Abstract
Zinc Finger nucleases (ZFNs) have been used to create precise genome modifications at frequencies that might be therapeutically useful in gene therapy. We created a mouse model of a generic recessive genetic disease to establish a preclinical system to develop the use of ZFN-mediated gene correction for gene therapy. We knocked a mutated GFP gene into the ROSA26 locus in murine embryonic stem (ES) cells and used these cells to create a transgenic mouse. We used ZFNs to determine the frequency of gene correction by gene targeting in different primary cells from this model. We achieved targeting frequencies from 0.17 to 6% in different cell types, including primary fibroblasts and astrocytes. We demonstrate that ex vivo gene-corrected fibroblasts can be transplanted back into a mouse where they retained the corrected phenotype. In addition, we achieved targeting frequencies of over 1% in ES cells, and the targeted ES cells retained the ability to differentiate into cell types from all three germline lineages. In summary, potentially therapeutically relevant frequencies of ZFN-mediated gene targeting can be achieved in a variety of primary cells and these cells can then be transplanted back into a recipient.
Introduction
Conceptually, the simplest application of gene therapy is for diseases caused by mutations in a single gene, the so-called monogenic diseases. Although millions of people suffer from monogenic diseases, a cure is only possible for a small fraction for whom either hematopoietic stem cell transplantation or organ transplantation is available. In contrast, gene therapy uses the patient's own cells and has the potential to cure many of these diseases. In the past decade, several clinical trials have been carried out that have highlighted both the promise of gene therapy (the benefit of tens of patients with severe combined immunodeficiency,1,2 a handful of patients with Leber's congenital amaurosis,3,4 and two patients with X-linked adrenoleukodystrophy5 from gene therapy based on viral delivery), and the potential harm from the uncontrolled integrations of the viral vectors used to deliver the therapeutic transgene.6 An alternative to using integrating viruses is to use gene targeting by homologous recombination to precisely control the genomic modification either through directly correcting a mutation or through controlling the site of transgene integration.7,8
The natural rate of gene targeting by homologous recombination, hereafter referred to as “gene targeting,” is 1 × 10−5 to 1 × 10−8 (refs. 9,10) and is too low to be therapeutically useful. This barrier has been overcome by the discovery that the creation of a gene-specific DNA double-strand break can stimulate gene targeting several thousand-fold,10,11,12,13,14 potentially to therapeutically relevant levels. To translate this finding to the field of gene therapy, it was necessary to devise a method to generate site-specific DNA double-strand breaks. There have been two major approaches to this problem. The first is to redesign homing endonucleases to recognize target sites in endogenous genes.15,16,17 The second is to design zinc finger nucleases (ZFNs) to recognize target sites in endogenous genes. ZFNs are artificial proteins in which the nonspecific nuclease domain from the FokI restriction endonuclease is fused to a zinc finger DNA-binding domain (reviewed in refs. 8,18). A ZFN can have 3–6 individual zinc-finger domains arranged in tandem recognizing a target site 9–18 base-pair long. Additionally, the FokI nuclease domain functions as a dimer.19,20 Therefore, a pair of ZFNs must be engineered to bind the target site in a way that permits the nuclease domain to dimerize and create the double-strand break. Thus, even with a pair of three-finger ZFNs, the full target site is 18 base pairs long. An 18-base-pair sequence should only occur once in the mammalian genome based on probability and can be empirically determined for any given sequence by BLAST searches. There are a number of different approaches to engineer ZFNs, each of which has their advantages and disadvantages (reviewed in Cathomen and Joung21). Nonetheless, ZFNs have been successfully engineered to a wide variety of different gene targets in a range of different species.10,22,23,24,25,26,27,28,29,30 These ZFNs have been used to generate high rates of precise genome modifications either by the use of mutagenic nonhomologous end-joining (in which short insertions or deletions are created at the site of the ZFN-induced double-strand break) or by the use of gene targeting, including creating genetically modified zebrafish and rats.24,25,26
In human cells, ZFNs have been used to stimulate gene targeting in a variety of different cell lines. The most recent advances demonstrated that ZFNs can stimulate gene targeting in human embryonic stem (ES) cells and induced pluripotent stem (iPS) cells.31,32 Moreover, Perez et al. demonstrated that human T cells modified at the CCR5 gene by mutagenic repair of a ZFN-induced double-strand break could survive when transplanted back into an immunodeficient mouse.33 Nonetheless, to date, there has been no easy way to model a therapeutic paradigm in which host-derived cells are precisely modified by ZFN-mediated gene targeting ex vivo and then transplanted back into a recipient, as might be done when trying to treat a patient with a genetic disease. To this end, we have created a mouse model of a generic recessive genetic disease in which a mutated GFP gene has been knocked-in to the ubiquitously expressed ROSA26 locus.34 Using this model, we show that gene correction of 0.17–6% of murine ES cells, novel ROSA-3T3 cell lines, primary embryonic fibroblasts, primary adult fibroblasts, and primary astrocytes can be achieved. We also demonstrate that gene-corrected cells can be transplanted back into a recipient mouse and the transplanted cells retain their gene-corrected phenotype.
