Abstract
Background and Aims
The angiosperm family Myrtaceae comprises 17 tribes with more than half of the estimated 5500 species being referred to the fleshy-fruited and predominantly rainforest associated Syzygieae and Myrteae. Previous studies suggest that fleshy fruits have evolved separately in these lineages, whereas generally shifts in fruit morphology have been variously implicated in diversification rate shifts among angiosperms. A phylogenetic hypothesis and estimate divergence times for Myrtaceae is developed as a basis to explore the evidence for, and drivers of, elevated diversification rates among the fleshy-fruited tribes of Myrtaceae.
Methods
Bayesian phylogenetic analyses of plastid and nuclear DNA sequences were used to estimate intertribal relationships and lineage divergence times in Myrtaceae. Focusing on the fleshy-fruited tribes, a variety of statistical approaches were used to assess diversification rates and diversification rate shifts across the family.
Key Results
Analyses of the sequence data provide a strongly supported phylogenetic hypothesis for Myrtaceae. Relative to previous studies, substantially younger ages for many of the clades are reported, and it is argued that the use of flexible calibrations to incorporate fossil data provides more realistic divergence estimates than the use of errorless point calibrations. It is found that Syzygieae and Myrteae have experienced elevated diversification rates relative to other lineages of Myrtaceae. Positive shifts in diversification rate have occurred separately in each lineage, associated with a shift from dry to fleshy fruit.
Conclusions
Fleshy fruits have evolved independently in Syzygieae and Myrteae, and this is accompanied by exceptional diversification rate shifts in both instances, suggesting that the evolution of fleshy fruits is a key innovation for rainforest Myrtaceae. Noting the scale dependency of this hypothesis, more complex explanations may be required to explain diversification rate shifts occurring within the fleshy-fruited tribes, and the suggested phylogenetic hypothesis provides an appropriate framework for this undertaking.
Keywords: Myrtaceae, Myrtoideae, Myrteae, Syzygieae, phylogeny, molecular dating, speciation, diversification rates
INTRODUCTION
A central question in evolutionary biology is the nature of processes that lead to accelerated rates of speciation relative to extinction, and species-rich groups provide some of the clearest examples of this phenomenon. Such groups are often significant in terms of taxonomic diversity, relative abundance and contribution to total biomass over broad geographic regions and can provide good model systems for interpreting the origins and maintenance of biotic diversity at the biome scale (Richardson et al., 2001; Ladiges et al., 2003; Crisp et al., 2004; Erkens et al., 2007). Recent methodological developments (e.g. Sanderson, 2002; Drummond and Rambaut, 2007) have seen the increasing availability of time-calibrated molecular phylogenetic trees, which provide a framework for evaluating the timing of, and correlations with, phenomena that have impacted on rates of lineage accumulation among clades (Moore and Donoghue, 2007; Rabosky et al., 2007). The recurrence of similar phenotypes in separate lineages provides an opportunity to assess the significance of the evolution of a trait, or traits, on diversification (Donoghue, 2005).
Myrtaceae are a moderate sized (approx. 5500 species in 140 genera), predominantly southern hemisphere family with a postulated origin in the Cretaceous (Briggs and Johnson, 1979; Wilson et al., 2001; Ladiges et al., 2003; Sytsma et al., 2004). A remarkable aspect is the ubiquity of the family within Australasia, and groups such as Eucalyptus s.l. have been considered a model for understanding the radiation of the Australian sclerophyll flora in response to Miocene–Pliocene aridification (e.g. Crisp et al., 2004; Ladiges et al., 2003). Myrtaceae are also an important component of humid tropical forests, including the tribe Myrteae (sensu Wilson et al., 2005; including approx. 2500 species), which is pantropical, although particularly well developed in Central and South America (McVaugh, 1968; Landrum and Kawasaki, 1997; Lucas et al., 2007), and Syzygieae (approx. 1000–1500 species; Parnell et al., 2006), which is widely distributed in the humid Palaeotropics, but with species richness and lineage diversity centred on the Australasian region (Craven, 2001). Traditionally, Syzygieae and Myrteae were included in Myrtaceae subfamily Myrtoideae, due to the shared possession of a succulent pericarp. Evidence from morphology (Schmid, 1972; Johnson and Briggs, 1984) and molecular phylogenetic studies (Wilson et al., 2001, 2005; Sytsma et al., 2004) suggests that the fleshy fruit of Myrtaceae has multiple origins, arising separately within Syzygieae, Myrteae and elsewhere in the family.
The relationships in Myrtaceae have been the focus of several recent studies (e.g. Johnson and Briggs, 1984; Gadek et al., 1996; Wilson et al., 2001, 2005; Sytsma et al., 2004). Wilson et al. (2001, 2005) presented an hypothesis, based upon plastid trnK-matK sequence data that forms the basis for the modern tribal classification of Myrtaceae (Wilson et al., 2005) whereas Sytsma et al. (2004) analysed a comparable family-wide taxon sample but included plastid matK and ndhF sequences. A limitation of the existing molecular-based hypotheses is the generally poor resolution of relationships among the tribes. Sytsma et al. (2004) used three fossil constraints, and Penalised Likelihood rate smoothing (Sanderson, 2002) to estimate divergence times among the lineages of Myrtaceae. Their approach employed fossil dates as fixed points and does not consider uncertainty, whereas for the same calibrations, Rutschmann et al. (2007) noted potentially large errors in divergence time estimates associated with alternative nodal placements (i.e. crown versus stem node) for the fossil dates. Recently developed Bayesian relaxed clock (BRC) methods allow calibration information to be incorporated in the form of parametric prior probability distributions (Yang and Rannala, 2006; Drummond et al., 2006) that, in contrast to point calibrations, can be designed to incorporate uncertainty associated with paleontological data (Yang and Rannala, 2006; Sanders and Lee, 2007).
Here, a reassessment of relationships within Myrtaceae are provided, based upon the Bayesian phylogenetic analysis of nuclear ribosomal internal transcribed spacer (ITS) sequences and plastid matK and ndhF sequences specifically aimed at improving resolution of relationships among the tribes. Using BRC methods and five fossil calibrations, lineage divergence times for the family were estimated. With this framework in place, the focus was on tribes Syzygieae and Myrteae. Using taxonomic and phylogenetic data, the hypothesis was tested that high species richness in either or both of these groups can be related to unusually high rates of lineage diversification and, specifically, that the recurrence of similar phenotypes in Syzygieae and Myrteae provides an opportunity to consider how shifts from dry to fleshy fruits has impacted on diversification rates among arborescent rainforest lineages.
METHODS
Sequence data
Sequences of the 18S–26S rDNA ITS and plastid regions matK and ndhF were assembled for 91, 96 and 84 taxa, respectively, Wherever possible, sequences for each of the three regions were sourced from a single accession, although in some cases the ITS data are from a different accession of the same taxon or a congeneric taxon (Appendix). Taxon sampling was designed to reflect the classification proposed by Wilson et al. (2005), including representatives of each of their 17 tribal groupings and a representative of the New Caledonian genus Cloezia, which was included but not placed by Wilson et al. (2005). The outgroup comprised representatives of Vochysiaceae, which in previous phylogenetic studies of both Myrtaceae and Myrtales, have been resolved (although with varying support) as sister to Myrtaceae s.l. (Gadek et al., 1996; Conti et al., 1997; Wilson et al., 2001, 2005; Sytsma et al., 2004; Rutschmann et al., 2007) but due to difficulties aligning the ITS regions, only plastid data for Vochysiaceae were included in analyses. Sequences were sourced from GenBank, or, for novel sequences (Appendix), primers, PCR and sequencing conditions were as outlined in Biffin et al. (2006), Harrington and Gadek (2004) and Lucas et al. (2007). The ITS alignment was performed using the structural partitioning scheme described by Biffin et al. (2007), which divides the spacers into ‘stem’ and ‘loop’ partitions based upon Minimum Free Energy predictions of putative RNA secondary structures. The plastid data were manually aligned.
Phylogenetic analysis
First, the nuclear and plastid data were analysed separately using MrBayes v. 3·1·2 (Ronquist and Huelsenbeck, 2003). For the ITS, the ‘doublet’ nucleotide substitution model with an HKY85-like rate matrix and gamma-distributed rate variation with an estimated proportion of invariant sites was applied to the stem-paired data. For the ITS ‘loop’ partition, and the matK and ndhF data, a General Time Reversible (GTR) + I + Γ substitution model was used, and each of the concatenated ITS and plastid data sets was analysed with model parameters estimated separately for each data partition (for details of model selection, refer to Biffin et al., 2007). All analyses were performed with uninformative priors on model parameters, and two independent runs (each with four chains, one cold, three heated) of 2 × 106 generations sampling every hundredth generation. Convergence between independent runs and the appropriate burn-in fraction were determined using the post-run ‘sump’ command in MrBayes and by analysing the output in Tracer v. 1·4 (Rambaut and Drummond, 2004).
A visual comparison of the topologies estimated for the ITS and plastid data was performed to identify strongly supported nodes [posterior probability (PP) ≥ 0·95] with conflicting resolutions amongst data sets. In subsequent analyses the separate data sets were concatenated and analysed in MrBayes with substitution model parameters estimated for each partition as above for the separate analyses of the nuclear and plastid data and analysis settings as previously described.
The concatenated data were also analysed using BRC methods as implemented in BEAST v. 1·4·7 (Drummond et al., 2006; Drummond and Rambaut, 2007) using a four-way partitioning scheme comprising ITS ‘stems’, ITS ‘loops’, matK and ndhF. A GTR + I + Γ substitution model was assumed with the model parameters unlinked across data partitions (note that BEAST v. 1·4·7 does not incorporate RNA-specific models). An uncorrelated log-normal model of rate variation among branches in the tree and a Yule prior on branching rates were used. Three independent MCMC runs were performed, each of 5 × 106 steps (sampling topology and parameter values every 250 steps) and Tracer was used to assess convergence between runs and estimate an appropriate burn-in proportion, estimate the mean and 95 % highest posterior density (HPD) of parameters sampled from the posterior distribution of the combined runs, and to ensure that the effective sample size was sufficient to provide reasonable estimates of model parameter variance (i.e. >200). After excluding an appropriate burn-in fraction (as described above), the topologies estimated from the three independent runs were combined and topology and parameter values were summarized (using TreeAnnotator; Drummond and Rambaut, 2007) on the ‘maximum credibility’ tree.
