and plated in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Samples were visualized by fluorescence microscopy. The “green fire blue” look up table in Image J (NIH software) was applied to all pictures shown in Figure 5b to better discriminate between background and highly fluorescent virus-associated regions. Tissues were weighed and homogenized in 1 ml of phosphate-buffered saline using a homogenizer (Kinematica AG-PCU-11, Staufen, Germany) and viral titers were quantified by standard plaque assay (see Supplementary Materials and Methods).
Statistics. Analysis of variance was used for multigroup comparisons where indicated. For viral titers, logarithm was computed before statistical analysis. Homogeneity of variance was verified using Levene's statistic. All tests were two-tailed.
SUPPLEMENTARY MATERIALFigure S1. VSe1 enhances spread of VSVΔ51 in a dose-dependent manner.Figure S2. Effect of VSe1 on HDAC enzymatic activity in vitro.Figure S3. Validation of microarray gene expression profiles by real-time PCR.Figure S4. VSe1 does not increase VSVΔ51 replication in normal mouse tissues and does not aggravate weight loss in response to virus.Table S1. Comparison of fold change in gene expression induced by VSVΔ51 in presence of VSe1 or SAHA.Table S2. Modulation of gene expression induced by SAHA.Table S3. Modulation of gene expression induced by VSe1.Materials and Methods.
Supplementary Material
VSe1 enhances spread of VSVΔ51 in a dose-dependent manner.
Effect of VSe1 on HDAC enzymatic activity in vitro.
Validation of microarray gene expression profiles by real-time PCR.
VSe1 does not increase VSVΔ51 replication in normal mouse tissues and does not aggravate weight loss in response to virus.
Comparison of fold change in gene expression induced by VSVΔ51 in presence of VSe1 or SAHA.
Modulation of gene expression induced by SAHA.
Modulation of gene expression induced by VSe1.
Acknowledgments
The authors acknowledge Alanah Kemp and Nichealla Keath for their technical assistance and Rebecca Auer, Johanne Weberpals, and Lisa Mackenzie for providing and processing clinical samples. We also thank Kelley Parato for critical review of the manuscript. J.-S.D is supported by a Fonds de Recherche en Santé du Québec fellowship. F.L.B is supported by a Canadian Institutes of Health Research industrial fellowship. The project was funded by the Ontario Institute for Cancer Research selective therapies program. The majority of this work was done in Ottawa, Ontario, Canada.
REFERENCES
- Parato KA, Senger D, Forsyth PA., and , Bell JC. Recent progress in the battle between oncolytic viruses and tumours. Nat Rev Cancer. 2005;5:965–976. doi: 10.1038/nrc1750. [DOI] [PubMed] [Google Scholar]
- Bushell M., and , Sarnow P. Hijacking the translation apparatus by RNA viruses. J Cell Biol. 2002;158:395–399. doi: 10.1083/jcb.200205044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria PA, Chakraborty P, Levay A, Barber GN, Ezelle HJ, Enninga J, et al. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol Cell. 2005;17:93–102. doi: 10.1016/j.molcel.2004.11.023. [DOI] [PubMed] [Google Scholar]
- Santoro MG, Rossi A., and , Amici C. NF-κB and virus infection: who controls whom. EMBO J. 2003;22:2552–2560. doi: 10.1093/emboj/cdg267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojdl DF, Lichty B, Knowles S, Marius R, Atkins H, Sonenberg N, et al. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat Med. 2000;6:821–825. doi: 10.1038/77558. [DOI] [PubMed] [Google Scholar]
- Stojdl DF, Lichty BD, tenOever BR, Paterson JM, Power AT, Knowles S, et al. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell. 2003;4:263–275. doi: 10.1016/s1535-6108(03)00241-1. [DOI] [PubMed] [Google Scholar]
- Hummel JL, Safroneeva E., and , Mossman KL. The role of ICP0-Null HSV-1 and interferon signaling defects in the effective treatment of breast adenocarcinoma. Mol Ther. 2005;12:1101–1110. doi: 10.1016/j.ymthe.2005.07.533. [DOI] [PubMed] [Google Scholar]
- Thorne SH, Hwang TH, O'Gorman WE, Bartlett DL, Sei S, Kanji F, et al. Rational strain selection and engineering creates a broad-spectrum, systemically effective oncolytic poxvirus, JX-963. J Clin Invest. 2007;117:3350–3358. doi: 10.1172/JCI32727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katze MG, Fornek JL, Palermo RE, Walters KA., and , Korth MJ. Innate immune modulation by RNA viruses: emerging insights from functional genomics. Nat Rev Immunol. 2008;8:644–654. doi: 10.1038/nri2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Nguyên TL, Abdelbary H, Arguello M, Breitbach C, Leveille S, Diallo JS, et al. Chemical targeting of the innate antiviral response by histone deacetylase inhibitors renders refractory cancers sensitive to viral oncolysis. Proc Natl Acad Sci USA. 2008;105:14981–14986. doi: 10.1073/pnas.0803988105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TC., and , Talaly P. A simple generalized equation for the analysis of multiple inhibitions of Michaelis-Menten kinetic systems. J Biol Chem. 1977;252:6438–6442. [PubMed] [Google Scholar]