Results
Generation of a mouse model of a generic recessive genetic disease
A critical aspect of translating the use of ZFN-mediated gene targeting to clinical use is to develop appropriate animal models to evaluate the feasibility of the strategy. In this work, we generated a mouse model that allows us to study the efficiency of ZFN-mediated gene targeting by homologous recombination in any primary cell type using a previously well-defined pair of ZFNs. This model provides a platform to directly determine the efficiency of gene targeting in a specific cell type and whether targeted cells can be successfully transplanted back into a host. We created a mouse model of a generic recessive genetic disease by knocking-in a mutated GFP gene into the ROSA26 locus using standard homologous recombination technology in murine ES cells (Figure 1a). From two of the targeted ES cell clones, we generated transgenic mouse lines in which either one (ROSA26GFP*/+) or both alleles (ROSA26GFP*/GFP*) of the ROSA26 locus contain a knock-in of the mutated GFP gene. The mutation in the GFP gene consists of an 85 nucleotide sequence that includes both an in-frame stop codon and the recognition site for the I-SceI homing endonuclease. This insertion is 12 bp downstream from the ZFN target site in the GFP gene that we have previously designed and validated several different pairs of “GFP”-ZFNs to recognize.35 This animal model mimics the GFP gene-targeting system that we have used to better understand ZFN-mediated gene targeting in tissue culture cells, and human ES and iPS cells.10,32,35,36 In this system, cells containing the integrated GFP* gene are transfected with two plasmids that each express a ZFN, and a third “donor” plasmid that carries sequence information needed to correct the mutation (by serving as a donor template during homologous recombination).10,18 Because the mutated GFP gene is knocked-in to the ubiquitously expressed ROSA26 locus, this model can be used to study the efficiency of gene correction by gene targeting in potentially every cell type in the mouse. There are two possible strategies to use gene targeting for gene therapy. The first is to attempt gene targeting in vivo and directly correct disease-causing mutations in cells without removing them from the animal beforehand. The second is to purify cells from the animal first, and correct the mutation ex vivo prior to transplanting back into the animal. Here, we report on our results at purifying primary cells from this generic genetic disease model and using ZFN-mediated gene targeting to directly correct the mutation in the GFP gene and then transplanting those cells back into a mouse.
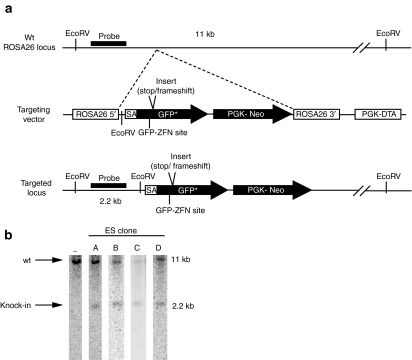
Figure 1.
Construction of targeting vector and screening of ES clones. (a) Schematic of targeting vector and screening strategy. Correct knock-in of the GFP* reporter cassette to the ROSA26 locus causes the addition of an upstream EcoRV site resulting in a 2.2 kb fragment upon digestion. (b) Southern blot analysis of correct knock-in ES clones. Genomic DNA was purified from ES clones, digested with EcoRV and used for Southern analysis. ES, embryonic stem; kb, kilobase; PGK-DTA, diphtheria toxin cassette; PGK-Neo, neomycin resistance cassette; SA, splice acceptor.
ZFN-mediated gene targeting in murine ES cells
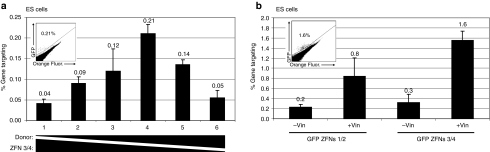
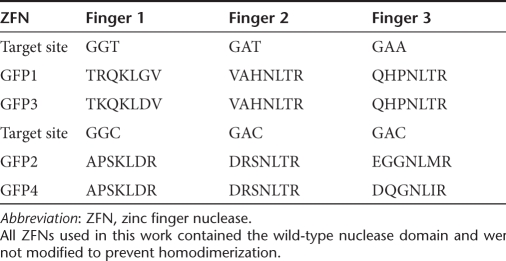
As we were generating the mouse line from the ROSA26GFP*/+-targeted ES cell clones, we performed a series of targeting experiments in the ES cell clones (Figure 2). To perform these experiments, we transfected ES cells with three plasmids—an EGFP donor vector that contains sequence information necessary to correct the mutation in the integrated target but is nonfluorescent because it is missing the first 37 nucleotides of the coding region and two ZFN expression plasmids where the ZFN is driven by the ubiquitin C promoter. In prior work, we have shown that titrating the amount of the donor plasmid and nuclease expression plasmid is important in maximizing the rate of gene targeting.35,36 In the gene-targeting titration experiment, we varied the amount of donor plasmid and ZFN expression plasmids while keeping the total amount of DNA transfected the same. We achieved the maximal rate of targeting when the transfection mix consisted of ~90% donor plasmid and ~10% of ZFN expression plasmids (Figure 2a). ROSA26GFP*/+ ES cells that were transfected with only the donor plasmid without ZFNs had an absolute targeting rate of <0.001% (data not shown). The targeting rate with ZFNs was 0.21%. An important concern involving the use of ZFNs to create DNA double-strand breaks is the potential for off-target cleavage. We have previously reported that the ZFNs used in this study demonstrate low levels of off-target toxicity in the murine ES cells used in this article,35 and no gross toxicity was observable in these experiments. We also tested whether exposing the cells to vinblastine would increase the rate of targeting in murine ES cells as has been demonstrated in other cell types arrested at the S/G2 phase of the cell cycle,28,37,38 and found that vinblastine exposure for the first 15 hours after transfection increased the rate of gene targeting in murine ES cells by approximately four- to fivefold to an overall rate of 1.6% (Figure 2b). At this concentration of vinblastine (100 nmol/l for 15 hours), no gross cytotoxic effects were readily apparent in this cell type. We also compared two different pairs of ZFNs that target the same sequence in the GFP gene (Table 1) and found that ZFN pair 3/4 was ~50% better than pair 1/2 (the pair we have previously published and characterized most extensively35). ZFN 3 only differs from ZFN 1 by a single amino acid in finger 1 and ZFN 4 has a different finger 3 than ZFN 2.