Molecular dating
Fossil dates were used to calibrate molecular evolutionary rates, including those derived from five myrtaceous fossils. In addition, the estimated age of the eudicots [approx. 121 million years ago (Ma) based upon the earliest appearance of tricolpate pollen in the fossil record; Drinnan et al., 1994; Magallón et al., 1999)] was used to constrain the upper age of the root. The Myrtaceae fossils include (a) the pollen taxon Myrtaceidites lisamae, which appears in the fossil record from the Cretaceous of Gabon (Santonian, 86 Ma; Herngreen, 1975; Boltenhagen, 1976; Muller, 1981), Borneo (Senonian, 89–83 Ma; Muller, 1968) and Colombia (Maastrichtian, 71–65 Ma; van der Hammen, 1954), and provides the earliest estimate for the radiation of Myrtaceae; (b) the eucalyptoid fruits from the Redbank Plains formation of eastern Australia, which are placed at 48 Ma (early Eocene; Rozenfelds, 1996), with postulated affinities to Eucalypteae; (c) Paleomyrtinaea princetonensis, from the Palaeocene (56 Ma; Crane et al., 1990; Pigg et al., 1993) to early Eocene (53 Ma; Manchester, 1999) of North America, comprising well-preserved fruits and seeds suggesting a relationship with guava (Psidium) and Mosiera (Pigg et al., 1993), or, more broadly, Myrtaceae subtribe Myrtineae (i.e. Myrteae genera with a pimentoid/myrtoid embryo; McVaugh, 1968; Landrum and Kawasaki, 1997); (d) the fossil leaves and fruits of Metrosideros from the Early Miocene (approx. 20 Ma) of New Zealand, which are considered to show close affinity to extant Metrosideros and are the stratigraphically oldest evidence of that genus in New Zealand (Pole et al., 2008); and (e) Tristaniandra alleyii from the Eocene of South Australia (41–46 Ma; Basinger et al., 2007), comprising flowers and fruits with characters that do not closely match an extant genus although the combination of flowers and fruit structures suggest an affinity to tribe Kanieae (Greenwood and Christophel, 2005; Basinger et al., 2007).
Lineage divergence times were estimated using BEAST v. 1·4·7, with model parameters and settings as outlined above. Where applicable, the fossils were used to provide a minimum age for the associated lineage by constraining the stem node (or the next deeper well-supported node) to be at least as old as the fossil-derived date. In these instances, the prior probability on the age of the node was assumed to follow a log-normal distribution with a ‘hard’ lower bound (i.e. there is a zero probability of dates much younger than the oldest known fossil assigned to that lineage) and a ‘soft’ upper bound (i.e. non-zero, but decreasing probability for dates that are older than the fossil constraint) on the age of the node (Sanders and Lee, 2007). Three fossils (Myrtaceidites, Paleomyrtinaea and the ‘eucalyptoid’ material) were used as minimum age constraints for the stem node of each corresponding lineage (Table 1 and Fig. 3). These calibrations were designed such that the ‘hard’ minimum node age (zero offset) was approx. 20 % younger than the age of the fossil, the lower limit of the 95 % confidence interval (CI) of the prior distribution approximates the fossil age, whereas for Paleomyrtinaea and the ‘eucalyptoid’ material, the peak probability was approx. 1·5 times the age of the fossil, allowing the possibility that the node is substantially older than the fossil constraint. A narrower prior probability distribution was used for Myrtacedeites, given that age estimates for the Myrtales crown node are generally younger than 121 Ma, i.e. the upper root constraint (e.g. Magallón and Sanderson, 2001; Sytsma et al., 2004; Davies et al., 2004). Therefore, the upper 95 % CI of the prior probability distribution corresponds to an age of approx. 100 Ma (Table 1). Using the stem node to calibrate molecular rates provides an objective criterion for fossil assignment where there is uncertainty in the correct nodal placement (Renner, 2005; for discussion of Myrtaceae fossils, see Rutschmann et al., 2007).
Table 1.
Calibrations used for the molecular dating analyses of Myrtaceae
| Calibration | Prior distribution | Prior (mean [95 % CI]) | Joint prior (mean [95 % CI]) | Posterior (mean [95 % HPD]) | |
|---|---|---|---|---|---|
| 1. Myrtaceae stem | Myrtacedietes86 Ma | Log-normal | 92 (86–100) | 93 (86–101) | 94 (87–102) |
| Myrtaceae | 87 (75–100) | 86 (74–96) | |||
| 2. Eucalypteae stem | Eucalyptus 48 Ma | Log-normal | 65 (52–86) | 58 (45–73) | 60 (53–68) |
| Eucalypteae | 36 (15–57) | 40 (28–50) | |||
| 3. Myrteae stem (BKMMST clade) | Paleomyrtineae56 Ma | Log-normal | 67 (55–89) | 69 (55–84) | 59 (53–67) |
| Myrteae | 50 (30–68) | 34 (25–43) | |||
| 4. Metrosidereae | Metrosideros20 Ma | Normal | 20 (10–31) | 20 (11–33) | 20 (13–28) |
| 5. Kanieae | Tristaniandra45 Ma | Normal | 45 (30–52) | 34 (21–48) | 39 (28–50) |
For each fossil, the calibration node is highlighted in bold, and alternative, reasonable fossil placements are indicated. The prior probability distribution for the constrained node, and the joint prior and posterior probability distributions of the constrained node and alternative fossil placements are indicated (millions of years). Numbering corresponds to node numbers in Fig. 3.
Fig. 3.
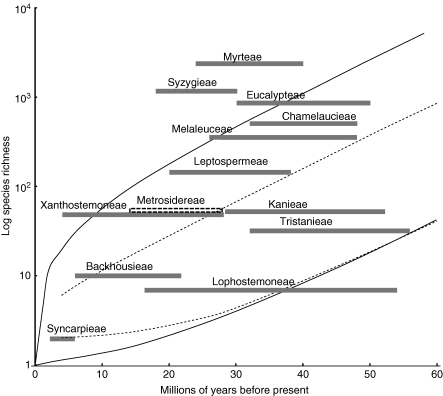
Confidence intervals of expected diversity (log scale) according to age of crown group, with the sets of lines representing the upper and lower limits of the 95 % CI of expected species numbers assuming a diversification rate of 0·084 (Myrtoideae median) and a relative extinction rate of 0 (broken lines) and 0·9 (continuous lines). Tribes of Myrtaceae are mapped according to estimated standing diversity and the age of the crown group (95 % HPD of divergence time estimates shown as horizontal bars).
For two fossils (Tristaniandra and Metrosideros), the associated lineages (Kanieae and Metrosidereae, respectively) are resolved within a well-supported polytomy that includes Myrteae (Fig. 3). The Paleomyrtinaea fossils provide the oldest known minimum age for that clade and the Tristaniandra and Metrosideros fossils were therefore used to constrain the crown group age of their associated lineage (Kanieae and Metrosidereae, respectively) using a normal prior probability distribution with the peak probability equivalent to the fossil age, and a broad CI with ‘soft’ lower and upper bounds allowing the possibility that the calibration node is older, or younger, than the fossil age (Table 1 and Fig. 3).
A specific concern with Bayesian statistical approaches is the influence of the priors on the posterior probabilities (e.g. Welch et al., 2005) and it is good practice to consider the distribution of the joint prior on posterior density estimates (Drummond et al., 2006). Here, it has been argued that a stem node placement is the most objective criterion for fossil nodal placement. Analyses were run (with calibrations and model settings as outlined above), sampling entirely from the prior, to assess the influence of the priors on the posterior estimate for alternative (more nested) fossil nodal placements and, also, to test whether the data are sufficiently informative to update the priors (Table 1).
Diversification rates of Myrtaceae
Based upon the phylogenetic data, species richness was estimated for clades of interest from the World Checklist of Myrtaceae (Govaerts et al., 2008). For taxa not included in the present analysis, the scheme of Wilson et al. (2005) was used to estimate the tribal affinities. Cloezia was not placed by Wilson et al. (2005), but in the present study it is resolved in a clade including Tristanieae (see Fig. 1).
Fig. 1.
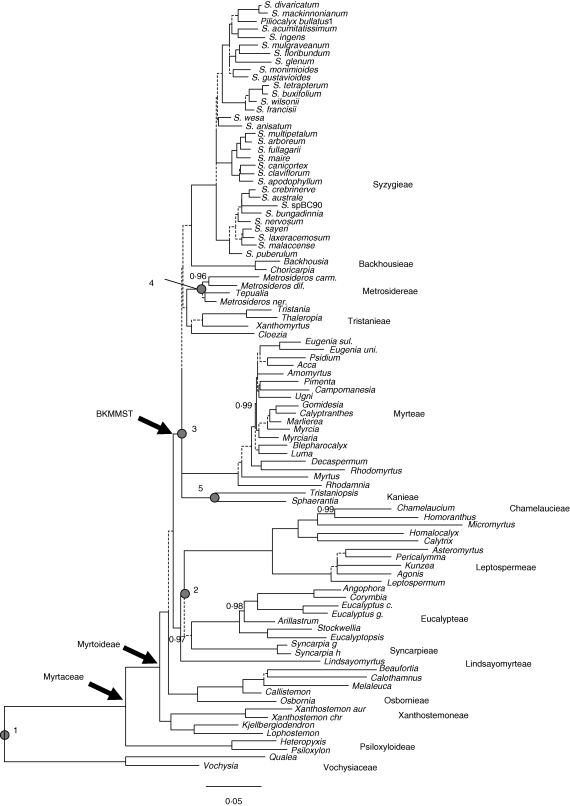
Relationships of Myrtaceae inferred from the Bayesian analysis of the concatenated ITS and plastid sequence data. Majority rule consensus topology, branch lengths proportional to the inferred number of changes along that branch. Dashed branches have a PP ≤ 0·95, PP values between 0·8 and 0·95 are indicated adjacent to the branch. Otherwise the PP = 1·0. Subfamilies and tribal groups follow Wilson et al. (2005), and the family Myrtaceae, subfamily Myrtoideae and BKMMST clade (this study) are indicated. Numbered nodes refer to fossil calibrations used for the molecular dating analyses: 1, Myrtaceae stem; 2, Eucalypteae stem; 3, Myrteae stem (BKMMST clade); 4, Metrosidereae; 5, Kanieae (refer to Table 1).
