Abstract
Backgrounds and Aims
Twenty-five genera having sterile inflorescence branches were recognized as the bristle clade within the x = 9 Paniceae (Panicoideae). Within the bristle clade, taxonomic circumscription of Cenchrus (20–25 species), Pennisetum (80–140) and the monotypic Odontelytrum is still unclear. Several criteria have been applied to characterize Cenchrus and Pennisetum, but none of these has proved satisfactory as the diagnostic characters, such as fusion of bristles in the inflorescences, show continuous variation.
Methods
A phylogenetic analysis based on morphological, plastid (trnL-F, ndhF) and nuclear (knotted) data is presented for a representative species sampling of the genera. All analyses were conducted under parsimony, using heuristic searches with TBR branch swapping. Branch support was assessed with parsimony jackknifing.
Key Results
Based on plastid and morphological data, Pennisetum, Cenchrus and Odontelytrum were supported as a monophyletic group: the PCO clade. Only one section of Pennisetum (Brevivalvula) was supported as monophyletic. The position of P. lanatum differed among data partitions, although the combined plastid and morphology and nuclear analyses showed this species to be a member of the PCO clade. The basic chromosome number x = 9 was found to be plesiomorphic, and x = 5, 7, 8, 10 and 17 were derived states. The nuclear phylogenetic analysis revealed a reticulate pattern of relationships among Pennisetum and Cenchrus, suggesting that there are at least three different genomes. Because apomixis can be transferred among species through hybridization, its history most likely reflects crossing relationships, rather than multiple independent appearances.
Conclusions
Due to the consistency between the present results and different phylogenetic hypotheses (including morphological, developmental and multilocus approaches), and the high support found for the PCO clade, also including the type species of the three genera, we propose unification of Pennisetum, Cenchrus and Odontelytrum. Species of Pennisetum and Odontelytrum are here transferred into Cenchrus, which has priority. Sixty-six new combinations are made here.
Keywords: Pennisetum, Cenchrus, Odontelytrum, Poaceae, phylogenetic analyses, ndhF, trnL-trnF, kn1, apomixis
INTRODUCTION
Morphological and molecular phylogenetic studies of the grass subfamily Panicoideae have shown that Pennisetum Rich. and Cenchrus L. are closely related genera within the bristle clade in tribe Paniceae (Gómez-Martínez and Culhman, 2000; Zuloaga et al., 2000; Duvall et al., 2001; Giussani et al., 2001; Kellogg et al., 2004; Bess et al., 2005; Doust et al., 2007; Donadío et al., 2009). This clade includes approximately 25 genera (Cenchrus, Ixophorus Schltdl., Paspalidium Stapf, Pennisetum and Setaria P.Beauv., among others), and is characterized by the presence of setae or bristles in the inflorescences, derived from inflorescence branch meristems (Doust and Kellogg, 2002; Bess et al., 2005).
Pennisetum and Cenchrus are distributed throughout tropical and subtropical regions of the Old and New World and contain 80–140 and 20–25 species, respectively (e.g. DeLisle, 1963; Türpe, 1983; Clayton and Renvoize, 1986; Crins, 1991; Watson and Dallwitz, 1992). Some species of Pennisetum are cultivated as cereal and forage grasses (e.g. P. purpureum Schumach. ‘elephant grass’, P. glaucum (L.) R.Br. ‘pearl millet’, P. clandestinum Hochst. ex Chiov. ‘kikuyu grass’) or ornamentals (e.g. P. setaceum (Forssk.) Chiov. ‘tender fountaingrass’, P. alopecuroides (L.) Spreng. ‘foxtail fountaingrass’), and some species of Cenchrus and Pennisetum are considered important weeds (e.g. C. ciliaris L. ‘buffel grass’, C. echinatus L. ‘southern sandbur’, C. myosuroides Kunth ‘big sandbur’ and P. polystachion (L.) Schult. ‘mission grass’) (DeLisle, 1963; Clayton and Renvoize, 1986; Watson and Dallwitz, 1992; Rúgolo de Agrasar and Puglia, 2004).
Pennisetum is not clearly distinguished from Cenchrus, and several species that are now included in Cenchrus have previously been assigned to Pennisetum. For example, P. ciliare is accepted by Chase (1921), Pohl (1980), Judziewicz (1990) and Wipff (2003), whereas it is treated under Cenchrus by DeLisle (1963), Clayton (1989), Pohl and Davidse (1994), Zuloaga and Morrone (2003), and Chen and Phillips (2006). The degree of fusion of the bristles is commonly used to separate these genera (Pilger, 1940; DeLisle, 1963; Clayton, 1972, 1989; Clayton and Renvoize, 1982, 1986; Filgueiras, 1984; Watson and Dallwitz, 1992). In most New World species of Cenchrus the degree of fusion is substantial, although there are several Old World species in which this distinction is less obvious (Crins, 1991). Other characters used to distinguish the genera are the presence of pedicellate spikelets and whether the bristles are flattened or stiff. However, none of these characters can be applied effectively to segregate the genera (Webster, 1988). Correll and Johnston (1970) treated Pennisetum under Cenchrus and presented several diagnostic features for the combined genus.
The species now included in Pennisetum (Stapf and Hubbard, 1934; Türpe, 1983; Wipff, 2001, 2003) have been previously placed in the genera Cenchrus, Gymnotrix P.Beauv., Holcus L., Panicum L., Penicillaria Willd. and Setaria. Most members of the genus are perennial. Distinctive characteristics of the genus include the shape and arrangement of the inflorescences, which are paniculate, contracted and spike-like, with fascicles of spikelets on reduced axes that disarticulate at maturity. The rachis terminates in a bristle; the bristles that subtend the spikelets are free, often plumose and disarticulate with the spikelets (Crins, 1991). Wipff (2003) characterized Pennisetum as having antrorsely scabrous bristles (not spiny) with fascicle axes that terminate in a bristle, and basic chromosome numbers of x = 5, 7, 8 or 9; he considered Cenchrus as having retrorsely scabrous, spiny bristles, fascicle axes that are terminated in a spikelet and a basic chromosome number of x = 7.
Authors have applied different criteria to subdivide Pennisetum. Stapf and Hubbard (1934) recognized five sections: Gymnotrix (P.Beauv.) Steud. (with two subsections, Acrostigma (Leeke) Stapf & C.E.Hubb. and Pleurostigma (Leeke) Stapf & C.E.Hubb.), Pennisetum, Penicillaria (Willd.) Benth & Hook.f. nom. superf., Heterostachya Schumach. and Brevivalvula Döll. Pilger (1940) recognized three subgenera: Dactylophora Leeke, Eriochaeta (Fig. & De Not.) Leeke (equivalent to section Brevivalvula) and Pennisetum; the latter was divided into sections Cenchropsis (Leeke) Pilg., Gymnotrix (with three subsections: Acrostigma, Beckeropsis (Fig. & De Not.) Pilg. and Pleurostigma) and Penicillaria. Brunken (1977) revised section Pennisetum and concluded that Penicillaria was a synonym of this type section. Finally, Clayton and Renvoize (1986) recognized only sections Brevivalvula, Dactylophora nom. inval., Gymnotrix (with the same subsections as Pilger), Heterostachya and Pennisetum. The differences between the sections are often weak (Schmelzer, 1997) and are based mainly on morphological characters of the inflorescence. Scholz (2006) established a new monotypic genus, Kikuyochloa, based on Pennisetum clandestinum. The inflorescences in Kikuyochloa are hidden in the leaf sheaths and the spikelets are arranged in simple units without a ring of basal, involucral bristles; the tiny bristles of the individual spikelets never completely encircle the spikelets.