Figure 2.
Gene targeting in ES cells. (a) Titration of donor plasmid and ZFNs in ES cells. Cells were transfected using Lipofectamine 2000 with different amounts of donor and ZFN plasmids. From left to right, amounts of donor plasmid increased, whereas ZFN amounts decreased. Transfection was performed with Lipofectamine 2000, and the amounts of donor plasmid and ZFNs in each lane are indicated as donor (ng), ZFN1 (ng)/ZFN2 (ng). (1) 100, 350/350; (2) 400, 200/200; (3) 600, 100/100; (4) 700, 50/50; (5) 750, 25/25; (6) 775, 13/13. Fifteen hours after transfection, media were changed to normal ESLX. Gene-targeting events were analyzed 4 days after transfection using flow cytometry. (b) Gene targeting in ES cells using two sets of GFP-ZFNs, with and without vinblastine treatment. Cells were plated in ESLX with and without vinblastine (100 nmol/l). Transfection mix was added, and 15 hours later, removed and replated with ESLX. Gene-targeting events were analyzed as in a. In the upper left of each graph is a representative flow cytometry plot after targeting in which GFP fluorescence is measured in the y axis and background orange fluorescence along the x axis. The number in the left corner of the flow plot is the percentage of GFP+ cells. ES, embryonic stem; ZFN, zinc finger nuclease.
Table 1.
ZFN along with the seven amino acid stretch that mediates DNA binding and the 3 base-pair DNA sequence to which it binds
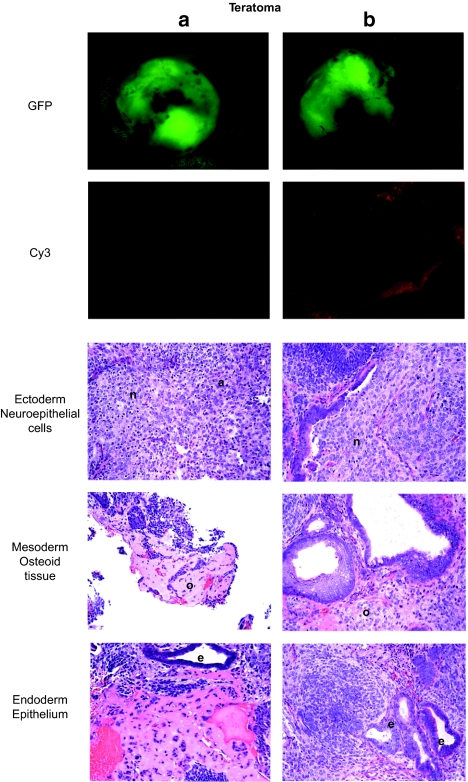
To determine whether the targeted ES cells retained pluripotency, we evaluated whether the targeted cells could differentiate into all three germline lineages using a teratoma formation assay. GFP+ targeted ES cells were purified using fluorescence-activated cell sorting (FACS), and these ES cells were injected subcutaneously into nude mice to create fluorescent teratomas (Figure 3). Histological examination of the teratomas showed that targeted ES cells formed tissues representative of all three germline lineages—endoderm, mesoderm, and ectoderm. In summary, ZFNs can stimulate gene targeting in murine ES cells to an absolute rate of 1.6% (>1,000-fold stimulation over the targeting rate without ZFNs), and these targeted cells retain the potential to differentiate into all of the major cell lineages.
Figure 3.
Teratoma formation assay. Two teratomas resulting from subcutaneous injection of targeted cells were harvested from nude mice. (a) Teratomas were initiated by gene-targeted ES cells as demonstrated by GFP fluorescence. Teratomas were also photographed under a Cy3 filter to show background fluorescence. (b) Sections were cut and stained with hematoxylin and eosin to identify tissue structures indicated in the figure. n, neuroepithelial cells; o, osteoid tissue; e, epithelium.
Gene targeting in ROSA-3T3 cells
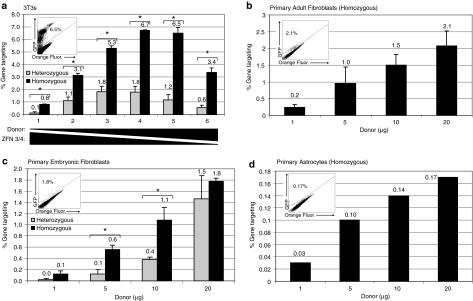
We generated immortalized fibroblast cell lines “ROSA-3T3s” from both ROSA26GFP*/+ and ROSA26GFP*/GFP* mice using a standard fibroblast immortalization protocol,39 and performed a gene targeting titration experiment with varying amounts of ZFN expression plasmids and donor plasmid (Figure 4a). As in the murine ES cells, we observed the maximal rate of targeting when the transfection mix contained ~90% donor plasmid and ~10% ZFN expression plasmids. Gene targeting rates reached a maximum of 1.8 and 6.7% in heterozygous and homozygous lines, respectively. Vinblastine exposure did not affect targeting rates in these lines (data not shown).