For selected clades, clade-specific diversification rates (i.e. the rate of lineage accumulation/unit time) were derived using the crown node method-of-moments estimators described by Magallón and Sanderson (2001) assuming a relative extinction rate of 0·9. For a given diversification rate, and relative extinction rate, Magallón and Sanderson (2001) describe a method by which it is possible to estimate a CI on the expected number of species included within a hypothetical crown group for each interval of time from its origin onwards. Following Magallón and Sanderson (2001), a 95 % CI (upper and lower boundary value within which 95 % of the results of the replicates of the stochastic process will fall) was calculated for intervals of 2 million years from time = 0 to time = 70 Ma, assuming the estimated diversification rate for subfamily Myrtoideae (sensu Wilson et al., 2005) and a relative extinction fraction of 0 and 0·9. Standing diversity for the tribes of Myrtoideae was then compared with these sets of critical values. The null hypothesis is that all lineages have diversified at a rate consistent with the overall Myrtoideae radiation. Standing diversities that exceed the upper or lower critical values can be considered unexpectedly species-rich or -poor, respectively, in the context of the Myrtoideae radiation. All calculations were performed using the R package GEIGER (Harmon et al., 2008).
Shifts in diversification rate within Myrtaceae were explored using the LASER package (Rabosky, 2006) for the R programming language, which implements the methods described by Sanderson and Wojciechowski (1996). Briefly, this approach uses phylogenetic (topology and branch length) and taxonomic (species richness) data first to test the null hypothesis that all lineages have diversified under a homogeneous rate and secondly, if a homogeneous rate is rejected, to identify the most likely node at which a diversification rate shift has occurred. Given an ultrametric topology (i.e. the BRC topologies; see Fig. 2) with species richness estimates for the terminals, LASER contrasts the likelihood of the data under a model that assumes that all lineages have diversified at a constant rate (one-rate model) with the likelihood of a model in which an ancestral diversification rate shifts at some point to a new diversification rate (flexible-rate model). The flexible-rate model estimates branch-specific diversification rates and the maximum likelihood (ML) shift point is the node with the highest combined likelihood determined by sequentially splitting the tree at each node and optimizing the diversification rate onto the two resulting subtrees (Rabosky et al., 2007).
Fig. 2.
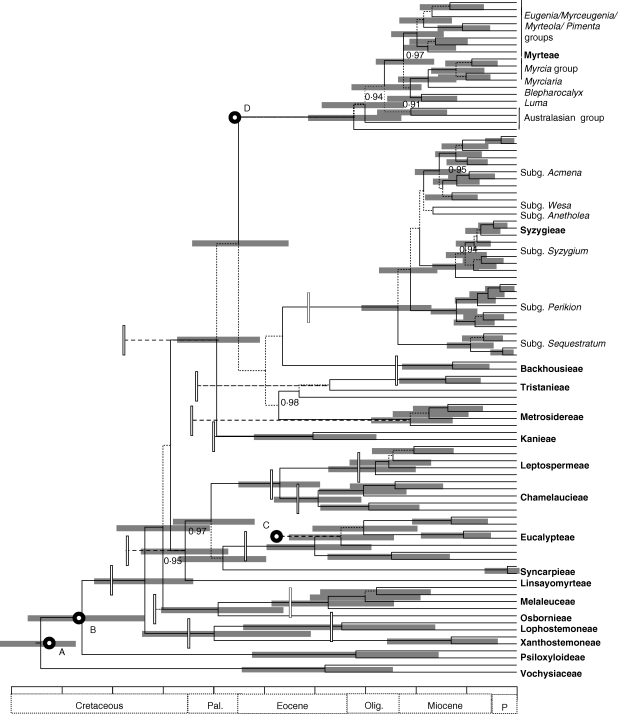
Maximum credibility chronogram inferred using BRC methods. The posterior probability of the branches is as in Fig. 1. Horizontal bars represent the 95 % HPD of divergence times for nodes receiving a PP ≥ 0·5. Vertical bars indicate the estimated divergence time for the equivalent node from Sytsma et al. (2004). Fossil calibrations from Sytsma et al. are also indicated (open circles: A, PHMV clade, 93 Ma; B, Myrtacedeites, 86 Ma; C, ‘eucalypt’ fruit, 48 Ma; D, Paleomyrtineae, 56 Ma). Groupings within Syzygieae and Myrteae are according to Craven and Biffin (2010) and Lucas et al. (2007), respectively. Each division in the scale bar is equal to 10 million years.
Tests for diversification rate shifts were conducted at the tribal level derived by pruning all but two taxa per tribe from ultrametric (BRC) topologies, and assigning half the estimated species richness per tribe to each terminal. To avoid conditioning results on a particular topology and branch lengths, trees were sampled from the 95 % HPD of the BRC analyses in proportion to the auto-correlation time (determined using Tracer v. 1·4), i.e. the number of steps (generations) between independent draws from the posterior probability distribution. Because taxon sampling density can influence divergence time estimates (Cook and Crisp, 2005; Linder et al., 2005), the effect of taxon pruning on branch lengths was assessed by summarizing divergence time estimates from the pooled sample of tribal level topologies for comparison with the estimates derived from the complete taxon sample.
Character evolution
Character states for ‘fruit-type’ (dry or fleshy pericarp) were scored from the literature for the complete taxon sample and a Bayesian approach was used to infer the ancestral states for fruit-type. BayesTraits V1·0 (Pagel and Meade, 2006) simultaneously accounts for phylogenetic uncertainty and uncertainty associated with the estimation of rate parameters upon alternative trees (Pagel and Lutzoni, 2002; Pagel et al., 2004).
Input trees were derived from the Bayesian ‘non-clock’ analyses of the combined concatenated Myrtaceae data, following ‘thinning’ to remove autocorrelated samples, as previously described (263 effectively independent topologies were retained). Prior to analyses, the ‘ratedev’ parameter, which controls the rate at which new states are accepted, was manually set so that the acceptance rate ranged between 20 and 50 % following the authors' recommendations. Priors of the rate parameters were estimated using a hyperprior approach (Pagel et al., 2004) with an exponential distribution, its mean seeded from a uniform distribution on the interval of 0–10·0. A reversible-jump MCMC method was used, in which the Markov chain searches the posterior distribution of different models of evolution and the posterior distributions of the parameters of these models (Pagel and Meade, 2006). Because not all of the trees necessarily contain the internal nodes of interest, reconstructions were performed using a ‘most recent common ancestor’ (MRCA) approach that identifies, for each tree, the MRCA to a group of species and reconstructs the state at the node, then combines this information across trees (Pagel et al., 2004). Three separate analyses were run over 107 generations. The ‘fossil’ command was used to contrast the level of support for each character state at a given node using Bayes factor comparisons (for an interpretation of Bayes factors, see Kass and Rafferty, 1995).
RESULTS
Phylogenetic relationships in Myrtaceae
The analyses of ITS (Fig. S1 in Supplementary data, available online) and the concatenated plastid data (Fig. S2 in Supplementary data) produced broadly consistent topologies, at least to the extent that well-supported nodes did not conflict among data sets, and consequently the data sets were analysed simultaneously. The 50 % majority-rule consensus topology inferred from the MrBayes analyses of the concatenated ITS and plastid data is shown in Fig. 1. The relationships inferred using BEAST, under a BRC and partitioned 4 × GTR + I + Γ substitution model, show little variation from the MrBayes consensus topology, with only minor differences in PP values among weakly supported nodes (Fig. 2).
Although the monophyly of Myrtaceae s.l. was not specifically assessed, the two subfamilies proposed by Wilson et al. (2005) (Psiloxyloideae and an expanded subfamily Myrtoideae) are resolved as monophyletic, as are their proposed tribal groupings within Myrtoideae (Wilson et al., 2005). The inferred relationships among the tribes are largely consistent with those reported by Sytsma et al. (2004) and Wilson et al. (2005), although relative to these, there is a higher level of confidence in some of the inter-tribal groupings. Examples include the grouping of Chamelaucieae, Eucalypteae, Leptospermeae and Syncarpieae (PP = 0·97) and a clade including Backhousieae, Kanieae, Metrosidereae, Myrteae, Syzygieae and Tristanieae (PP = 1·0) (BKMMST clade, Figs 1 and 2). Inter-tribal relationships within the BKMMST clade are generally poorly resolved, although a novel resolution is the inclusion of the New Caledonian-centred Cloezia within the BKMMST clade, and a strong association of Cloezia with Tristanieae (Figs 1 and 2).
Molecular dating
Figure 2 shows the maximum credibility topology derived from the three independent MCMC runs in BEAST, with median node heights and the 95 % HPD of divergence times illustrated. Analyses were performed without the sequence data (i.e. sampling from the prior) in order to examine the influence of the priors on the posterior probability of divergence time estimates. Table 1 compares the 95 % CI of prior probability distributions and 95 % HPD of posterior probability densities of the Myrtaceae calibrations, and where stem group nodes were used, the prior and posterior densities of the crown group age associated with each of the fossil calibrations. Note that in each instance, the prior distribution for the crown node includes the associated fossil age for that lineage, i.e. a crown node placement is not ruled out a priori. Comparison of the 95 % CI of the prior and 95 % HPD estimates suggests that the data are sufficiently informative to update the priors, and, for example, the prior mean for the Myrteae crown node is some 20 Ma older than the median value estimated from the posterior (Table 1).