- Minucci S., and , Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- Sadler AJ., and , Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HM, Paulson M, Holko M, Rice CM, Williams BR, Marié I, et al. Induction of interferon-stimulated gene expression and antiviral responses require protein deacetylase activity. Proc Natl Acad Sci USA. 2004;101:9578–9583. doi: 10.1073/pnas.0400567101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford MM, Shaban M, Barrett JW, Werden SJ, Gilbert PA, Bondy-Denomy J, et al. Myxoma virus oncolysis of primary and metastatic B16F10 mouse tumors in vivo. Mol Ther. 2008;16:52–59. doi: 10.1038/sj.mt.6300348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda H, Yin J, Mueller S, Wimmer E., and , Cello J. Oncolytic treatment and cure of neuroblastoma by a novel attenuated poliovirus in a novel poliovirus-susceptible animal model. Cancer Res. 2007;67:2857–2864. doi: 10.1158/0008-5472.CAN-06-3713. [DOI] [PubMed] [Google Scholar]
- Watanabe D, Goshima F, Mori I, Tamada Y, Matsumoto Y., and , Nishiyama Y. Oncolytic virotherapy for malignant melanoma with herpes simplex virus type 1 mutant HF10. J Dermatol Sci. 2008;50:185–196. doi: 10.1016/j.jdermsci.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Hirasawa K, Nishikawa SG, Norman KL, Coffey MC, Thompson BG, Yoon CS, et al. Systemic reovirus therapy of metastatic cancer in immune-competent mice. Cancer Res. 2003;63:348–353. [PubMed] [Google Scholar]
- Kaplan JM. Adenovirus-based cancer gene therapy. Curr Gene Ther. 2005;5:595–605. doi: 10.2174/156652305774964677. [DOI] [PubMed] [Google Scholar]
- Lorence RM, Roberts MS, O'Neil JD, Groene WS, Miller JA, Mueller SN, et al. Phase 1 clinical experience using intravenous administration of PV701, an oncolytic Newcastle disease virus. Curr Cancer Drug Targets. 2007;7:157–167. doi: 10.2174/156800907780058853. [DOI] [PubMed] [Google Scholar]
- Mace AT, Ganly I, Soutar DS., and , Brown SM. Potential for efficacy of the oncolytic Herpes simplex virus 1716 in patients with oral squamous cell carcinoma. Head Neck. 2008;30:1045–1051. doi: 10.1002/hed.20840. [DOI] [PubMed] [Google Scholar]
- Liu TC, Hwang TH, Bell JC., and , Kirn DH. Development of targeted oncolytic virotherapeutics through translational research. Expert Opin Biol Ther. 2008;8:1381–1391. doi: 10.1517/14712598.8.9.1381. [DOI] [PubMed] [Google Scholar]
- Kaur B, Cripe TP., and , Chiocca EA. “Buy one get one free”: armed viruses for the treatment of cancer cells and their microenvironment. Curr Gene Ther. 2009;9:341–355. doi: 10.2174/156652309789753329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichty BD, McBride H, Hanson S., and , Bell JC.2006Matrix protein of Vesicular stomatitis virus harbours a cryptic mitochondrial-targeting motif J Gen Virol 87(Pt 11): 3379–3384. [DOI] [PubMed] [Google Scholar]
- Lyles DS., and , McKenzie MO. Activity of vesicular stomatitis virus M protein mutants in cell rounding is correlated with the ability to inhibit host gene expression and is not correlated with virus assembly function. Virology. 1997;229:77–89. doi: 10.1006/viro.1996.8415. [DOI] [PubMed] [Google Scholar]
- Lyles DS, McKenzie MO, Kaptur PE, Grant KW., and , Jerome WG. Complementation of M gene mutants of vesicular stomatitis virus by plasmid-derived M protein converts spherical extracellular particles into native bullet shapes. Virology. 1996;217:76–87. doi: 10.1006/viro.1996.0095. [DOI] [PubMed] [Google Scholar]
- Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- Lengyel P. Tumor-suppressor genes: news about the interferon connection. Proc Natl Acad Sci USA. 1993;90:5893–5895. doi: 10.1073/pnas.90.13.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmann G, Robek MD., and , van den Pol AN. Variable deficiencies in the interferon response enhance susceptibility to vesicular stomatitis virus oncolytic actions in glioblastoma cells but not in normal human glial cells. J Virol. 2007;81:1479–1491. doi: 10.1128/JVI.01861-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Kazdhan N., and , Lichty BD. Using G-deleted vesicular stomatitis virus to probe the innate anti-viral response. J Virol Methods. 2008;153:276–279. doi: 10.1016/j.jviromet.2008.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
VSe1 enhances spread of VSVΔ51 in a dose-dependent manner.
Effect of VSe1 on HDAC enzymatic activity in vitro.
Validation of microarray gene expression profiles by real-time PCR.
VSe1 does not increase VSVΔ51 replication in normal mouse tissues and does not aggravate weight loss in response to virus.
Comparison of fold change in gene expression induced by VSVΔ51 in presence of VSe1 or SAHA.
Modulation of gene expression induced by SAHA.
Modulation of gene expression induced by VSe1.