Figure 4.
ZFN-mediated gene targeting in primary cells. (a) Gene targeting in homozygous and heterozygous ROSA-3T3s: transfections were performed using Lipofectamine 2000 with the indicated amounts of donor plasmid and ZFNs. The next day, media were changed, and on day 4, gene-targeting events were analyzed. (b,c) Gene targeting in MAF/MEFs: transfection of plasmids was performed by nucleofection using 2 µg of each ZFN and the indicated amounts of donor plasmid. Gene-targeting events were analyzed 4 days after transfection. (d) Gene targeting in astrocytes: targeting was performed in the same manner as MAFs/MEFs. In the upper left of each graph is a representative flow cytometry plot after targeting in which GFP fluorescence is measured in the y axis and background orange fluorescence along the x axis. The number in the left corner of the flow plot is the percentage of GFP+ cells. The corrected ROSA-3T3 cells show much higher GFP fluorescence than the primary cells demonstrating that although the ROSA26 locus is ubiquitously expressed, expression levels from the locus vary significantly depending on the cell type. Data are presented as mean ± SEM (*P < 0.05). ZFN, zinc finger nuclease.
Gene targeting in primary embryonic and adult fibroblasts
We also studied gene-targeting rates in fibroblasts derived from both embryonic (E13.5) and adult (3–6 months old) mice. In these experiments, we used nucleofection rather than Lipofectamine 2000 as the method of transfection and found that increasing the amount of donor plasmid while keeping the amount of ZFN expression plasmid constant increased the frequency of gene targeting (Figure 4b,c). The maximal gene-targeting rate in primary adult fibroblasts from ROSA26GFP*/GFP* mice was over 2% (Figure 4b). In murine embryonic fibroblasts, the maximal rate of gene targeting was ~1.8%. Interestingly, the targeting rate in murine embryonic fibroblasts from heterozygous mice was significantly different from fibroblasts derived from homozygous mice when low amounts of donor were transfected, but this difference disappeared when the highest amount of donor plasmid was transfected (Figure 4c). In summary, ZFN-mediated gene-targeting rates of ~2% can be achieved in primary fibroblasts of either adult or embryonic origin.
Gene targeting in primary astrocytes
The final primary somatic cell type that we examined for ZFN-mediated gene targeting was astrocytes. Using nucleofection with a constant amount of ZFN expression plasmid and increasing amounts of donor plasmid, we found that 0.03–0.17% of primary astrocytes could be targeted (Figure 4d). As in all of the other cell types examined, the rate of targeting increased as the amount of donor plasmid transfected increased. Overall, this rate of targeting is lower than what we found in primary fibroblasts. This difference may reflect an intrinsic difference between astrocytes and fibroblasts but may also reflect an underestimation of the rate of targeting in astrocytes because of decreased ability to detect targeted cells secondary to the low level of GFP expression from the ROSA26 promoter in astrocytes compared to ROSA-3T3s and primary fibroblasts (Figure 4a,c,d). As in the immortalized cells and primary fibroblasts, there was no effect of vinblastine exposure on the rate of targeting in primary astrocytes (data not shown).
Transplantation of gene-corrected primary mouse fibroblasts into an immunocompetent recipient
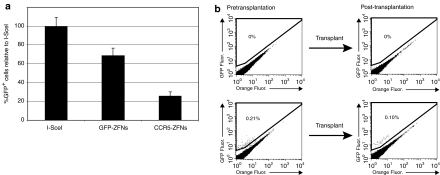
To perform cell-based gene correction therapy clinically, gene-corrected cells must be able to survive transplantation back into a recipient. A potential treatment for hemophilia is to modify fibroblasts ex vivo to secrete factor VIII or factor IX, followed by transplanting the cells back into recipients. This has already been performed with transient but significant clinical benefits observed in mice, rats, and human patients.40,41,42 As mentioned above, ZFNs may be toxic to cells and affect their transplantability. Although no toxic effects were readily apparent upon microscopic examination of treated fibroblasts, we further looked for toxic effects using a previously described toxicity assay that measures how well cells survive after expression of the ZFNs. When fibroblasts were subjected to this assay, a small degree of toxicity was measured (Figure 5a). This was minor in comparison with a previously published pair of ZFNs designed to target the CCR5 locus,33 and which exhibit noticeable toxic effects on cells. Both pairs of ZFNs contained the wild-type nuclease domain and were not modified to prevent homodimerization.35,43,44 To test whether our gene-corrected fibroblasts were capable of transplantation, we isolated primary adult fibroblasts and performed another round of gene targeting. On day 6 after nucleofection, we analyzed a portion of the total cell population and found that 0.21% of the total cells had undergone gene correction. We next took the remaining population (without selecting for GFP+ cells), embedded the cells in Matrigel, and transplanted the cells subcutaneously into an immunocompetent, isogenic mouse. Two weeks after transplantation, we excised the Matrigel plug, cultured the isolated cells for 6 days, analyzed the transplanted cells by flow cytometry, and found that 0.10% of the isolated cells were GFP+. The lower frequency of targeted cells may be due to contaminating host cells that were excised with the plug (Figure 5b). This study demonstrated that ex vivo ZFN gene-corrected fibroblasts could be successfully transplanted.
Figure 5.