Diversification rates and diversification rate shifts
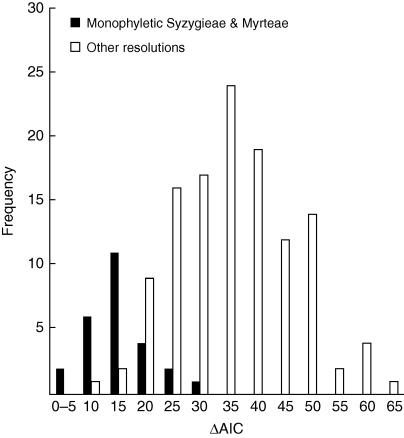
The hypothesis that the extant diversities of the tribes of Myrtaceae are not unexpected was tested under the assumption that the entire Myrtoideae radiation has diversified at a constant rate [0·084 net species/million years, assuming 5656 species, an age of 75 million years for the Myrtoideae crown (Fig. 2), and a relative extinction rate of 0·9]. Of the tribal lineages included in the comparisons (Fig. 3), only Syzygieae (1189 species) and Myrteae (2379 species) have standing diversities significantly exceeding the upper 95 % CI of the expected number of species given the assumptions used, regardless of the relative extinction fraction (e = 0 and 0·9) and for all estimates included within the 95 % HPD of divergence time estimates for that clade (P < 0·00015 and P < 0·0006, respectively, at e = 0·9, using the upper age estimate for the crown group included within the 95 % HPD of divergence times). In support of these findings, the analyses using LASER (repeated over 300 sampled tribal-level topologies; Fig. 4) strongly reject a homogeneous diversification rate for Myrtaceae in favour of a flexible-rate model (ΔAIC > 30·7; ΔLH < 2·91 × 10−8).
Fig. 4.
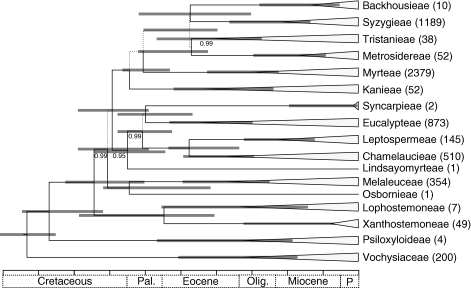
Consensus topology of 300 trees obtained from the BRC analyses (see Fig. 2) and pruned to represent tribal diversity in Myrtaceae. Diversity per tribe according to Govaerts et al. (2008) is indicated adjacent to the terminal. Clade support is as in Fig. 1. Vertical bars represent the range of divergence times obtained from the 300 sampled topologies for all nodes receiving a PP ≥ 0·5. Each division in the scale bar is equal to 10 million years.
There is some ambiguity in the location of the reconstructed ML shift point under the flexible-rate model, although a few generalizations can be made (Table 2). First, the ML shift point is associated with, or nested within, the BKMMST clade across all sampled topologies, and secondly, the clade associated with the shift always includes the tribes Syzygieae and Myrteae, or only Syzygieae or Myrteae. Generally, this ambiguity is a reflection of the poor resolution of relationships among the BKMMST tribes (Figs 1 and 2).
Table 2.
The distribution of reconstructed ML shift point under the flexible-rate model amongst nodes from 300 sampled tribal level topologies, the posterior probability of each node, and the proportion of topologies containing the ML shift point on that node (used to derive a corrected ML shift statistic by dividing the two values)
| Node* |
||||||||
|---|---|---|---|---|---|---|---|---|
| BKMeMST | BMeMST | MeMST | MeMS | SMT | SM | M | S | |
| Posterior probability | 1·0 | 0·61 | 0·02 | 0·02 | 0·03 | 0·18 | 1·0 | 1·0 |
| Proportion of topologies containing ML shift | 0·06 | 0·43 | 0·13 | 0·13 | 0·27 | 0·18 | 0·17 | 0·1 |
| Corrected ML shift | 0·06 | 0·71 | 0·66 | 0·66 | 0·89 | 1·0 | 0·17 | 0·1 |
* B, Backhousieae; K, Kanieae; Me, Metrosidereae; M, Myrteae; S, Syzygieae; T, Tristanieae.
In Table 2, the ML shift point is represented as the proportion of the 300 sampled topologies associated with that clade. Perhaps a more meaningful measure is a ‘corrected’ clade-specific ML shift-point proportion, derived by dividing the proportion of sampled topologies by the posterior probability of that clade, i.e. the proportion of topologies that contain that clade and also include the ML shift point at that node. For this value, only the topologies that include a monophyletic Syzygieae + Myrteae contain the ML shift point in 100 % of the sampled trees (Table 2). Figure 5 shows the differences in AIC (ΔAIC) values obtained by subtracting the AIC score for the reconstructed ML shift point on each of the included topologies with the best AIC score for a rate shift estimated from the overall sample. The best AIC (576·6) was associated with the MRCA of Syzygieae + Myrteae, and in Fig. 5, the ΔAIC scores for topologies with this resolution are highlighted for comparison with all other reconstructed ML shift points. In general, the lowest ΔAIC values are associated with the former and, for instance, the mean (± s.d.) AIC score for shifts on Syzygieae + Myrteae was 589·1 (6·2) versus a mean of 610·2 (10·7) for the AIC score derived from all alternative ML shift points.
Fig. 5.
Frequency distribution of AIC differences obtained by subtracting the AIC score for the ML shift point estimated under a flexible-rate diversification model on each of 300 sampled topologies (see Fig. 4) with the best AIC score from the overall sample. The black columns represent the ΔAIC for sampled topologies that resolve monophyletic Syzygieae and Myrteae while the white columns represent the ΔAIC for all other resolutions (see Table 2).
Character evolution
From our Bayesian reconstructions of ancestral fruit types, the probability of a single origin for the fleshy pericarp (i.e. the MRCA of Myrteae, Syzygieae and Tristanieae had fleshy fruit) is <0·19, and the probability that a fleshy pericarp has arisen twice within the BKMMST clade (i.e. once within Xanthomyrtus and once for the MRCA of Syzygieae and Myrteae) has a probability of <0·21. Bayes factor comparisons was used to test the relative support for one state over the other for each of these groupings. For the MRCA of Myrteae, Syzygieae and Xanthomyrtus, there is strong support for the dry fruit state [2loge(B0) = 8·8] whereas for the MRCA Syzygieae + Myrteae, the Bayes factor comparison has a 2loge(B0) of 7·3, again favouring the dry-fruited state at this node.
DISCUSSION
Phylogenetic relationships
Our analyses of the concatenated data (Figs 1 and 2) produced a well-supported hypothesis of relationships in Myrtaceae and, in several instances, a high level of confidence for groupings that were not strongly supported in previous molecular studies (Sytsma et al., 2004; Wilson et al., 2001, 2005). This may be, in part, a consequence of different analytical approaches and, in particular, several studies have found that the Bayesian posterior probabilities of clades tend to be higher than support from non-parametric bootstrapping (BS) for the same set of data (e.g. Erixon et al., 2003; Simmons et al., 2004). The studies of Sytsma et al. and Wilson et al. used the latter approach to assess statistical support for clades. However, considering the plastid-only Bayesian analysis (Fig. S2 in Supplementary data, available online), which is most comparable to the study of Sytsma et al., there are several nodes receiving relatively weak support (PP < 0·95) that are robustly supported in the analysis of the concatenated data. For instance, the grouping of Chamelaucieae, Eucalypteae, Leptospermeae, Lindsayomyrteae and Syncarpieae has a PP = 0·83 in the analysis of the plastid data, and for the ML estimate of Sytsma et al. receives a BS of <70 %. In the Bayesian analyses of the concatenated dataset, this clade is statistically well supported (Fig. 1). Similarly, Sytsma et al. (2004) identified a clade including Backhousieae, Kanieae, Metrosidereae, Myrteae, Syzygieae and Tristanieae, but with low statistical support. An equivalent grouping (Fig. S2 in Supplementary data) is weakly supported by the Bayesian analysis of the plastid data (PP = 0·89), but is strongly supported in the concatenated data analyses (BKMMST clade; Fig. 1). These findings suggest a direct positive effect from the addition of the ITS data rather than, or in addition to, the contrasting measures of statistical support employed in this versus previous molecular studies of Myrtaceae.
Molecular dating
In their comparable study, Sytsma et al. (2004) used rate-variable molecular dating analyses to estimate divergence times among Myrtaceae, and, in addition to fixing the age of the root using a ‘derived’ date (93 Ma), the ‘eucalypt’ crown group (including Eucalyptus, Angophora and Arillastrum) was fixed at 48 Ma [based upon the older of two possible ages that have been suggested for the Nelly Creek Formation fossils from central Australia described in Lange (1978) and Ambrose et al. (1979); see Rozenfelds (1996)]; the Myrteae crown node was fixed at 56 Ma (based on the age of the Paleomyrtinaea fossil fruit); and the Myrtaceidites pollen taxon was used to provide a maximum age of 86 Ma for the Myrtaceae crown node.
The calibration points used by Sytsma et al. (2004) are compared with the divergence time estimates from the BEAST analyses, for the equivalent node, in Fig. 2. Of these, the age of the root and the Myrtaceae crown node are highly consistent with the fossil constraints of Sytsma et al., whereas the estimated ‘eucalypt’ crown group age is at least comparable with their fossil placement (95 % HPD 35–45 Ma). However, there is considerable discrepancy with respect to the age of the Myrteae crown node, which in the present study is approx. 10–30 Ma younger (95 % HPD) than the age implied by the placement of the Paleomyrtinaea fossil fruit by Sytsma et al. More generally, the age estimates from the two studies [compare Fig. 5 (S93), Sytsma et al. (2004) and Fig. 2] are reasonably consistent across much of the Myrtaceae phylogeny, although there are large discrepancies with respect to the BKMMST clade with Sytsma et al. reporting divergence time estimates for the relevant taxa that are generally older than those estimated here. We hypothesize that these differences are driven primarily by the contrasting treatment of the Paleomyrtinaea fossil constraint among studies. On the one hand, constraining the stem group age with the fossil constraint could force a younger age on more nested nodes or, alternatively, using a fixed age for the crown node rules out the possibility that the associated fossil lineage may in fact be older than the extant crown radiation (e.g. an extent stem lineage).