Several cereals or forage species of Pennisetum are very important resources for food (e.g. P. glaucum). A phylogenetic approach and information relating to the ploidy of the species could prove useful in clarifying the relatedness of species without the necessity of crossing, in particular when species are difficult to cultivate. Using internal transcribed spacer (ITS) data, Martel et al. (2004) placed two wild forms of Pennisetum glaucum (L.) R.Br. subsp. monodii (Maire) Brunken in the primary gene pool of domesticated P. glaucum (diploid, x = 7), where the primary gene pool is defined as taxa that are as easy to cross as within the same species (Harlan and de Wet, 1971). In the secondary gene pool (coenospecies where gene transfer is possible, but fertile hybrids are difficult to obtain) they placed P. purpureum (tetraploid, x = 7). All remaining species of Pennisetum were in the tertiary gene pool (species for which hybrids with the crop would be difficult to obtain and maintain, representing the extreme outer limit of potential genetic exchange; cf. Harlan and de Wet, 1971).
Several basic chromosome numbers have been reported for Cenchrus (x = 9, 10, 17) and Pennisetum (x = 5, 7, 8, 9, 17; Table 1). The genera are included in the x = 9 Paniceae clade, and the basic chromosome number x = 9 is plesiomorphic within the bristle clade and the Pennisetum–Cenchrus clade (Giussani et al., 2001; Martel et al., 2004; Donadío et al., 2009). Although diploids are not rare among species of Cenchrus and Pennisetum, polyploid and aneuploid numbers are common within both genera (Table 1). A high frequency of univalents or multivalent associations in metaphase I (e.g. Sisodia, 1970), lagging chromosomes in anaphase I (Dujardin and Hanna, 1984a) and anomalies in meiosis provide evidence of outbreeding in polyploid species of Pennisetum (P. polystachion, P. pedicellatum Trin., P. squamulatum Fresen.) and Cenchrus (C. ciliaris, C. incertus M.A.Curtis, C. setigerus Vahl.). In this context, consistency and incongruence among plastid (maternal) and nuclear (biparental) phylogenetic analyses should help to elucidate the origin of allopolyploids (e.g. P. pedicellatum, P. polystachion).
Table 1.
Chromosome numbers and reproductive behaviour reported for the species included in the analyses
| Species | n | 2n | x | Reproductive behaviour |
|---|---|---|---|---|
| Cenchrus agrimonioides | – | – | – | – |
| C. brownii | 34(14) | 34(48), 36(26)(24), 70(23) | 9*, 17(14)(14) | – |
| C. caliculatus | 34(30) | – | 17* | – |
| C. ciliaris | 16(18), 17(43), 18(18)(22) | 34(23), 36(16)(24), 45(32), 54(26), 63(32), 78(23), 90(32) | 9(43) | APO, SEX(53) |
| C. echinatus | 34(14)(18) | 68 (27)(45)(25)(23), 70(23) | 17(18)(22) | – |
| C. incertus | 16(22), 17(18)(14) | 30(22), 34(24)(14)(48) | 17(14)(18) | SEX(59) |
| C. myosuroides | 27(6), 35(18) | 54(6), 70(3)(23)(50) | 9, 10(17) | SEX(59) |
| C. pilosus | 17(14) | 34(14)(48) | 17(14)(14) | – |
| C. setigerus | 17(43), 18(47) | 34(47), 36(46), 37(47) | 9(43), 17(43) | APO(46) |
| Ixophorus unisetus | – | 34(49) | 17* | – |
| Odontelytrum abyssinicum | – | – | – | – |
| Paspalidium geminatum | 9(43) | 18(56), 36(24) | 9(43) | – |
| Pennisetum sect. Brevivalvula† | ||||
| P. hordeoides | 9(51) | 36(51), 54(51) | 9(1) | APO(53) |
| P. pedicellatum | – | 24(60), 30(60), 32(60), 35(60), 36(60), 42(60), 45(51), 48(60), 53(60), 54(36)(51) | 9(36)(51) | APO(36)(53) |
| P. polystachion subsp. polystachion | 18(13), 36(12) | 18(51), 24(60), 32(60), 36(51), 45(51), 48(60), 52(60), 53(48), 54(36)(51)(13)(27), 56(60), 63(60), 78(23) | 9(36)(51) | APO(20)(53) |
| P. polystachion subsp. atrichum | – | 36(7) | 9* | – |
| Pennisetum sect. Dactylophora† | ||||
| P. lanatum | 18(43) | – | 9(43) | – |
| Pennisetum sect. Gymnotrix subsect. Acrostigma† | ||||
| P. alopecuroides | – | 18(36)(54) | 9(36) | SEX(36) |
| P. basedowii | – | 54(20) | 9* | – |
| P. chilense | – | – | – | – |
| P. frutescens | – | 63(45) | 9(45) | APO(53) |
| P. glaucocladum | – | – | – | – |
| P. macrourum | – | 36(20) | 9(46) | APO(20)(53) |
| P. massaicum | – | 16(46), 32(55) | 8(46) | APO, SEX(53) |
| P. mezianum | – | 16(52), 32(36) | 8(36, 52) | APO(36) |
| P. natalense | – | – | – | – |
| P. nervosum | – | 36(45), 72(48) | 9(45) | – |
| P. ramosum | – | 10(36)(52)(55) | 5(36)(52)(55) | APO, SEX(36) |
| P. sphacelatum | 18(32) | – | 9* | – |
| P. thunbergii | – | 18(16) | 9* | – |
| Pennisetum sect. Gymnotrix subsect. Beckeropsis† | ||||
| P. montanum | 16(31) | – | 8(31) | – |
| P. unisetum | – | 18(16) | 9* | – |
| Pennisetum sect. Gymnotrix subsect. Pleurostigma† | ||||
| P. latifolium | – | 36(45) | 9(45) | APO(53) |
| P. trachyphyllum | – | – | – | – |
| P. tristachyum | – | – | – | – |
| Pennisetum sect. Heterostachya† | ||||
| P. schweinfurthii | – | 14(36)(52) | 7(36)(52) | SEX(36) |
| P. squamulatum | – | 54(36), 56(2) | 7(2), 9(20)(36) | APO(20)(36)(53) |
| Pennisetum sect. Pennisetum† | ||||
| P. clandestinum | 18(38), 27(34) | 36(29)(45)(56) | 9(29) | APO, SEX(46) |
| P. flaccidum | 9(34), 27(43) | 18(46), 36(9)(46) | 9(43) | APO(46), SEX(53) |
| P. foermeranum | – | – | – | – |
| P. glaucum | 7(43) | 14(8)(36) | 7(36)(43) | SEX(36) |
| P. orientale | 9, 18(43) | 36(36)(29), 45(40) | 9(36) | APO(36)(53), SEX(53) |
| P. purpureum | 7(1), 28(34) | 27(26), 28(36)(8)(33), | 7(36)(45) | APO(59), SEX(36) |
| P. setaceum | – | 27(36)(29), 54(36), 68(58) | 9(36), 17(58) | APO(53) |
| P. sieberianum | – | – | – | – |
| P. villosum | – | 18(46), 27(46), 36(36), 45(45), 54(45) | 9(36) | APO(36)(53) |
| P. violaceum | – | 14(36) | 7(36) | SEX(36) |
| Pseudoraphis paradoxa | – | – | – | – |
| P. spinescens | – | 32(27) | 8* | – |
| Rupichloa acuminata | 13(42) | – | 13(42) | – |
| Setaria palmifolia | 27(41)(38) | 54(27) | 9* | – |
| S. sphacelata | 9(28), 18(28), 27(28) | 36(5), 54(28) | 9* | SEX(59) |
| S. parviflora | 18(28)(38) | 36(25)(15) | 9* | – |
| Spinifex sericeus | – | 18(57) | 9(57) | – |