Toxicity assay and transplantation of gene-targeted adult fibroblasts. (a) Toxicity of GFP-ZFNs in adult fibroblasts was measured using a fluorescence reporter assay. Cells were transfected with a tdTomato reporter along with an I-SceI, GFP-ZFN, or CCR5-ZFN expression plasmids. ZFN toxicity in cells is reported as fluorescence lost (due to cytotoxic effect) compared to cells transfected with the I-SceI expression plasmid. (b) For transplantation, adult fibroblasts underwent gene correction by nucleofection of 2 µg of each ZFN expression plasmid and 10 µg of donor plasmid. Gene targeting was measured by flow cytometry immediately before transplantation 6 days after nucleofection. Fibroblasts were then injected subcutaneously in a Matrigel matrix. Two weeks after transplantation, the Matrigel plug and surrounding skin were excised, cells dissociated, and plated to allow for fibroblast enrichment from other host-derived infiltrating cell types. On day 6, cells were harvested and analyzed using flow cytometry. ZFN, zinc finger nuclease.
Discussion
ZFNs have now been used in a wide variety of situations to create precise genome modifications.23 The precise genome modifications caused by ZFNs fall into two general classes. The first is to create small insertions/deletions at a specific locus by the mutagenic repair of a ZFN-induced double-strand break. Although the exact mutation cannot be controlled using this strategy, the precise location of the mutation is controlled by the specificity of the ZFN-induced double-strand break. This strategy is being increasingly used to create knockout cell lines and organisms in which the ability to efficiently create such knockouts was not previously available.24,25,26,27,29,30,45,46,47 In addition, this strategy has been used to create targeted mutations in the CCR5 gene in human T cells, thereby creating a population of T cells that are resistant to human immunodeficiency virus infection33—a strategy that has now entered a phase I clinical trial (ClinicalTrials.gov identifier NCT00842634). Theoretically, this approach could also be used to treat dominant genetic diseases by selectively mutating the dominantly acting allele. The second way to create precise genome modifications (gene targeting) is to use the ZFNs to create a gene-specific double-strand break and then have the cell repair that break by homologous recombination using an introduced donor sequence as the template for repair. Using this strategy, both the specific site of the genome change and the specific sequence change can be controlled. In addition to the double-precision of gene targeting, it also has the advantage that one can precisely create both small (single nucleotide) or large (the insertion of full transgene cassette) changes in the genome36,48 and reviewed in ref. 8. In addition to being able to create inactivating mutations as the first strategy does, this strategy also allows the controlled integration of a transgene or the direct correction of a disease-causing mutation and could theoretically be used to treat recessive genetic diseases. ZFN-mediated gene targeting has now been used in a wide variety of different cell types at a large number of different loci. These include the modification of the IL2RG gene in various cell lines,28 the modification of the CCR5 gene in a variety of cell lines including primary human ES cells and hematopoietic stem cells,33 the correction of GFP reporter genes and the modification of the PIG-A gene in human ES (hES) and human iPS cells,32 and the modification of the OCT4 (POU5F1) and AAVS1 locus in hES and iPS cells.31
As part of the effort to translate the use of ZFN-mediated gene targeting to clinical use, we have generated a mouse model of a generic recessive disease by knocking-in a mutated GFP gene into the murine ROSA26 locus. Our goal is to generate a model in which one can either test the ability to use ZFN-mediated gene targeting to correct mutations directly in vivo (similar to work performed by Miller et al. to assess AAV gene targeting49), or to mimic a paradigm in which patient cells are purified and then precisely modified ex vivo before transplanting the modified cells back into the patient. Here, we report a first step in establishing the ex vivo cell modification paradigm by demonstrating that primary embryonic fibroblasts, primary adult fibroblasts, and primary astrocytes can all be isolated from the transgenic mouse line and the mutation in the GFP gene efficiently corrected using ZFN-mediated gene targeting ex vivo. The rates of correction in primary fibroblasts (~2%) are of a sufficient magnitude to suggest clinical utility in the appropriate situation. Moreover, the gene-corrected adult fibroblasts could be successfully transplanted back into an immunocompetent mouse where they both survived and retained their corrected phenotype. Unlike previous studies where virally modified fibroblasts were transplanted back into a recipient mouse, gene correction allows for the corrected gene to be driven by its own promoter, thus preventing the silencing that can occur with transgenes that are driven by viral elements.
The use of gene targeting by homologous recombination without the use of nucleases in murine ES cells is a well-established procedure. Here, we demonstrate that ZFNs can stimulate gene targeting in >1% of cells without selection that is an order of magnitude or more than the rate of targeting in murine ES cells using I-SceI,11 or in human ES or iPS cells.31,32 Thus, this efficiency is high enough that one might be able to use ZFNs to create ES cells with extremely precise genetic modifications without using selectable markers.
This work also highlights an alternative strategy to creating gene-corrected iPS cells. In prior work, investigators have first converted fibroblasts into iPS cells and then used gene targeting by homologous recombination in the iPS cells to correct a mutation.50 The relatively high rates of gene correction stimulated by ZFNs in primary fibroblasts that we demonstrate here suggests that one could first correct disease-causing mutations in patient fibroblasts, and subsequently convert those gene-corrected fibroblasts into iPS cells. The advantages and disadvantages of whether to correct disease-causing mutations at the primary cell stage or after conversion to iPS cells will likely be assessed in future studies.