To explore this hypothesis, BRC analyses were performed with settings, as described above, and the following age constraints: using uniform priors, the age of the root was fixed at 92–94 Ma, the Myrtaceae crown node at 87–85 Ma, and the ‘eucalypt’ crown at 49–47 Ma. All values within these bounds are equally probable, whereas there is zero probability associated with values outside of the bounds, i.e. approximating a ‘fixed’ node age, as used by Sytsma et al. (2004). From this analysis (full results not shown), the age of the Myrteae crown group was estimated at 32 (95 % HPD 23–39) Ma, and the age of the BKMMST clade was 55 (95 % HPD 44–66) Ma, which are highly consistent with the divergence time estimates obtained for these nodes using a log-normal prior to constrain the stem group age (Table 1). This suggests that an early stem group lineage is the most appropriate placement for Paleomyrtinaea, and that Sytsma et al. may have substantially overestimated the age of the Myrteae and lineages within the BKMMST clade by constraining the crown node age.
The ‘likely vicariance’ hypothesis of Sytsma et al. (2004) to explain intercontinental disjunctions in Myrteae needs reassessment in light of the estimated Oligocene–Miocene (95 % HPD 34–22 Ma) radiation of the American Myrteae (Table 1 and Fig. 2), which cannot rule out an early widespread South American distribution and subsequent extinction of (an) ancestor(s) of modern Myrteae (represented by Paleomyrtinaea). Modern Myrteae may then have recolonized South America from Australasia, possibly post-dating the opening of the Drake Passage (approx. 28 Ma; McLoughlin, 2001).
Diversification rates
Strong evidence was found for a positive diversification rate shift within Myrtaceae that was consistently associated with the BKMMST clade (Table 2 and Fig. 4), and in particular with Syzygieae and Myrteae (Table 2 and Figs 3 and 5). The vast majority of lineages within the BKMMST clade are woody rainforest trees whereas all of Backhousieae, Kanieae, Metrosidereae and Tristanieae (excluding Xanthomyrtus) have dry capsular fruits indicative of abiotic dispersal. All of Myrteae, Syzygieae (excluding the monotypic subgenus Anetholea of Syzygium) and Xanthomyrtus develop a fleshy pericarp, which is here considered indicative of biotic dispersal. Biotic dispersal has been variously proposed as a mechanism promoting elevated rates of cladogenesis among angiosperms, through adaptive divergence in response to different dispersal vectors and/or by promoting allopatry among plant populations due to the movement behaviour of biotic dispersers (e.g. Tiffney and Mazer, 1995; Smith, 2001). Several studies have failed to find a general effect of biotic dispersal on extant species numbers within angiosperms (Herrera, 1989; Midgley and Bond, 1991; Eriksson and Bremer, 1992; Davies et al., 2004), but there does appear to be significant interaction between diversification rates, dispersal syndromes and the specific ecological context (Tiffney and Mazer, 1995; Smith, 2001; de Quieroz, 2002). Tiffney and Mazer (1995), for example, argue that among angiosperms, an arborescent habit favours large seed size and biotic dispersal. In spatially unpredictable closed forest communities, lineages with these characteristics may experience higher recruitment success and lower extinction rates, relative to woody arborescent lineages with unassisted dispersal.
In light of the present findings (Table 2 and Figs 3 and 5), it is tempting to suggest that the possession of fleshy fruits per se may be related to elevated diversification rates within Syzygieae and Myrteae relative to the other lineages of Myrtoideae. This hypothesis would gain support if there were multiple shifts from dry to fleshy fruits associated with significant positive diversification shifts. Although the relationships among Syzygieae and Myrteae are not well resolved, the separation of Xanthomyrtus (Tristanieae) from traditional Myrtoideae is strongly supported (Wilson et al., 2005; Figs 1 and 2), suggesting at least paraphyly of the fleshy fruited lineages. The condition for paraphyly of Myrtoideae s.s. would require that that Tristanieae, Syzygieae and Myrteae form a clade (PP = 0·03, Table 2) and that Xanthomyrtus is sister to the capsular-fruited Tristanieae, whereas alternatively there could be two or three separate origins of fleshy fruits if Syzygieae and Myrteae are not resolved as sister lineages (PP = 0·18 for this monophyly; Table 2). The evolution of fruit type in Myrtaceae was estimated using a method that simultaneously accounts for phylogenetic and branch-length uncertainty (Pagel et al., 2004). The ancestral state reconstructions of fruit morphology strongly support at least two, and probably three, separate origins of fleshy fruits within the BKMMST clade suggesting that Syzygieae and Myrteae have undergone independent shifts from dry to fleshy-fruited states. Furthermore, it was found that the highest AIC shift scores were consistently associated with a monophyletic Syzygieae + Myrteae (Table 2 and Fig. 5), suggesting that both of these lineages have experienced higher diversification rates than Myrtaceae in general compared with a less parsimonious hypothesis that a rate shift has occurred at a deeper node and Syzygieae and Myrteae have merely retained a high ancestral rate (see Rabosky et al., 2007). While these findings imply causality, further sampling of Xanthomyrtus (here, represented by a single terminal but including 23 species; Govaerts et al., 2008) would help to clarify the association between fleshy fruits and diversification rate shifts in the BKMMST clade and Myrtaceae.
A further consideration is the diversity within both Syzygieae and Myrteae, as both lineages include species-rich groups. In Myrteae, Eugenia has an estimated 1050+ species, and Myrcia s.l. (Lucas et al., 2007) includes approx. 750 species (Govaerts et al., 2008). For Syzygieae, comprising the single genus Syzygium (Craven et al., 2006), the six subgenera proposed by Craven and Biffin (2010) include two monotypic lineages, and it has been suggested that subgenus Syzygium includes approx. 90–95 % (or approx. 1000 species) of the total species richness of Syzygieae (Parnell et al., 2006). Given these estimates, and in light of the timing of radiation of the corresponding crown groups (95 % HPD; Fig. 2), the estimated diversification rates are 0·29–0·93, 0·25–0·61 and 0·27–0·58 net speciation events per million years (assuming e = 0·9) for Eugenia, Myrcia s.l. and subgenus Syzygium, respectively. These values are high in the context of angiosperms in general (Magallón and Sanderson, 2001) and rival the values inferred for island plant radiations (e.g. Baldwin and Sanderson, 1998). They are also significant in light of predictions that woody arborescent angiosperm lineages in general should have low rates of diversification, for example, relative to herbaceous lineages (reviewed by Petit and Hampe, 2006). In terms of the present study, it is plausible that the high diversification rates reported for the Syzygieae and Myrteae crown groups are a consequence of ‘trickle down’ effects (Moore et al., 2004) driven by these species-rich, more deeply nested lineages. Therefore, a simple correlation between high species richness and the evolution of fleshy fruit is contingent on more detailed studies of the evolutionary relationships and timing of lineage diversification events within Syzygieae and Myrteae.
Additional factors for consideration (alone or in conjunction) as potential drivers of rapid speciation in large genera of Syzygieae and Myrteae are pollination strategy and embryo specialization. Syzygium and most genera of Myrteae display relative homogeneity of primarily bee-pollinated flower types with flexibility in flowering strategy. Flowering, particularly in the three largest genera, is showy but timing varies from mass-flowering in a few days, to pulsed or steady flowering lasting up to 3 months (Proença and Gibbs, 1994). This faithful but flexible bee pollination may allow species to exploit a wide variety of bee species and behaviour (e.g. trap-lining and buzz pollination) and may be a shared source of success with large genera in other families (e.g. Melastomataceae and Solanaceae). Morphological divergence in seeds types from small, wind-dispersed or hard, C-shaped seeds embedded in fleshy pulp to larger, more complex architectures may also convey evolutionary benefit to the genera in question. The embryonic cotyledons of Myrcia s.l. are folded, green and ready to photosynthesize, the embryos of Eugenia and Syzygium s.l. are energy-rich homogeneous structures derived from swollen and fused cotyledons. Presence of the putative evolutionary drivers described above, including those linked to fleshy fruits such as endozoochory, are strongly correlated with large genera of Myrtaceae established in the moist lowland tropics. Fruit dispersal by birds, bats or larger marsupials is reported in Syzygium (Nic Lughadha and Proença, 1996), whereas Eugenia fruits are mainly dispersed by birds (Snow, 1981). The implication then is that independent ancestral lineages of Myrtaceae encountered similar niches available in separately developing tropical forest and where once established, and seemingly in parallel, their shared potential for speciation on a remarkable scale was exploited.
Conclusions
Traditionally, the fleshy-fruited Myrtaceae have been treated as a monophyletic group, although phylogenetic studies suggest that para- or polyphyly is likely, with Xanthomyrtus, the predominantly American Myrteae and the predominantly Australasian Syzygieae forming distinct, well-supported lineages. The results of this study suggest a relatively recent (Oligocene–Miocene) radiation of these tribes and multiple origins of fleshy fruits within the more inclusive BKMMST clade. There is strong support for exceptionally high diversification rates for both Syzygieae and Myrteae, and a highly significant positive shift in diversification rates associated with both of these lineages relative to the overall radiation of Myrtaceae. Taken together, these factors suggest a link between the evolution of fleshy fruits and elevated rates of lineage accumulation within the BKMMST clade. An alternative hypothesis is that the high species richness within Syzygieae and Myrteae is driving the elevated speciation rates, i.e. comparisons at the tribal level are insensitive to bona fide shifts at more nested nodes and, in terms of formulating causal hypotheses, are potentially misleading (Moore et al., 2004). Future studies are required, focusing specifically on the evolutionary relationships and the timings of diversification in Syzygieae and Myrteae. For instance, significant shifts in diversification rate within these groups would suggest that, at a minimum, more complex hypotheses than those suggested above are required to account for the disparate lineage diversities among Myrtaceae, although these elevated diversification rates may be contingent on the evolution of fleshy fruits in rainforest tree lineages.