| Stenotaphrum secundatum | – | 18(45)(6)(24), 36(27) | 9(45) | SEX(59) |
References: (1) Akenova and Chehheda (1981), (2) Akiyama et al. (2006), (3) Avdulov (1931), (4) Bir and Sahni (1986), (5) Bir and Sahni (1987), (6) Brown (1950), (7) Brunken (1979), (8) Burton (1942), (9) Chatterji and Timothy (1969), (10) Chopanov and Yurtsev (1976), (11) Christopher and Abraham (1976), (12) Davidse and Pohl (1972), (13) Davidse and Pohl (1978), (14) Davidse and Pohl (1974), (15) de Wet (1954), (16) de Wet (1960), (17) DeLisle (1963), (18) DeLisle (1964), (19) Dujardin (1979), (20) Dujardin and Hanna (1984b), (21) Emery (1957), (22) Gould (1958), (23) Gould (1965), (24) Gould (1968), (25) Gould and Soderstom (1967), (26) Gould and Soderstrom (1970), (27) Gould and Soderstrom (1974), (28) Gupta and Singh (1977), (29) Hrishi (1952), (30) Hsu (1972), (31) Hunziker et al. (1998), (32) Jensen et al. (1989), (33) Kammacher et al. (1973), (34) Khosla and Mehra (1973), (35) Khosla and Sharma (1973), (36) Martel et al. (1997), (37) Miège (1962), (38) Mehra (1982), (39) Mehra and Rememanandan (1973), (40) Mehra and Sharma (1973), (41) Mehra and Sharma (1975), (42) Morrone et al. (1995), (43) Ahsan et al. (1994), (44) Norrmann et al. (1994), (45) Núñez (1952), (46) Ozias-Akins et al. (2003), (47) Crins (1991), (48) Pohl and Davidse (1971), (49) Reeder (1967), (50) Reeder (1968), (51) Renno et al. (1995), (52) Rao et al. (1989), (53) Schmelzer (1997), (54) Sinha et al. (1990), (55) Swaminathan and Nath (1956), (56) Tateoka (1965), (57) Connor (1984), (58) Shanthamma (1979), (59) Brown and Emery (1958), (60) Sisodia (1970).
SEX = sexual; APO = apomictic.
* Inferred basic chromosome number.
† Sectional and subsectional treatment sensu Clayton and Renvoize (1986).
Apospory is a mode of asexual reproduction in which a gametophyte develops directly from diploid cells of the sporophyte without meiosis (Gustafsson, 1946). This apomictic mode is well documented in Panicoideae (Brown and Emery, 1958), being frequent in several species of Cenchrus and Pennisetum (Ozias-Akins et al., 2003; Ozias-Akins, 2006), and its developmental pattern has been well studied in Cenchrus ciliaris and Pennisetum squamulatum (Dujardin and Hanna, 1984a; Ozias-Akins et al., 1998; Wen et al., 1998). Evidence for transmission of apomixis by a single chromosome was reported by Ozias-Akins et al. (1993), and subsequent studies have identified molecular markers linked to apomixis (Ozias-Akins et al., 2003). An apospory-specific genomic region (ASGR) (Ozias-Akins et al., 1998), located on a single chromosome, is necessary and sufficient for the expression of apomixis in polyploid taxa (Goel et al., 2003). Also, at least two genes, Pca21 and Pca24, were identified to play a role during apomictic development in Pennisetum ciliare or Cenchrus ciliaris (Singh et al., 2007), although they can be inherited independently of the ASGR.
Previous phylogenetic studies showed that Pennisetum and Cenchrus form a strongly supported monophyletic group (Martel et al., 2004; Doust et al., 2007; Donadío et al., 2009). Donadío et al. (2009), using two plastid markers and including almost 20 and seven species of Pennisetum and Cenchrus, respectively, found the former to be polyphyletic. However, if P. lanatum is excluded, Pennisetum is paraphyletic with all the species of Cenchrus nested within it. Doust et al. (2007) developed primers for a novel single copy nuclear marker that comprises two introns and three exons of the knotted1 (kn1) gene, and also obtained sequences of ndhF for four species of Pennisetum, six species of Cenchrus and some other members of the bristle clade. In their analyses, Cenchrus formed a monophyletic group derived from within a paraphyletic Pennisetum. Martel et al. (2004), using ITS sequences of nuclear ribosomal DNA and including 13 species of Pennisetum and only one species of Cenchrus, found C. ciliaris embedded within Pennisetum. Recently, when studying the phylogenetic relationships of Setaria and related genera of the bristle clade, Kellogg et al. (2009) also placed Odontelytrum abyssinicum Hack. in a clade with Pennisetum and Cenchrus. Odontelytrum has only a single species, in which an herbaceous involucre that subtends the spikelet may be homologous to the bract-like cupule in Cenchrus and Pennisetum.
Based on the resolution previously reported for various genetic markers (Doust et al., 2007; Donadío et al., 2009), two plastid markers (the trnL-F region, comprising the trnL intron and trnL-F spacer, and the ndhF gene), and one nuclear marker (knotted1) were selected here to study relationships among Cenchrus, Pennisetum and genera of the bristle clade. In addition, a morphological matrix was used in a combined analysis. Basic chromosome numbers and reproductive characters were optimized on the resulting phylogenetic trees. The present goals were (1) to test the monophyly and circumscription of Pennisetum, Cenchrus and allied genera; (2) to assess the validity of the traditional taxonomic divisions of the genus Pennisetum; and (3) to interpret the role that apomixis, polyploidy and hybridization may have played in the evolution of Pennisetum and Cenchrus.
MATERIALS AND METHODS
Sampling
For the plastid markers, 51 species (a total of 53 specimens) were sampled, representing nine species of Cenchrus (56 % of the total number of species according to Clayton and Renvoize, 1986), 32 of Pennisetum (43 %) and Odontelytrum abyssinicum. Species representing different genera of the bristle clade were used as outgroups: Ixophorus unisetus, Paspalidium geminatum, Setaria palmifolia, S. parviflora, S. sphacelata and Stenotaphrum secundatum; Rupichloa acuminata was used as functional outgroup (Appendix 1). Of the 49 sequences for the trnL-F region, 23 were generated and 26 were downloaded from GenBank. For ndhF, 34 sequences of the 53 were generated for this study and 19 were added from GenBank (Appendix 1).