This work shows that in primary cells, just as in cell lines, that the best gene-targeting rates are obtained when the optimal mixture of ZFN expression plasmids and donor plasmid are introduced. Our finding that optimal targeting rates are achieved when the donor plasmid is at least tenfold more abundant than the ZFN expression plasmid suggests that keeping the two elements separate so that each can be introduced at their optimal amounts will be important in achieving optimal targeting rates in different cell types.
In summary, we have developed a mouse model of a generic recessive disease in which ZFN-mediated gene targeting can be studied in any cell of the mouse. A particular advantage of this model is that correctly targeted cells can easily be quantified and isolated for subsequent experiments using flow cytometry. In this article, we demonstrated gene targeting and quantified the rates within several cell types from this mouse model and that targeted adult fibroblasts, a potentially clinically relevant cell type, can be transplanted back into a mouse. Our current work focuses on isolating stem cells (such as hematopoietic stem cells, mesenchymal stem cells, muscle progenitor cells, and adipocyte precursor cells) from the mouse followed by performing gene targeting and subsequently transplanting the targeted stem cells back into a mouse, thus extending our current results of correcting and then transplanting a somatic cell. This study represents an important step in developing a paradigm for gene correction–based gene therapy in an animal model prior to human clinical trials.
Materials and Methods
Generation of ROSA26-GFP* targeting construct. We constructed the ROSA26-GFP* targeting construct by first destroying the XhoI and XbaI sites present in the insert of the published GFP* reporter gene. This was done by digesting GFP* with XhoI followed by blunting of ends with Klenow. Next, the XbaI site was destroyed in the same manner creating GFP*-B. We then used PCR to fuse an XhoI-XbaI-ClaI linker to GFP* using the follower primers: GFP*-XhoI-XbaI-ClaI-forward: 5′-GTCTCGAGTCTAGAATCGATATGGTGAGCAAGGGCGAGG-3′ and GFP*-XhoI-reverse: 5′-GACTCGAGTTACTTGTACAGCTCGTCCATGCCG-3′. The vector GFP*-B-Pgk-Neo was created by ligating GFP*-B into the XhoI site of Pgk-Neo-Pgk-PA, giving GFP*-B-Neo. Next, a splice acceptor with ClaI ends was generated by PCR of pSAβGeo with primers SAClaI-forward: 5′-GGATCGATATCTGTAGGGCGCAGTAGTCCAG-3′ and SAClaI-reverse: 5′-GTATCGATACCGTCGATCCCCACTG-3′. We digested the PCR product with ClaI and ligated it into the ClaI site of GFP*-B-Neo, generating SA-GFP*-B-Neo. Finally, this vector was digested with XbaI, and ligated into the XbaI site of the ROSA26-1-targeting vector. All restriction enzymes were ordered from New England Biolabs, Ipswich, MA. The vectors pSAβGeo and pROSA26-1 were generous gifts from Philippe Soriano (Mount Sinai, New York, NY).
Generation of reporter mice. All experiments involving mice were approved by the IACUC at the University of Texas Southwestern Medical Center. Targeting of the GFP* reporter construct to the ROSA26 locus was performed by the UT Southwestern Transgenic Core Facility. Briefly, 50 µg of targeting vector was linearized with KpnI and electroporated into 1 × 107 129/SvEvTac (SM-1) ES cells. Selection for targeted clones was performed using G418 at 250 µg/ml. Resistant clones were picked and screened using Southern analysis (Figure 1). Correctly targeted ES cells were injected into pseudopregnant C57/Bl6 females. Chimeric offspring were bred to identify mice transmitting the ROSA26-GFP* allele to their progeny.
Gene targeting in ES cells. ES cells were cultured in ESLX media consisting of ES-DMEM, 20% ES qualified FBS, 1× nonessential amino acids, 1× nucleosides, 1,000 U/ml ESGRO LIF, all purchased from Chemicon (Billerica, MA), 2 mmol/l L-glutamine, 1× Pen/Strep (Invitrogen, Carlsbad, CA), and 0.12 mg/ml sodium pyruvate and 0.1 mmol/l BME (Sigma-Aldrich, St Louis, MO). Before reaching confluency, cells were harvested and plated 100,000 cells per well in a 24-well gelatinized plate with and without 100 nmol/l vinblastine. We then transfected ES cells using Lipofectamine 2000 (Invitrogen) with the indicated amount of donor and ZFN plasmids. Two sets of ZFNs designed to target GFP were compared. Both sets bind to the same recognition site; however, their amino acid sequences differ (Table 1). Fifteen hours after transfection, media were changed to ESLX without vinblastine. Gene-targeting events were analyzed 4 days after transfection using a FACSCalibur (Becton Dickinson, Franklin Lakes, NJ).
Gene targeting and teratoma formation in ES cells. Gene-targeting experiments in ES cells were performed by nucleofection using the Mouse ES cell kit (cat. no. VPH-1001; Lonza, Basel, Switzerland). Briefly, 1.5 × 106 cells were nucleofected with 1.8 µg donor substrate and 1.6 µg of each GFP-ZFN using program A-30, giving an initial targeting rate of 0.07%. Cells underwent two rounds of sorting for targeted GFP+ cells. For teratoma analysis, nude mice were sublethally irradiated with 350 rads using a cesium-137 source. The following day, mice received subcutaneous injects in the hind flank with 1.3 × 106 ES cells suspended in phosphate-buffered saline. Teratomas were harvested 16–20 days after injections, photographed for GFP and Cy3 fluorescence using a Zeiss Stemi-11 stereoscope (Carl Zeiss, Thornwood, NY) equipped with an epifluorescence illuminator and Optronics Macrofire CCD camera (Optronics, Goleta, CA). Teratomas were subsequently fixed in 4% paraformaldehyde for 48 hours and paraffin processed for histopathologic analysis. Resulting H&E stains were photographed on a Leica DM2000 upright microscope (Leica Microsystems, Bannockburn, IL) with standard bright-field optics and Optronics Microfire CCD camera.