SUPPLEMENTARY DATA
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank Dr Peter G. Wilson for access to DNA samples and Prof. Paul Gadek for use of laboratory facilities at James Cook University, Cairns. We thank Dr Michael Fay and two anonymous reviewers for their comments on the manuscript. E.B. was supported by an Australian Biological Resources Study Postgraduate scholarship
APPENDIX
Taxa studied, GenBank accessions and voucher information
| Taxon | matK | ndhF | ITS | Voucher | |
|---|---|---|---|---|---|
| Acca sellowiana (O.Berg) Burret | Myrteae | AY525128 | AY498783 | AM234067 | |
| Agonis flexuosa (Muhl. ex Willd.) Sweet | Leptospermeae | AF184711 | AY498762 | DQ499115 | |
| Amomyrtus meli (Phil.) D.Legrand & Kausel | Myrteae | AM489976 | AM234069 | ||
| Angophora hispida (Sm.) Blaxell | Eucalypteae | AF368196 | AY498763 | AF190357 | |
| Arillastrum gummiferum (Brong. & Gris) Panch. ex Baill. | Eucalypteae | AF368198 | AY498765 | AF058454 | |
| Asteromyrtus arnhemica (Byrnes) Craven | Chamelaucieae | EF026603 | |||
| Asteromyrtus lysicephala (F.Muell. & F.M.Bailey) Craven | Chamelaucieae | AF184718 | AY498766 | ||
| Backhousia myrtifolia Hook. & Harv. | Backhousieae | AF368200 | DQ088472 | DQ088408 | |
| Beaufortia orbifolia F.Muell. | Melaleuceae | AY521530 | AY498771 | AF048888 | |
| Blepharocalyx tweediei (Hook. & Arn.) O.Berg | Myrteae | AY521531 | AY498772 | AM234084 | |
| Callistemon polandii F.M.Bailey | Melaleuceae | AF184705 | AY498773 | ||
| Callistemon viminalis (Sol. ex Gaertn.) G.Don ex Loudon. | Melaleuceae | EF041510 | |||
| Calothamnus quadrifidus R.Br. ex W.T.Aiton | Melaleuceae | EF041511 | |||
| Calothamnus validus S.Moore | Melaleuceae | AF184704 | AY498774 | ||
| Calyptranthes concinna DC. | Myrteae | AF368201 | AY498775 | AM234103 | |
| Calytrix tetragona Labill. | Chamelaucieae | AF489396 | AY498776 | HM160102/03 | UNSW21772 |
| Campomanesia guazumifolia (Cambess.) O.Berg | Myrteae | AY521532 | AY498777 | AM234076 | |
| Chamelaucium uncinatum Schauer | Chamelaucieae | AY259816 | EF026605 | ||
| Choricarpia subargentea (C.T.White) L.A.S.Johnson | Backhousieae | AF368202 | DQ088473 | DQ088409 | |
| Cloezia floribunda Brongn. & Gris | Unplaced | AY521533 | AF172767 | ||
| Corymbia variegata (F.Muell.) K.D.Hill & L.A.S.Johnson | Eucalypteae | AF368203 | DQ993141 | ||
| Decaspermum humile (G.Don) A.J.Scott | Myrteae | AY521534 | AY498780 | AM234128 | |
| Eucalyptopsis papuana C.T.White | Eucalypteae | AF368205 | AF190354 | ||
| Eucalyptus curtisii Blakely & C.T.White | Eucalypteae | AF368206 | AY498781 | AF390525 | |
| Eucalypus globulus Labill. | Eucalypteae | AY521535 | AY780259 | AF058467 | |
| Eugenia sulcata Spring ex Mart. | Myrteae | AM489987 | HM160097 | AM234089 | Lucas 68 |
| Eugenia uniflora L. | Myrteae | AF368207 | DQ088457 | AY487284 | |
| Gomidesia flagellaris D.Legrand | Myrteae | AM489989 | HM16009 | AM234113 | |
| Heteropyxis natalensis Harv. | Heteropyxideae | AF368208 | AY498824 | HM160104/05 | PGW 1475- NSW |
| Homalocalyx aureus (C.A.Gardner) Craven | Chamelaucieae | AF489398 | AY498785 | HM160106/07 | UNSW22947 |
| Homoranthus darwinioides Cheel | Chamelaucieae | AF489399 | AY498786 | HM160108 | UNSW23267 |
| Kjellbergiodendron celebicum (Koord.) Merr. | Lophostemoneae | AF368209 | AY498788 | HM160109/10 | Zich s.n. |
| Kunzea capitata (Sm.) Heynh. | Leptospermeae | AF184723 | AY498790 | ||
| Kunzea sinclairii (Kirk) W.Harris | Leptospermeae | AY772399 | |||
| Leptospermum scoparium J.R.Forst & G.Forst | Leptospermeae | AM489991 | AM235423 | AY772398 | |
| Lindsayomyrtus racemoides (Greves) Craven | Lindsayomyrteae | AF184706 | AY498793 | HM160111/12 | |
| Lophostemon confertus (R.Br.) P.G.Wilson & J.T.Waterh. | Lophostemoneae | AF184707 | AY498794 | AF390444 | |
| Luma apiculata (DC.) Burret | Myrteae | AY521540 | AY498795 | AY463105 | |
| Marlierea eugeniopsoides (D.Legrand & Kausel) D.Legrand | Myrteae | AM489996 | HM160099 | AM234107 | Lucas 61 |
| Melaleuca viridiflora Sol. ex Gaertn. | Melaleuceae | AY498798 | AF184708 | AF294611 | |
| Metrosideros carminea W.R.B.Oliv. | Metrosidereae | AY521541 | AY498799 | AF211498 | |
| Metrosideros macropus Hook. & Arn. | Metrosidereae | AF368212 | AY498801 | AF172745 | |
| Metrosideros nervulosa C.Moore & F.Muell. | Metrosidereae | Q088535 | DQ088458 | DQ088395 | |
| Micromyrtus ciliata (Sm.) Druce | Chamelaucieae | AF489400 | HM160113/14 | UNSW23860 | |
| Myrcia saxatilis (Amshoff) McVaugh | Myrteae | AM490004 | HM160100 | AM234119 | Lucas 98 |
| Myrciaria cauliflora (Mart.) O.Berg | Myrteae | AM234093 | |||
| Myrciaria vexator McVaugh | Myrteae | AY521544 | AY498804 | ||
| Myrtus communis L. | Myrteae | AY525136 | AF215593 | AF215628 | |
| Osbornia octodonta F.Muell. | Osbornieae | AF368213 | AY498805 | EF041844 | |
| Pericalymma ellipticum (Endl.) Schauer | Melaleuceae | AF184740 | AY498806 | EF026604 | |
| Piliocalyx bullatus Brong. & Gris | Syzygieae | DQ088552 | DQ088478 | DQ088413 | |
| Pimenta racemosa (Mill.) J.W.Moore | Myrteae | DQ088554 | AY498808 | EF026631 | |
| Psidium cattleianum Afzel. ex Sabine | Myrteae | AM490014 | HM160101 | AM234080 | Lucas 213 |
| Psiloxylon mauritianum (Bouton ex Hook.f.) Baill. | Psiloxyloideae | AF368215 | AY498825 | EF026606 | |
| Qualea grandiflora Mart. | Vochysiaceae (outgroup) | AF368216 | |||
| Qualea sp. | Vochysiaceae (outgroup) | AY498829 | |||
| Rhodamnia argentea Benth. | Myrteae | AF368217 | AY498810 | AY487302 | |
| Rhodomyrtus macrocarpa Benth. | Myrteae | AY498811 | AY525137 | ||
| Rhodomyrtus psidioides (G.Don) Benth. | Myrteae | AM234134 | |||
| Sphaerantia chartacea P.G.Wilson & B.Hyland | Kanieae | AY521547 | HM160115/16 | PGW 1348 | |
| Stockwellia quadrifida D.J.Carr, S.G.M.Carr & B.Hyland | Eucalypteae | AY498812 | AY525138 | AF390445 | |
| Syncarpia glomulifera (Sm.) Nied. | Syncarpieae | AY498813 | AF368220 | HM160117/18 | UNSW23246 |
| Syncarpia hillii F.M.Bailey | Syncarpieae | AY525139 | |||
| Syzygium acuminatissimum DC | Syzygieae | DQ088537 | DQ088462 | EF026611 | |
| Syzygium anisatum (Vickery) Craven & Biffin | Syzygieae | AF368195 | DQ088471 | DQ088407 | |
| Syzygium apodophyllum (F.Muell.) B.Hyland | Syzygieae | DQ088558 | DQ088482 | DQ088417 | |
| Syzygium arboreum (Baker f.) J.W.Dawson | Syzygieae | DQ088560 | DQ088484 | DQ088418 | |
| Syzygium bungadinnia (F.M.Bail.) B.Hyland | Syzygieae | DQ088568 | DQ088490 | DQ088423 | |
| Syzygium buxifolium Hook. & Arn. | Syzygieae | DQ088569 | DQ088491 | DQ088424 | |
| Syzygium canicortex B.Hyland | Syzygieae | DQ088570 | DQ088492 | DQ088425 | |
| Syzygium claviflorum (Roxb.) Wall. ex Stued. | Syzygieae | DQ088546 | DQ088470 | DQ088406 | |
| Syzygium crebrinerve (C.T.White) L.Johnson | Syzygieae | DQ088574 | DQ088495 | DQ088428 | |
| Syzygium divaricatum (Merr. & L.M.Perry) Craven & Biffin | Syzygieae | DQ088538 | DQ088463 | ||
| Syzygium floribundum F.Muell. | Syzygieae | DQ088620 | DQ088531 | DQ088453 | |
| Syzygium francisii (F.M.Bail.) L.Johnson | Syzygieae | DQ088578 | DQ088498 | DQ088430 | |
| Syzygium fullagarii (F.Muell.) Craven | Syzygieae | DQ088579 | DQ088499 | DQ088431 | |
| Syzygium glenum Craven | Syzygieae | DQ088539 | DQ088464 | AY187162 | |
| Syzygium gustavioides (F.M.Bail.) B.Hyland | Syzygieae | DQ088582 | DQ088501 | DQ088433 | |
| Syzygium ingens (F.Muell. ex C.Moore) Craven & Biffin | Syzygieae | DQ088542 | DQ088466 | DQ088402 | |
| Syzygium laxeracemosum (Guillaumin) J.W.Dawson | Syzygieae | DQ088586 | DQ088505 | DQ088436 | |
| Syzygium mackinnonianum (B. Hyland) Craven & Biffin | Syzygieae | DQ088543 | DQ088467 | DQ088403 | |
| Syzygium maire (A.Cunn.) Sykes & P.J.Garnock-Jones | Syzygieae | DQ088589 | DQ088508 | DQ088438 | |
| Syzygium malacense (L.) Merr. & Perry | Syzygieae | DQ088590 | DQ088509 | AY187199 | |
| Syzygium monimioides Craven | Syzygieae | DQ088544 | DQ088468 | DQ088404 | |
| Syzygium mulgraveanum (B.Hyland) Craven & Biffin | Syzygieae | DQ088622 | DQ088533 | DQ088455 | |
| Syzygium multipetalum Pancher ex Brongn. & Gris | Syzygieae | DQ088594 | DQ088512 | DQ088440 | |
| Syzygium nervosum D.C. | Syzygieae | DQ088595 | DQ088513 | EF026636 | |
| Syzygium paniculatum Gaertner | Syzygieae | DQ088598 | DQ088515 | AY187204 | |