For the nuclear marker (knotted1), 42 taxa were studied: eight species belonged to Cenchrus and 24 to Pennisetum. Ixophorus unisetus, Paspalidium jubiflorum, Pseudoraphis paradoxa, P. spinescens, Setaria palmifolia, S. parviflora, S. sphacelata, Spinifex sericeus and Stenotaphrum secundatum were used as outgroups. Trees were rooted using Panicum miliaceum. Seventy-one sequences were generated in this study, and 33 sequences belonging to the ingroup and 27 to the outgroups were obtained from GenBank (Appendix 1).
DNA extraction and sequencing
The material used for DNA extraction was obtained from plants collected in wild populations and dried in silica gel or grown from seeds provided by Plant Gene Resources of Canada (CN), US Department of Agriculture (PI) and the Missouri Botanical Garden (MO) (Appendix 1). DNA extraction was via the modified CTAB protocol of Doyle and Doyle (1987), adapted for small amounts of plant material. When fresh material was not available, DNA was extracted from herbarium specimens using a Dneasy Plant Mini Kit (QIAGEN Inc., Hilden, Germany).
Genomic DNA was used as a template to amplify (by PCR) a plastid fragment containing the trnL (UAA) intron and the intergenic spacer between the trnL (UAA) 3′ exon and the trnF (GAA) gene (trnL-F region) and the ndhF gene encoding a subunit of the respiratory-chain NADH dehydrogenase. A fragment of the single copy nuclear marker knotted1 (kn1), comprising two introns and three exons, was also amplified (see fig. 2 in Doust et al., 2007). ndhF was amplified in three to five overlapping fragments, depending on the difficulty of amplification and the quality of the leaf material, using the following primers: 5F, 536F, 536R, 972F, 972R, 1318F, 1318R and 3R from Olmstead and Sweere (1994), 1660F and 1660R from Aliscioni et al. (2003), and 1821F and 1821R from Clark et al. (1995). The intron and the intergenic spacer from the trnL-F region were amplified using the primers c and f from Taberlet et al. (1991) and primers Cii and Fdw from Giussani et al. (2009). kn1 was amplified in one fragment with a nested PCR approach, using as a first set of primers kn1-345F and kn1-622R from Doust et al. (2007) and a second set of primers, designed specifically for this paper, kn1-nestF (YGAGTGCCRGAAGGCAAGTA), kn1-nest3R (ATRTTGGCGCAGCGATCTG) and kn1-nestR (YCTCGTCRGYTCCTCYCTGA).
Fig. 2.
A single tree depicting relative branch lengths for one of the 2316 MPTs based on a combined analysis using two plastid markers: ndhF and trnL-F.
PCR reactions were performed in a final volume of 25 µL. Each reaction contained between 50 and 100 ng of DNA, 1·5 units of Taq polymerase (Invitrogen Life Technologies, São Paulo, Brazil), 1× PCR buffer, 5 mm MgCl2, 0·2 µm of each primer and 0·025 mm dNTP each. In species for which these protocols were unsuccessful, 0·4 % bovine serum albumin and 1·6 % dimethyl sulfoxide were included as additives and enhancing agents to increase the yield of PCR reactions. PCR amplifications followed the following programme: a first denaturation period at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 48 °C for 1 min and extension at 72 °C for 90 s. Final extension at 72 °C for 6 min terminated the reactions. The annealing temperature was varied in some cases to improve amplification.
For kn1, sequences from direct PCR products were used only when they had <3 % polymorphisms as indicated by double peaks in the chromatograms. However, for most species, kn1 was cloned before sequencing. When possible, at least five clones were sequenced per accession. PCR reactions were run out on a 1 % Tris-borate-EDTA (TBE) agarose gel, the bands of DNA were excised, purified using the QIAquick Gel Extraction Kit Protocol (QIAGEN Inc.) and cloned using the PGEM-T Easy Vector system (Promega Corp., Madison, WI, USA). Colonies were picked and incubated overnight in liquid Luria–Bertani (LB) medium. For checking the insert, plasmids were extracted and incubated with EcoRI at 37 °C for 2 h. Digestions were electrophoresed on a 1 % TBE agarose gel stained with ethidium bromide, and colonies that had incorporated the plasmid were re-grown in liquid LB medium. Plasmids for sequencing were extracted using the QIAprep Miniprep protocol (QIAGEN Inc.).
PCR products were sequenced by Macrogen, Inc. (Seoul, Korea). PCR products were cleaned using a Montage PCR purification kit from Millipore following the manufacturer's protocol, and sequencing reactions used ABI PRISM BigDyeTM Terminator Cycle Sequencing Kits with AmpliTaq DNA polymerase (Applied Biosystems, Seoul, Korea). Single-pass sequencing was performed on each template using the same primers used for PCR reactions (see above), as well as primers e and d from Taberlet et al. (1991) for the trnL-F region. Unincorporated terminators were removed by ethanol precipitation. The samples were resuspended in distilled water and subjected to electrophoresis in an ABI PRISM 3730XL sequencer (96-capillary type; Applied Biosystems).
Assembly and editing of sequences used the program Chromas Pro ver. 1·34 (Technelysium Pty, Ltd, Tewantin, Australia). Sequences of ndhF and kn1 were aligned manually following alignments performed by Aliscioni et al. (2003) and Doust et al. (2007), respectively. Sequences of the trnL-F region were aligned using the program DIALIGN at BiBiServ (http://bibiserv.techfak.uni-bielefeld.de/dialign/) (Morgenstern, 2004). DIALIGN is a program that compares complete segments of sequences, instead of relying on the sum of individual similarity values or on gap penalties as optimization criteria; it is thus able to establish small conserved regions that cannot be detected by other alignment programs (Morgenstern et al., 1998). The alignment was then adjusted manually. Voucher information and GenBank accession numbers are provided in Appendix 1. Alignments and phylogenetic trees were submitted to TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S10252).
Morphological characters
Fifty-one morphological characters were scored and used in the phylogenetic analyses (Appendix 2). Characters were taken from direct examination of herbarium specimens (AAU, CAMB, K, PRE and SI; Appendix 3), and in some cases information was obtained from the literature (Türpe, 1983; Clayton et al., 2006). Three to 11 specimens (average, five) were measured per species including, when possible, the specimen used to obtain DNA. The matrix is presented as Supplementary Data, available online, and has been submitted to TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S10252).
Phylogenetic analyses
Molecular analyses of the plastid markers (trnL-F region and ndhF) were performed separately and in combination. A morphological analysis was conducted separately, and then a combined analysis of morphology, trnL-F and ndhF was also performed. The nuclear marker (kn1) was analysed independently using several clones per species.
All analyses were conducted using the program TNT version 1·1 (Goloboff et al., 2008), with all characters equally weighted and considered unordered. Gaps were scored as missing data. In all analyses, parsimony-uninformative characters were deactivated. Heuristic searches were performed using 1000 random addition replicates and tree bisection–reconnection (TBR) branch swapping, saving ten trees per replicate. Thereafter, a new search with TBR branch swapping was performed using the shortest trees saved in memory. A strict consensus tree was obtained with all shortest trees found during searches.
Branch support was assessed with 10 000 parsimony jackknife (JK) replicates (Farris et al., 1996), using ten series of random addition sequences, swapped using TBR and holding two optimal trees per series. Clades were considered to have strong branch support when JK ≥ 90 %; moderate support, JK ≥ 75 % to <90 %; low support, JK ≤74 %.