Gene targeting in primary cells
Mouse embryonic fibroblasts: Mouse embryonic fibroblasts were isolated from E13.5 embryos using the WiCell protocol (WiCell Research Institute, Madison, WI). Cells were cultured in DMEM, 10% FBS, Pen/Strep, and L-Glut. Before senescence occurred, cells were harvested and nucleofected in triplicate using the Basic Fibroblast kit (cat. no. VPI-1002; Lonza) with program U-23. Each nucleofection consisted of 4 × 105 cells plus 2 µg of each GFP-ZFN, and 1, 5, 10, or 20 µg of donor plasmid. We analyzed gene-targeting events on day 4 after transfection.
Mouse adult fibroblasts: Mouse adult fibroblasts were isolated from the ears of 3- to 6-month-old mice and cultured in DMEM, 20% FBS, Pen/Strep, L-Glut, Fungizone, and 1× nonessential amino acids. 5 × 105 cells per sample were nucleofected per sample using the basic fibroblast kit (cat. no. VPI-1002) with program U-23 and analysis performed on day 4 after transfection.
Astrocytes: Astrocytes were isolated from newborn mice and cultured in the same media used for MAFs. Nucleofections were performed on 5 × 105 cells per sample using the mouse astrocyte kit (cat. no. VPG-1006; Lonza) with program T-20.
Transplantation of gene-targeted primary adult fibroblasts and cell survival assay. Toxicity of GFP-ZFNs was measured as previously described.35 Briefly, 1 × 106 primary fibroblasts were nucleofected with 4 µg of a tdTomato expression plasmid along with 4 µg of an I-SceI expression plasmid or 2 µg of each GFP-ZFN, and replated in a 12-well plate. On day 2, a portion of the cells was analyzed using flow cytometry and the remainder of cells replated in a 12-well plate. On day 6, cells were harvested and analyzed for tdTomato expression using flow cytometry. Toxicity compared to I-SceI was determined by first calculating the change in GFP expression from day 6 to day 2 of the I-SceI-transfected cells and then the GFP-ZFN-transfected cells (ΔGFPnuclease = %GFP+day6/%GFP+day2). Toxicity compared to I-SceI was then calculated as Toxicity = ΔGFPZFNs/ΔGFPI-SceI. Analysis of toxicity for ZFNs targeting the CCR5 gene “CCR5 ZFNs” was performed in parallel using 2 µg of each CCR5 ZFN.
For transplantation experiments, fibroblasts underwent gene targeting by nucleofection as described above, using 2 µg of each ZFN expression plasmid and 10 µg of donor plasmid. Cells were analyzed by flow cytometry 6 days after nucleofection, immediately before transplantation. 9.3 × 105 fibroblasts, of which 0.21% were GFP+, were injected subcutaneously in a Matrigel (BD Biosciences, San Jose, CA) matrix in the back of a wild-type, immunocompetent mouse not containing our ROSA26-GFP* transgene. The Matrigel plug and surrounding tissue were excised 2 weeks later. Cells were dissociated by incubation in collagenase/dispase (25 mg/ml) (Roche Diagnostics, Indianapolis, IN) for 1 hour at 37 °C. One milliliter MAF media was then added and cells incubated overnight at 37 °C. The next morning, cells were triturated, filtered with a 70 µmol/l cell strainer (BD Biosciences) and plated in 24-well plates. On day 6, cells were harvested and analyzed using flow cytometry.
Acknowledgments
We thank Robert Hammer and Robin Nguyen at the UT Southwestern Transgenic Core for creation of the ROSA26 knock-in ES cells, derivation of the transgenic mouse and ES cell culture protocols. We also thank James Richardson and John Shelton at the JAR Molecular Pathology Core for help with photography and performing histological analysis of the teratomas. We thank Matthew Foglia for his careful reading and editing of the manuscript and Robert Bachoo for astrocyte isolation instruction. Finally, we thank Philippe Soriano for the pROSA26-1 and pSAβGeo vectors. The work in the Porteus Lab is supported by National Institutes of Health (NIH) grants PN2EY018244 (a Nanomedicine Development Center grant as part of the Director's initiative), R01 HL079295, and K08 H1070268; a career development award from the Burroughs Wellcome Fund, and State of Texas funding through the University of Texas Southwestern Medical Center. J.C.B. was supported by the UTSW Medical Scientist Training Program. S.P.-M. was supported by a National Institutes of Health pharmacology training grant at the University of Texas Southwestern Medical Center.