| Syzygium puberulum Hartley & Perry | Syzygieae | DQ088601 | DQ088517 | AY187207 | |
| Syzygium sp. ‘Sulawesi 2’ | Syzygieae | DQ088611 | DQ088524 | EF026649 | |
| Syzygium sayeri (F.Muell.) B.Hyland | Syzygieae | DQ088607 | AY187209 | ||
| Syzygium tetrapterum (Miq.) Chantaranothai & J.Parn. | Syzygieae | DQ088615 | DQ088527 | DQ088448 | |
| Syzygium wesa B.Hyland | Syzygieae | DQ088617 | DQ088529 | DQ088450 | |
| Syzygium wilsonii (F.Muell.) B.Hyland subsp. wilsonii | Syzygieae | DQ088618 | DQ088530 | DQ088451 | |
| Tepualia stipularis Griseb. | Metrosidereae | AF368222 | AM234071 | ||
| Thaleropia queenslandica (L.S.Sm.) Peter G.Wilson | Tristanieae | AF368223 | DQ088460 | DQ088397 | |
| Tristania neriifolia (Sims) R.Br. | Tristanieae | AF368224 | DQ088461 | EF026608 | |
| Tristaniopsis laurina (Sm.) Peter G.Wilson & J.T.Waterh. | Kanieae | AF184710 | AY498818 | EF041514 | |
| Ugni molinae Turcz. | Myrteae | AM490018 | AY498819 | AM234143 | |
| Vochysia hondurensis Sprague | Vochysiaceae (outgroup) | AY572446 | AY498832 | ||
| Xanthomyrtus montivaga A.J.Scott | Tristanieae | AM234147 | |||
| Xanthomyrtus papuana Merr. & L.M.Perry | Tristanieae | AF368226 | AY498822 | ||
| Xanthostemon aurantiacus (Brongn. & Gris) Schltr. | Xanthostemoneae | AY525144 | |||
| Xanthostemon chrysanthus (F.Muell.) Benth. | Xanthostemoneae | AF368227 | AY498823 | EF041515 |
LITERATURE CITED
- Ambrose GJ, Callen RA, Flint RB, Lange RT. Eucalyptus fruits in stratigraphic context in Australia. Nature. 1979;280:387–389. [Google Scholar]
- Baldwin B, Sanderson MJ. Age and rate of diversification of the Hawaiian silversword alliance. Proceedings of the National Academy of Sciences of the USA. 1998;95:9402–9406. doi: 10.1073/pnas.95.16.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basinger JF, Greenwood DR, Wilson PG, Christophel DC. Fossil flowers and fruit of capsular Myrtaceae from the Eocene of South Australia. Canadian Journal of Botany. 2007;85:204–215. [Google Scholar]
- Biffin E, Craven LA, Crisp MD, Gadek PA. Molecular systematics of Syzygium and allied genera (Myrtaceae): evidence from the chloroplast genome. Taxon. 2006;55:79–94. [Google Scholar]
- Biffin E, Harrington MG, Crisp MD, Craven LA, Gadek PA. Structural partitioning, paired-sites models and the evolution of the rDNA ITS transcripts in Syzygium and Myrtaceae. Molecular Phylogenetics and Evolution. 2007;43:134–139. doi: 10.1016/j.ympev.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Boltenhagen E. Pollens et spores sénoniennes du Gabon. Cahiers de Micropaléontologie. 1976;17:1–21. [Google Scholar]
- Briggs BG, Johnson LAS. Evolution in the Myrtaceae: evidence from inflorescence structure. Proceedings of the Linnaean Society of New South Wales. 1979;102:157–256. [Google Scholar]
- Conti E, Litt A, Wilson PG, et al. Interfamilial relationships in Myrtales: molecular phylogeny and patterns of morphological evolution. Systematic Botany. 1997;22:629–647. [Google Scholar]
- Cook LG, Crisp MD. Not so ancient: the extant crown group of Nothofagus represents a post-Gondwanan radiation. Proceedings of the Royal Society B: Biological Sciences. 2005;272:2535–2544. doi: 10.1098/rspb.2005.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane PR, Manchester SR, Dilcher DL. A preliminary survey of fossil leaves and well-preserved reproductive structures from the Sentinel Butte Formation (Paleocene) near Almont, North Dakota. Fieldiana Geology. 1990;1418:1–63. [Google Scholar]
- Craven LA. Unravelling knots or platting rope: what are the major taxonomic strands in Syzygium sens. lat. (Myrtaceae) and what should be done with them? In. In: Saw LG, Chua LS, Khoo KC, editors. Taxonomy: the cornerstone of biodiversity. Proceedings of the Fourth International Flora Malesiana Symposium, 1998. Kuala Lumpur: Institut Penyelidikan Perhutanan Malaysia; 2001. pp. 75–85. [Google Scholar]
- Craven LA, Biffin E. An infrageneric classification for Syzygium (Myrtaceae) Blumea. 2010;55:94–99. [Google Scholar]
- Craven LA, Biffin E, Ashton PS. Acmena, Acmenosperma, Cleistocalyx, Piliocalyx and Waterhousea formally transferred to Syzygium (Myrtaceae) Blumea. 2006;51:131–142. [Google Scholar]
- Crisp MD, Cook LG, Steane DA. Radiation of the Australian flora: what can comparisons of molecular phylogenies across multiple taxa tell us about the evolution of diversity in present-day communities? Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2004;359:1551–1571. doi: 10.1098/rstb.2004.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies TJ, Barraclough TG, Chase MW, Soltis PS, Soltis DE, Savolainen V. Darwin's abominable mystery: insights from a supertree of the angiosperms. Proceedings of the National Academy of Sciences of the USA. 2004;101:1904–1909. doi: 10.1073/pnas.0308127100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue MJ. Key innovations, convergence, and success: macroevolutionary lessons from plant phylogeny. Palaeobiology. 2005;31:77–93. [Google Scholar]
- Drinnan AN, Crane PR, Hoot SB. Patterns of floral evolution in the early diversification of non-magnoliid dicotyledons (eudicots) Plant Systematics and Evolution. 1994;8:93–122. [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ho SWY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating confidence. PLoS Biology. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. doi:10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson O, Bremer B. Pollination systems, dispersal modes, life forms, and diversification rates in angiosperm families. Evolution. 1992;46:258–266. doi: 10.1111/j.1558-5646.1992.tb02000.x. [DOI] [PubMed] [Google Scholar]
- Erixon P, Svennblad B, Britton T, Oxelman B. Reliability of Bayesian posterior probabilities and bootstrap frequencies in phylogenetics. Systematic Biology. 2003;52:665–673. doi: 10.1080/10635150390235485. [DOI] [PubMed] [Google Scholar]
- Erkens RHJ, Chatrou LW, Maas JW, van der Niet T, Savolainen V. A rapid diversification of rainforest trees (Guatteria; Annonaceae) following dispersal from Central into South America. Molecular Phylogenetics and Evolution. 2007;44:399–411. doi: 10.1016/j.ympev.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Gadek PA, Wilson PG, Quinn CJ. Phylogenetic reconstruction in Myrtaceae using matK, with particular reference to the position of Psiloxylon and Heteropyxis. Australian Systematic Botany. 1996;9:283–290. [Google Scholar]
- Govaerts R, Sobral M, Ashton P, et al. World checklist of Myrtaceae. London: Board of Trustees of the Royal Botanic Gardens, Kew; 2008. [Google Scholar]
- Greenwood DL, Christophel DC. The origins and Tertiary history of Australian tropical rainforests. In: Bermingham E, Dick C, Moritz C, editors. Tropical rainforests: past, present and future. Chicago, IL: Chicago University Press; 2005. pp. 336–373. [Google Scholar]
- van der Hammen T. El desarrollo de la flora colombiana en los periodos geológicos. I. Maestrichtiano hasta Terciario más inferior. Boletín de Geología. 1954;2:49–106. [Google Scholar]
- Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W. GEIGER: investigating evolutionary radiations. Bioinformatics. 2008;24:129–131. doi: 10.1093/bioinformatics/btm538. [DOI] [PubMed] [Google Scholar]
- Harrington MG, Gadek PA. Molecular systematics of the Acmena alliance (Myrtaceae): phylogenetic analyses and evolutionary implications with respect to Australian taxa. Australian Systematic Botany. 2004;17:63–72. [Google Scholar]
- Hererra CM. Frugivory and seed dispersal by carnivorous mammals, and associated fruit characteristics, in undisturbed Mediterranean habitats. OIKOS. 1989;55:250–262. [Google Scholar]
- Herngreen GFW. An Upper Senonian pollen assemblage of borehole 3-PIA-10-AL, State of Alagoas, Brazil. Pollen Spores. 1975;17:93–140. [Google Scholar]
- Johnson LAS, Briggs BG. Myrtales and Myrtaceae: a phylogenetic analysis. Annals of the Missouri Botanic Gardens. 1984;71:700–756. [Google Scholar]
- Kass RE, Rafferty AE. Bayes factors. Journal of the American Statistical Association. 1995;90:773–795. [Google Scholar]
- Ladiges PY, Udovicic F, Nelson G. Australian biogeographical connections and the phylogeny of large genera in the plant family Myrtaceae. Journal of Biogeography. 2003;30:989–998. [Google Scholar]