Optimization of morphological, cytological and reproductive characters was performed on all most-parsimonious trees (MPTs) obtained in the plastid analyses and combined analysis, using the command ‘Common Synapomorphies’ of TNT (Goloboff et al., 2008), by which the optimization shared by all MPTs is represented in the consensus diagram.
RESULTS
Plastid analysis
The total length of the amplified trnL-F region ranged from 803 to 888 bp. The aligned matrix consisted of 941 characters, only 39 of which were potentially phylogenetically informative. The matrix included 3·22 % missing data (gaps not included). Four species had >10 % missing data; of these P. lanatum had 25 %. When aligning the 53 ndhF sequences, only one gap of 6 bp was introduced by Setaria sphacelata, producing a matrix length of 2055 characters. The total proportion of missing positions was 2·79 %. Only Pennisetum glaucocladum and P. hordeoides had >20 % missing data. A total of 116 characters were potentially parsimony-informative.
There was no contradiction in the placement of the ingroup taxa between the ndhF and the trnL-F datasets when analysed separately, and several moderately and strongly supported clades were recovered in both analyses. Hence, partitions were assumed to be congruent and they were analysed together. The combined data matrix consisted of 53 specimens and 2996 characters in total, with 155 potentially parsimony-informative characters. Only four species could not be amplified for trnL-F (Cenchrus agrimonioides, C. caliculatus, Pennisetum natalense and P. sphacelatum); however, when excluding these species from the analysis, neither the topology of the consensus tree nor the branch support varied significantly, and hence these species were included in the combined analysis.
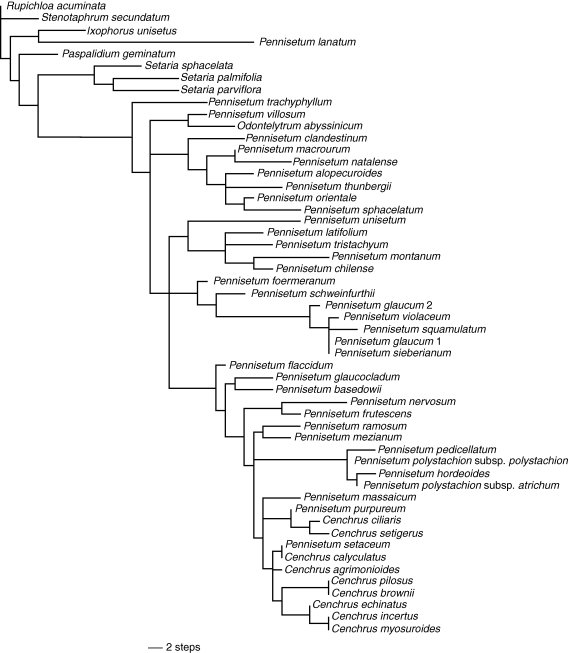
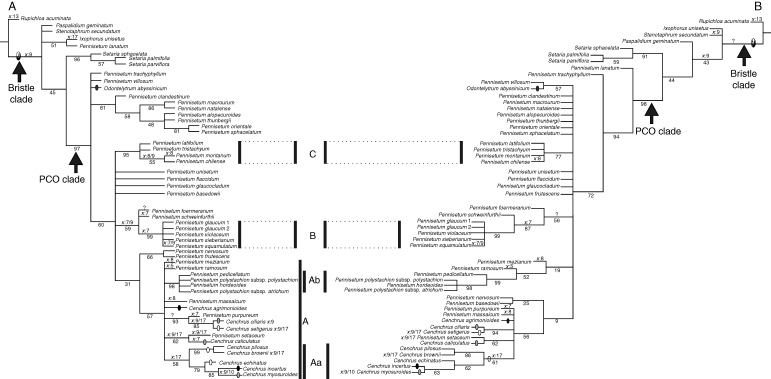
The combined plastid analysis produced 2316 trees of 342 steps (CI: 0·53, RI: 0·80); the trees from the combined analysis are congruent with those from the independent analyses (Fig. 1A). Figure 2 shows one of the MPTs with branch lengths drawn to scale. The consensus tree shows Pennisetum, Cenchrus and Odontelytrum in a strongly supported clade, the ‘PCO clade’ with JK = 97 % (Figs 1A and 2). Only P. lanatum is excluded from the PCO clade; it is placed in a clade with Ixophorus unisetus (Figs 1A and 2) with only weak support (JK = 51 %). This clade does not appear in the trnL-F analysis, in which P. lanatum is placed in a polytomy outside the PCO clade (tree not shown). Within the PCO clade, Odontelytrum, P. villosum, P. trachyphyllum and a clade of seven species of Pennisetum (P. alopecuroides, P. clandestinum, P. macrourum, P. natalense, P. orientale, P. sphacelatum and P. thunbergii) form a polytomy with the remaining species of Pennisetum and Cenchrus (Fig. 1A).
Fig. 1.
Phylogenetic relationships of Pennisetum, Cenchrus and Odontelytrum. (A) Strict consensus tree of 2316 MPTs from the combined analysis of the plastid markers (ndhF + trnL-F). (B) Strict consensus tree of the 3264 MPTs from the combined analysis (ndhF + trnL-F + cytology and morphology). Numbers below branches represent jackknife branch support. Optimization of the basic chromosome number (Appendix 2, character 1) is shown above the branches and, when necessary, next to the species names. Optimization of the degree of fusion of the bristles (Appendix 2, character 21) is shown as follows: black and white oval, not connate; grey oval, connate below; white oval, connate up to half the total length; black oval, connate up to two-thirds the total length. Bars represent principal clades as discussed in the text. Pennisetum glaucum 1 corresponds to voucher PI 326520 (sequences downloaded from GenBank) and P. glaucum 2 to voucher Caxambu 375 (see Appendix 1).
The relationships among the species of Cenchrus are weakly supported in clade A (JK = 57 %) together with several species of Pennisetum. Clade A includes all sampled species of Cenchrus, plus P. hordeoides, P. massaicum, P. mezianum, P. pedicellatum, P. polystachion subsp. atrichum, P. polystachion subsp. polystachion, P. purpureum, P. ramosum and P. setaceum (Fig. 1A). Within clade A, five species of Cenchrus form a clade (subclade Aa) that comprises C. pilosus and C. brownii (strongly supported as sister taxa: JK = 99 %), which are sister to a clade with C. echinatus, C. myosuroides and C. incertus (JK = 79 %). Cenchrus caliculatus is related to P. setaceum (JK = 62 %), and C. ciliaris and C. setigerus are closely related (JK = 85 %) with P. purpureum as their sister group (JK = 93 %). The other strongly supported group within clade A comprises P. hordeoides, P. pedicellatum, and both subspecies of P. polystachion (subclade Ab; JK = 98 %) (Fig. 1A).
Clade B is here represented by P. glaucum (both specimens), P. sieberianum, P. squamulatum and P. violaceum; all these species form a strongly supported group (JK = 99 %). Clade C includes P. chilense, P. latifolium, P. montanum and P. tristachyum (JK = 95 %).
Morphological analysis
The parsimony analysis of 51 morphological characters yielded 249 trees of 345 steps (CI: 0·27, RI: 0·59). The strict consensus tree showed little resolution, and nodes were poorly supported (tree not shown). However, the PCO clade itself is not strongly contradicted and has moderate support (JK = 81 %). Pennisetum lanatum, contrary to the molecular results, is situated in a polytomy near the base of the PCO clade.