REFERENCES
- Hacein-Bey-Abina S, Le Deist F, Carlier F, Bouneaud C, Hue C, De Villartay JP, et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N Engl J Med. 2002;346:1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- Aiuti A, Cattaneo F, Galimberti S, Benninghoff U, Cassani B, Callegaro L, et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N Engl J Med. 2009;360:447–458. doi: 10.1056/NEJMoa0805817. [DOI] [PubMed] [Google Scholar]
- Bainbridge JW, Smith AJ, Barker SS, Robbie S, Henderson R, Balaggan K, et al. Effect of gene therapy on visual function in Leber's congenital amaurosis. N Engl J Med. 2008;358:2231–2239. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- Maguire AM, High KA, Auricchio A, Wright JF, Pierce EA, Testa F, et al. Age-dependent effects of RPE65 gene therapy for Leber's congenital amaurosis: a phase 1 dose-escalation trial. Lancet. 2009;374:1597–1605. doi: 10.1016/S0140-6736(09)61836-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- Porteus MH., and , Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- Doetschman T, Gregg RG, Maeda N, Hooper ML, Melton DW, Thompson S, et al. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature. 1987;330:576–578. doi: 10.1038/330576a0. [DOI] [PubMed] [Google Scholar]
- Porteus MH., and , Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- Donoho G, Jasin M., and , Berg P. Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cells. Mol Cell Biol. 1998;18:4070–4078. doi: 10.1128/mcb.18.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet P, Smih F., and , Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choulika A, Perrin A, Dujon B., and , Nicolas JF. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenneman M, Gimble FS., and , Wilson JH. Stimulation of intrachromosomal homologous recombination in human cells by electroporation with site-specific endonucleases. Proc Natl Acad Sci USA. 1996;93:3608–3612. doi: 10.1073/pnas.93.8.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnould S, Chames P, Perez C, Lacroix E, Duclert A, Epinat JC, et al. Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J Mol Biol. 2006;355:443–458. doi: 10.1016/j.jmb.2005.10.065. [DOI] [PubMed] [Google Scholar]
- Pâques F., and , Duchateau P. Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr Gene Ther. 2007;7:49–66. doi: 10.2174/156652307779940216. [DOI] [PubMed] [Google Scholar]
- Ashworth J, Havranek JJ, Duarte CM, Sussman D, Monnat RJ, Jr, Stoddard BL, et al. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durai S, Mani M, Kandavelou K, Wu J, Porteus MH., and , Chandrasegaran S. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005;33:5978–5990. doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Bibikova M, Whitby FG, Reddy AR, Chandrasegaran S., and , Carroll D. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000;28:3361–3369. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitinaite J, Wah DA, Aggarwal AK., and , Schildkraut I. FokI dimerization is required for DNA cleavage. Proc Natl Acad Sci USA. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathomen T., and , Joung JK. Zinc-finger nucleases: the next generation emerges. Mol Ther. 2008;16:1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Beumer K, Trautman JK., and , Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- Carroll D. Progress and prospects: zinc-finger nucleases as gene therapy agents. Gene Ther. 2008;15:1463–1468. doi: 10.1038/gt.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Noyes MB, Zhu LJ, Lawson ND., and , Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton J, Davis MW, Jorgensen EM., and , Carroll D. Induction and repair of zinc-finger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells. Proc Natl Acad Sci USA. 2006;103:16370–16375. doi: 10.1073/pnas.0605633103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, et al. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature. 2009;459:437–441. doi: 10.1038/nature07992. [DOI] [PubMed] [Google Scholar]
- Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, et al. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature. 2009;459:442–445. doi: 10.1038/nature07845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrowicz BP, Imamoto A, Fiering S, Herzenberg LA, Kerr WG., and , Soriano P. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc Natl Acad Sci USA. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruett-Miller SM, Connelly JP, Maeder ML, Joung JK., and , Porteus MH. Comparison of zinc finger nucleases for use in gene targeting in mammalian cells. Mol Ther. 2008;16:707–717. doi: 10.1038/mt.2008.20. [DOI] [PubMed] [Google Scholar]
- Porteus MH. Mammalian gene targeting with designed zinc finger nucleases. Mol Ther. 2006;13:438–446. doi: 10.1016/j.ymthe.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts PR, Porteus MH., and , Yu H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 2006;25:3377–3388. doi: 10.1038/sj.emboj.7601218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro GJ., and , Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Lu D, Zhou J, Wang J, Yang J, Meng P, et al. Implantation of autologous skin fibroblast genetically modified to secrete clotting factor IX partially corrects the hemorrhagic tendencies in two hemophilia B patients. Chin Med J. 1996;109:832–839. [PubMed] [Google Scholar]
- Roth DA, Tawa NE, Jr, O'Brien JM, Treco DA., and , Selden RF, Factor VIII Transkaryotic Therapy Study Group Nonviral transfer of the gene encoding coagulation factor VIII in patients with severe hemophilia A. N Engl J Med. 2001;344:1735–1742. doi: 10.1056/NEJM200106073442301. [DOI] [PubMed] [Google Scholar]
- Palmer TD, Thompson AR., and , Miller AD. Production of human factor IX in animals by genetically modified skin fibroblasts: potential therapy for hemophilia B. Blood. 1989;73:438–445. [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- Szczepek M, Brondani V, Büchel J, Serrano L, Segal DJ., and , Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Golic M, Golic KG., and , Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A, Plaisier CL, Carroll D., and , Drews GN. Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc Natl Acad Sci USA. 2005;102:2232–2237. doi: 10.1073/pnas.0409339102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci USA. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle EA, Moehle EA, Rock JM, Rock JM, Lee YL, Lee YL, et al. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci USA. 2007;104:3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DG, Wang PR, Petek LM, Hirata RK, Sands MS., and , Russell DW. Gene targeting in vivo by adeno-associated virus vectors. Nat Biotechnol. 2006;24:1022–1026. doi: 10.1038/nbt1231. [DOI] [PubMed] [Google Scholar]
- Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]