- Landrum LR, Kawasaki ML. The genera of Myrtaceae in Brazil: an illustrated synoptic treatment and identification keys. Brittonia. 1997;49:508–536. [Google Scholar]
- Lange RT. Carpological evidence for fossil Eucalyptus and other Leptospermeae (subfamily Leptospermoideae of Myrtaceae) from a Tertiary Deposit in the South Australian arid zone. Australian Journal of Botany. 1978;26:221–233. [Google Scholar]
- Linder HP, Hardy CR, Rutschmann F. Taxon sampling effects in molecular clock dating: an example from the African Restionaceae. Molecular Phylogenetics and Evolution. 2005;35:569–582. doi: 10.1016/j.ympev.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Lucas EJ, Harris SA, Mazine FF, et al. Suprageneric phylogenetics of Myrteae, the generically richest tribe in Myrtaceae (Myrtales) Taxon. 2007;56:1105–1128. [Google Scholar]
- McLoughlin S. The breakup history of Gondwana and its impact on pre-Cenozoic floristic provincialism. Australian Journal of Botany. 2001;49:271–300. [Google Scholar]
- McVaugh R. The genera of American Myrtaceae: an interim report. Taxon. 1968;17:354–418. [Google Scholar]
- Magallón S, Sanderson MJ. Absolute diversification rates in angiosperm clades. Evolution. 2001;55:1762–1780. doi: 10.1111/j.0014-3820.2001.tb00826.x. [DOI] [PubMed] [Google Scholar]
- Magallón S, Crane PR, Herendeen PS. Phylogenetic pattern, diversity, and diversification of eudicots. Annals of the Missouri Botanic Garden. 1999;86:297–372. [Google Scholar]
- Manchester SR. Biogeographical relationships of North American Tertiary floras. Annals of the Missouri Botanical Garden. 1999;86:472–522. [Google Scholar]
- Midgley JJ, Bond WJ. Ecological aspects of the rise of angiosperms: a challenge to the reproductive superiority hypotheses. Biological Journal of the Linnaean Society. 1991;44:81–92. [Google Scholar]
- Moore BR, Donoghue MJ. Correlates of diversification in the plant clade Dipsacales: geographic movement and evolutionary innovation. American Naturalist. 2007;170:S28–S55. doi: 10.1086/519460. [DOI] [PubMed] [Google Scholar]
- Moore BR, Chan K, Donoghue MJ. Detecting diversification rate variation in supertrees. In: Bininda-Emonds O, editor. Phylogenetic supertrees: combining information to reveal the Tree of Life. New York, NY: Kluwer Academic Publishers; 2004. pp. 487–533. [Google Scholar]
- Muller J. Palynology of the Pedawan and plateau sandstone formations (Cretaceous-Eocene) in Sarawak, Malaysia. Micropaleontology. 1968;14:1–37. [Google Scholar]
- Muller J. Fossil pollen records of extant angiosperms. Botanical Review. 1981;47:1–142. [Google Scholar]
- Nic Lughadha E, Proença C. A survey of reproductive biology of the Myrtoideae (Myrtaceae) Annals of the Missouri Botanic Garden. 1996;83:480–503. [Google Scholar]
- Pagel M, Lutzoni F. Accounting for phylogenetic uncertainty in comparative studies of evolution and adaptation. In: Lässig M, Valleriani A, editors. Biological evolution and statistical physics. Berlin: Springer; 2002. pp. 148–161. [Google Scholar]
- Pagel M, Meade A. Bayesian analysis of correlated evolution of discrete characters by reversible-jump Markov chain Monte Carlo. American Naturalist. 2006;167:808–825. doi: 10.1086/503444. [DOI] [PubMed] [Google Scholar]
- Pagel M, Meade A, Barker D. Bayesian estimation of ancestral character states on phylogenies. Systematic Biology. 2004;53:673–684. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- Parnell JAN, Craven LA, Biffin E. Matters of scale: dealing with one of the largest genera of angiosperms. In: Hodkinson TR, Parnell JAN, editors. Reconstructing the Tree of Life: taxonomy and systematics of species rich taxa. London: CRC Press; 2006. pp. 251–274. [Google Scholar]
- Petit RJ, Hampe A. Some evolutionary consequences of being a tree. Annual Review of Ecology and Systematics. 2006;37:187–214. [Google Scholar]
- Pigg KB, Stockey RA, Maxwell SL. Paleomyrtinaea, a new genus of permineralized myrtaceous fruits and seeds from the Eocene of British Columbia and Paleocene of North Dakota. Canadian Journal of Botany. 1993;71:1–9. [Google Scholar]
- Pole M, Dawson J, Denton T. Fossil Myrtaceae from the early Miocene of southern New Zealand. Australian Journal of Botany. 2008;56:67–81. [Google Scholar]
- Proença C, Gibbs PE. Reproductive biology of eight sympatric Myrtaceae from central Brazil. New Phytologist. 1994;126:343–354. [Google Scholar]
- de Queiroz A. Contingent predictability in evolution: key traits and diversification. Systematic Biology. 2002;51:917–929. [PubMed] [Google Scholar]
- Rabosky DL. LASER: a maximum likelihood toolkit for detecting temporal shifts in diversification rates. Evolutionary Bioinformatics Online. 2006;2:257–260. [PMC free article] [PubMed] [Google Scholar]
- Rabosky DL, Donnellan SC, Tallaba AL, Lovette IJ. Exceptional among-lineage variation in diversification rates during the radiation of Australia's most diverse vertebrate clade. Proceedings of the Royal Society B: Biological Sciences. 2007;274:2915–2923. doi: 10.1098/rspb.2007.0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. Tracer. 2004 version 1·3. Available from http://beast.bio.ed.ac.uk/Tracer . [Google Scholar]
- Renner SS. Relaxed molecular clocks for dating historical plant dispersal events. Trends in Plant Science. 2005;10:550–558. doi: 10.1016/j.tplants.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Richardson JE, Pennington RT, Pennington TD, Hollingsworth PM. Rapid diversification of a species-rich genus of neotropical rain forest trees. Science. 2001;293:2242–2245. doi: 10.1126/science.1061421. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rozefelds AC. Eucalyptus phylogeny and history: a brief summary. Tasforests. 1996;8:15–26. [Google Scholar]
- Rutschmann F, Eriksson T, Abu Salim K, Conti E. Assessing calibration uncertainty in molecular dating: the assignment of fossils to alternative calibration points. Systematic Biology. 2007;56:591–608. doi: 10.1080/10635150701491156. [DOI] [PubMed] [Google Scholar]
- Sanders KL, Lee MSY. Evaluating molecular clock calibrations using Bayesian analyses with soft and hard bounds. Biology Letters. 2007;3:275–279. doi: 10.1098/rsbl.2007.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Molecular Biology and Evolution. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ, Wojciechowski MF. Diversification rates in a temperate legume clade: are there ‘so many species’ of Astragalus (Fabaceae)? American Journal of Botany. 1996;83:1488–1502. [Google Scholar]
- Schmid R. A resolution of the Eugenia-Syzygium controversy (Myrtaceae) American Journal of Botany. 1972;59:423–436. [Google Scholar]
- Simmons MP, Pickett KM, Miya M. How meaningful are Bayesian support values? Molecular Biology and Evolution. 2004;21:188–199. doi: 10.1093/molbev/msh014. [DOI] [PubMed] [Google Scholar]
- Smith JF. High species diversity in fleshy-fruited tropical understorey plants. American Naturalist. 2001;147:646–653. doi: 10.1086/320625. [DOI] [PubMed] [Google Scholar]
- Snow DW. Tropical frugivorous birds and their food plants: a world survey. Biotropica. 1981;13:1–14. [Google Scholar]
- Sytsma KJ, Litt A, Zjhra ML, et al. Clades, clocks, and continents: historical and biogeographical analysis of Myrtaceae, Vochysiaceae, and relatives in the Southern Hemisphere. International Journal of Plant Sciences. 2004;165:S85–S105. [Google Scholar]
- Tiffney BH, Mazer SJ. Angiosperm growth habit, dispersal and diversification reconsidered Evolutionary. Ecology. 1995;9:93–117. [Google Scholar]
- Welch JJ, Fontanillas E, Bromham L. Molecular dates for the Cambrian explosion: the influence of prior assumptions. Systematic Biology. 2005;54:672–678. doi: 10.1080/10635150590947212. [DOI] [PubMed] [Google Scholar]
- Wilson PG, O'Brien MM, Gadek PA, Quinn CJ. Myrtaceae revisited: a reassessment of infrafamilial groups. American Journal of Botany. 2001;88:2013–2025. [PubMed] [Google Scholar]
- Wilson PG, O'Brien MM, Heslewood MM, Quinn CJ. Relationships within Myrtaceae sensu lato based on a matK phylogeny. Plant Systematics and Evolution. 2005;251:3–19. [Google Scholar]
- Yang Z, Rannala B. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Molecular Biology and Evolution. 2006;23:212–226. doi: 10.1093/molbev/msj024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.