Combined analysis of morphology and plastid data
Combining the three datasets (morphology, trnL-F and ndhF), the analysis yielded 3264 trees of 763 steps with CI = 0·35 and RI = 0·65 (Fig. 1B). When compared with the results from the plastid data alone, support for most of the branches is diminished, probably due to the conflict added by the morphological characters (Fig. 1A, B). The molecular characters dominate the analysis, as the consensus tree of the combined analysis has more nodes in common with the molecular analysis than with the morphological one. However, P. lanatum, similar to the morphological analysis, is included as an early branching taxon in the PCO clade (JK = 98 %).
Lettered clades (A, Aa, Ab, etc.) correspond to clades as identified in the combined plastid trees. The combined analysis recovered subclade Aa (JK = 61 %). This subclade is supported by a basic chromosome number of x = 17, although this character reversed in C. brownii and C. myosuroides (x = 9, 10; Table 1). Two morphological characters support this clade: the bristles are fused (char 21, 0 > 2) and the upper glume is almost as long as the spikelet (char 31, 2 > 3).
Subclade Ab (P. hordeoides, P. pedicellatum and both subspecies of P. polystachion), clade B (both specimens of Pennisetum glaucum, P. sieberianum, P. squamulatum and P. violaceum) and clade C (P. chilense, P. latifolium, P. montanum and P. tristachyum), as in the combined plastid analysis, are also moderately to strongly supported in the combined analysis (Fig. 1A, B). Subclade Ab is supported by a single morphological synapomorphy which is the coriaceous consistency of the upper lemma (char 38, 1 > 2). Species of clade B are characterized by eight morphological synapomorphies: the upper glume is vestigial, and consequently is reduced in size (char 30, 2 > 1; char 31, 1 > 0), the apex of the lower lemma is scaberulous (char 37, 1 > 2), the apex of the upper lemma is not acuminate (char 39, 1 > 0/2) and is ciliate (char 40, 1 > 0), the apex of the upper palea is tridentate (char 44, 1 > 4) and ciliate (char 45, 1 > 0), and anther tips are penicillate (char 49, 1 > 0). Pennisetum schweinfurthii is the sister taxon to clade B (JK = 87 %); this relationship is supported by a basic chromosome number of x = 7, and two morphological characters: a rounded apex of the lower lemma (char 36, 1 > 2) and connate styles (char 48, 1 > 0).
Clade C is characterized by two morphological characters: a membranous-ciliate ligule (char 2, 1 > 2) and an acute apex of the upper palea (char 44, 1 > 0).
Knotted1 – nuclear data
In all, 131 sequences were used. Pennisetum clandestinum and P. sieberianum were sequenced directly, whereas for most other species three to five clones were recovered; only one or two clones were obtained for P. frutescens, P. montanum and P. violaceum (Appendix 1). It was not possible to amplify kn1 from the available herbarium material of Odontelytrum, so its placement is unknown. The total length of the aligned matrix was 817 bp; sequences varied from 602 bp in one clone of Pennisetum chilense to 747 bp in a clone of Pseudoraphis spinescens.
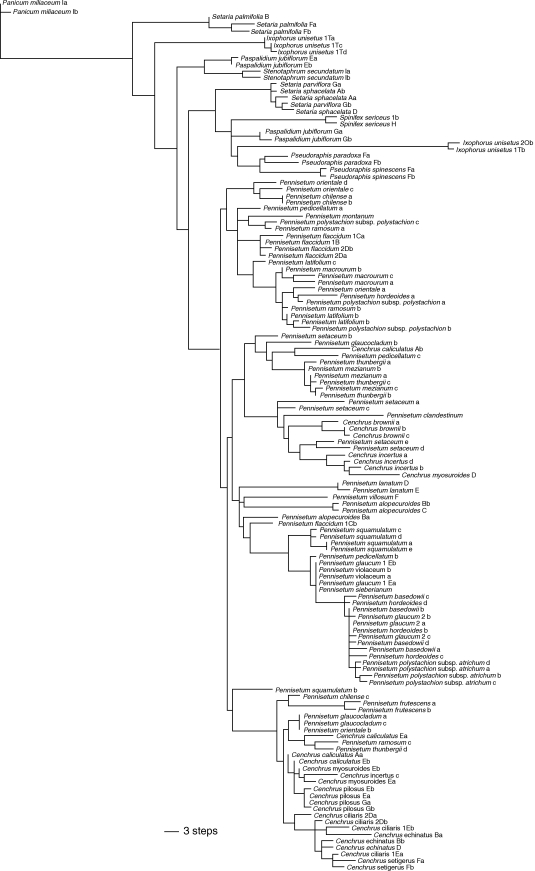
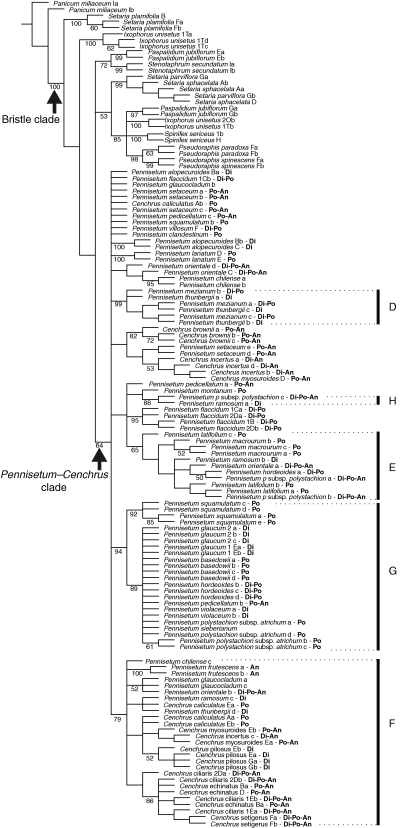
In total, 315 characters were potentially parsimony-informative, and the analysis was stopped when it found 30 000 trees (maximum saved) of 833 steps (CI = 0·53, RI = 0·82); one of the MPTs with branch lengths drawn to scale is shown in Fig. 3. Although the consensus of the 30 000 MPTs revealed several basal polytomies including individual clones, or some clones of the same species were represented in more than one clade, it was possible to recover several major strongly supported clades (Fig. 4).
Fig. 3.
A single tree depicting relative branch lengths for one of the 30 000 MPTs based on the nuclear marker knotted1.
Fig. 4.
Strict consensus tree of 30 000 MPTs obtained from parsimony analysis of the nuclear marker knotted1. Numbers below the branches represent jackknife branch support. Letters following the species names represent different clones; letters in bold represent the reported ploidy/ies: Di, diploid; An, aneuploid; Po, polyploid. Bars represent principal clades as discussed in the text. Pennisetum glaucum 1 corresponds to voucher PI 326520 (sequences downloaded from GenBank) and P. glaucum 2 to voucher Caxambu 375 (see Appendix 1).
All species of Cenchrus and Pennisetum, including P. lanatum, form a monophyletic group (JK = 64 %). Other major well-supported clades revealed by the analysis are: clade D (JK = 99 %), which includes three of four clones of P. thunbergii (3/4) and all clones of P. mezianum (3/3); clade E (JK = 65 %) with all clones of P. latifolium (3/3) and P. macrourum (3/3) and separate clones of P. ramosum (1/3), P. orientale (1/4), P. hordeoides (1/4) and P. polystachion (2/3); clade F (JK = 79 %), which represents a subset of the species included in clade A (Fig. 1A), with C. caliculatus (3/4), C. ciliaris (4/4), C. echinatus (3/3), C. incertus (1/4), C. myosuroides (2/3), C. pilosus (4/4), C. setigerus (2/2), P. frutescens (2/2) and P. ramosum (1/3), but also including clones of P. chilense (1/3), P. glaucocladum (2/3), P. orientale (1/4) and P. thunbergii (1/4). Clade G (JK = 94 %) grouped all species included in clade B (Fig. 1A, B): P. glaucum (PI 326520, sequences downloaded from GenBank) (2/2) + P. glaucum (Caxambu 375) (3/3), P. squamulatum (4/5), P. violaceum (2/2) and P. sieberianum (1) and representatives of subclade Ab: P. hordeoides (3/4), P. polystachion subsp. atrichum (4/4), P. pedicellatum (1/3) and P. basedowii (4/4). A minor clade, but strongly to moderately supported, included only two clones, P. polystachion (1/3) and P. ramosum (1/3), and will be referred herein as clade H (JK = 88 %).
Comparison of nuclear vs. plastid trees
Pennisetum lanatum
The position of P. lanatum was in disagreement among data partitions. When using the morphological partition alone, P. lanatum is included in the PCO clade (although with little support), whereas the plastid markers place P. lanatum outside. However, the combined analysis included P. lanatum within the PCO clade (Fig. 1A, B).
Clades A–F (Figs 1 and 4)
Many of the species in the plastid clade A (Fig. 1) appear in the nuclear clade F (Fig. 4). Clade F includes almost all species of Cenchrus, except C. brownii, and P. frutescens, P. glaucocladum, P. thunbergii, P. ramosum, P. chilense and P. orientale. Clones of these species are also related to other groups: P. thunbergii shares at least one genome with species of clade D, whereas P. ramosum is allied to species of clades E and H. Meanwhile, clones of P. chilense and P. orientale fall in a polytomy at the base of the Pennisetum–Cenchrus clade.
Cenchrus incertus and C. myosuroides are part of clade F, although several clones are related to C. brownii and Pennisetum setaceum in a polytomy separated from clade F. The plastid and combined phylogenetic analyses show four x = 17 Cenchrus species (C. pilosus, C. brownii, C. echinatus and C. incertus) and C. myosuroides (x = 9, 10) together in subclade Aa. Two clones of P. setaceum are closely related to clones of C. brownii, C. incertus and C. myosuroides in the consensus tree of kn1.
Four of six taxa of Pennisetum section Brevivalvula were grouped in subclade Ab in the plastid and combined analyses: P. polystachion subsp. polystachion, P. polystachion subsp. atrichum, P. hordeoides and P. pedicellatum (Fig. 1A, B). The relationship among these species suggests that they share a common genome, as shown by the plastid markers. However, species of subclade Ab present a reticulate pattern of relationships in the nuclear analysis (kn1). Different clones of P. polystachion subsp. polystachion linked this taxon to P. ramosum (clade H, Fig. 4) and to species of clade E (P. hordeoides, P. latifolium, P. macrourum and P. orientale).
Clade D (Fig. 4)
All clones of Pennisetum mezianum and most clones of P. thunbergii (3/4) are closely related in the nuclear phylogenetic tree in clade D, whereas a single clone of P. thunbergii is included in clade F. However, in the plastid analysis, P. mezianum and P. massaicum are included in clade A and P. thunbergii is distantly related and included in a weakly supported clade with P. alopecuroides, P. macrourum, P. orientale and P. sphacelatum (Fig. 1A). Pennisetum mezianum and P. massaicum are morphologically similar and have a basic chromosome count of x = 8 (Table 1).
Clades C–E (Figs 1 and 4)
All kn1 clones of P. macrourum (3/3), P. latifolium (3/3) and the majority of clones of P. polystachion subsp. polystachion (2/3) are present in clade E together with single clones of P. hordeoides, P. orientale and P. ramosum (Fig. 4). However, these species have apparently acquired their plastids from disparate sources and are distantly related in the plastid phylogenetic tree (Fig. 1A): P. macrorum is sister to P. natalense, the latter not included in the nuclear tree, and P. chilense, P. latifolium, P. montanum, and P. tristachyum (the last of these not included in kn1) are closely related in the strongly supported clade C. Meanwhile, P. polystachion is included in the strongly supported subclade Ab with P. polystachion subsp. atrichum, P. hordeoides and P. pedicellatum.
Clades B–G (Figs 1 and 4)
Clade G includes all species of clade B from the plastid and combined phylogenetic analyses: P. glaucum, P. sieberianum, P. violaceum and P. squamulatum, plus species corresponding to subclade Ab in part: P. hordeoides, P. pedicellatum, P. polystachion subsp. atrichum and P. basedowii.
Clade H (Fig. 4)
This is a small clade that includes individual kn1 clones of P. ramosum and P. polystachion subsp. polystachion; other clones of both taxa are also related in clade E. The relationship between these two species is not well resolved by the plastid and morphological data, although they are closely related to species of clade A (Fig. 1A, B).
Ploidy
Most species in the Pennisetum/Cenchrus clade are polyploid. In the present sample of species, P. alopecuroides, P. glaucum, P. ramosum, P. schweinfurthii, P. thunbergii, P. unisetum and P. violaceum are only known as diploids, although polyploidy cannot be ruled out. At least two copies of kn1 were retrieved for P. alopecuroides, P. ramosum and P. thunbergii, indicating polyploidy or duplication of this gene. In addition, diploid, polyploid and aneuploid plants have been reported for P. flaccidum, P. hordeoides, P. massaicum, P. mezianum, P. orientale, P. polystachion subsp. polystachion, P. purpureum and P. villosum, although the exact chromosome count for the individual plants sampled here is unknown. Although P. ramosum and P. thunbergii are reported to be diploids only (Table 1), clones appear in at least two places in the phylogenetic tree (Fig. 4), indicating a polyploid history. Also, P. ramosum is reported to be apomictic (Table 1), and apomixis is almost always associated with polyploidy. All species of Cenchrus are probably of allopolyploid origin, as the lowest chromosome numbers reported are n = 17. All remaining Pennisetum species are also polyploid.
DISCUSSION
The monophyly of the Pennisetum, Cenchrus and Odontelytrum clade (PCO clade) is strongly supported by each marker separately or combined (trnL-F and ndhF), and by the combined analysis (trnL-F, ndhF and morphology). The position of P. lanatum is unclear, depending on the evidence included in the analyses; if considering morphology, or plastid and morphological data together, it is also included in the PCO clade. From the partition analyses, six unambiguous morphological synapomorphies support the group: spikelet not disarticulating from the pedicel and falling together with the bristles as a unit; the pedicel glabrous; the apices of the lemmas acuminate; the margins of the lemma flat; and a membranaceous–cartilaginous upper anthecium. Doust and Kellogg (2002) also identified several developmental synapomorphies for the clade: the reduction of the internode on the secondary axis and on other axes, differential elongation of the bristles at maturity, and more bristles than spikelets being initiated in early development. Previous molecular studies proposed the inclusion of Cenchrus within Pennisetum (Giussani et al., 2001; Doust and Kellogg, 2002; Aliscioni et al., 2003; Bess et al., 2005; Doust et al., 2007; Donadío et al., 2009) and also Odontelytrum within Pennisetum (Kellogg et al., 2009). The position of P. clandestinum within the PCO clade does not support recognition of the genus Kikuyochloa proposed by Scholz (2006).
Relationships among major clades within Pennisetum–Cenchrus
The ingroup includes a representative sample of the species treated in the sectional treatment (Stapf and Hubbard, 1934; Pilger, 1940; Clayton and Renvoize, 1986), but only Pennisetum section Brevivalvula is supported as monophyletic by the morphological and plastid analyses (subclade Ab, Fig. 1A, B). Likewise, Martel et al. (2004), in their ITS analysis, only found sections Brevivalvula and Penicillaria to be monophyletic, although most of the sections were under-represented.
The phylogenetic results from independent data sets (nuclear, plastid and morphology) help to elucidate interspecific relationships within the complex Pennisetum–Cenchrus–Odontelytrum. From the plastid phylogenetic trees it was possible to identify monophyletic groups that relate species via maternal inheritance. Evidence of genetic exchange among species via biparental inheritance has been provided by the nuclear marker (kn1). The optimization of morphological, cytological and reproductive characters helps to interpret the principal pattern of relationships among species. In most taxa investigated, kn1 is a single-copy gene. In polyploids, one copy per genome is expected, and the gene is thus useful for dissecting the evolutionary history of polyploid species. Sequences of clones from allopolyploid species are expected to fall in multiple positions in a gene tree, with a set of sequences corresponding to each of the parental genomes.
The position of P. lanatum within the Pennisetum–Cenchrus clade is reinforced by results of the nuclear marker (kn1; Fig. 4). Because P. lanatum is a tetraploid, it is possible that it represents a particularly wide hybridization event, with the pistillate parent being outside the PCO clade.
Clade A (Fig. 1) was first reported by Donadío et al. (2009), based on data from trnL-F and rpl16, with minor subclades within it. Both their work and the present study includes C. brownii, C. ciliaris, C. echinatus, C. incertus, C. myosuroides, C. pilosus, C. setigerus, P. ramosum, P. polystachion subsp. polystachion, P. purpureum and P. setaceum within the clade. In addition, the data here place C. agrimonioides, C. caliculatus, P. hordeoides, P. massaicum, P. mezianum, P. pedicellatum and P. polystachion subsp. atrichum within the clade. Donadío et al. (2009) placed P. frutescens and P. flaccidum within clade A, whereas the here data suggest that they are outside it. Placements of those two species are not strongly supported here, so we cannot rule out the possibility that they belong in the clade. The present results and those from Donadío et al. (2009) use one of the same markers (trnL-F); discrepancies may be due to differences in phylogenetic signal between ndhF and rpl16, and/or to the addition of more species.
Cenchrus brownii is clearly related, in plastid and morphological analyses, to other x = 17 Cenchrus taxa. The grouping of four x = 17 Cenchrus species (C. pilosus, C. brownii, C. echinatus and C. incertus) has been reported by Donadío et al. (2009). Pennisetum setaceum has been reported as having a basic chromosome number of x = 9 and 17 (under P. macrostachyon, Table 1). The x = 17 basic chromosome number and its phylogenetic position revealed that P. setaceum shares at least one genome with x = 17 species of Cenchrus.
The nuclear phylogenetic analysis supports a larger sampling for the secondary gene pool of P. glaucum (Martel et al., 2004). Clade G includes all species of clade B from the plastid and combined phylogenetic analyses (Figs 1 and 4) with a basic chromosome number of x = 7 (P. glaucum, P. violaceum and P. squamulatum), P. sieberianum (basic chromosome number unknown), plus species corresponding to subclade Ab with a basic chromosome number of x = 9 (P. hordeoides, P. pedicellatum and P. polystachion subsp. atrichum) and P. basedowii (Table 1). Following the gene pool classification of Harlan and de Wet (1971), species of clade G would be part of the secondary gene pool of P. glaucum, as well as others previously cited for clade B (P. purpureum and P. nervosum; Donadío et al., 2009).
Reticulation pattern, polyploidy and apomixis
Figures 3 and 4 show that the entire Pennisetum/Cenchrus clade is a large polyploid complex. The kn1 tree does not provide enough resolution to determine the ancestry of all the polyploids, but a few observations can be made. First, the history of allopolyploidization is intricate. Cenchrus is probably the result of an ancient cross between an ancestor similar to P. ramosum or P. orientale and another species of Pennisetum. The tree does not identify the other parent with certainty, but one possibility might be a diploid species related to P. setaceum.
Speciation has occurred at the polyploid level, as shown by the mix of polyploids in each of the major clades of the phylogenetic tree. It is possible that gene flow is occurring among the species. Because crossing barriers are often reduced in polyploids, this may be expected.
The strongly variable chromosome number found in species of subclade Ab or Pennisetum section Brevivalvula is a remarkable fact in favour of reticulation among species of Cenchrus and Pennisetum. Although the basic chromosome number was reported as x = 9, P. polystachion was found to be a hexaploid with, possibly, three different genomes and little or no pairing among them (Sisodia, 1970). Pennisetum polystachion subsp. polystachion was reported as 2n = 18, 36, 45, 48, 52, 53, 54 and 63, 2n = 24 under P. subangustum, and 2n = 32, 56 and 78 under P. setosum (the last two names now synonyms of P. polystachion). Two or three distinct kn1 sequences were found in our accession of P. polystachion subsp. polystachion, suggesting that the individual plant sequenced was either tetraploid or hexaploid. In addition, sequences of kn1 for P. polystachion subsp. polystachion are phylogenetically unrelated to those for P. polystachion subsp. atrichum, even though their plastid sequences are quite similar. This suggests that the morphological similarity of the two may reflect ancestral gene flow, despite the distinct nuclear genomes.
Similarly, P. pedicellatum was found to be an allohexaploid characterized by a low frequency of multivalent formation (tri-, tetra- and hexavalents), a large number of uni- and bivalents, and 75 % anomalies in anaphase I (Naithani and Sisodia, 1966; Sisodia, 1970). Ploidy in P. pedicellatum was variable: 2n = 36, 45 and 54 with several aneuploid numbers: 24, 30, 32, 35, 42, 48, 52 and 53. Whereas P. pedicellatum is shown to be related by the plastid and combined analyses to subclade Ab (Fig. 1A, B), the nuclear marker (Fig. 4) shows it to be related to clade G (which also includes all x = 7 species of clade B and representatives of subclade Ab, together with P. basedowii).
When optimizing the basic chromosome number in the plastid and combined consensus trees, x = 9 was found to be plesiomorphic and x = 5, 7, 8, 10 and 17 were derived (Fig. 1). Similarly, when optimizing the basic chromosome number on the nuclear phylogenetic trees (kn1), x = 9 is plesiomorphic although several clones of x = 9 species were included with x = 7 species in clade G.
The nuclear phylogenetic analysis revealed a reticulate pattern of relationships among species of Pennisetum and Cenchrus, which is also supported by evidence from cytogenetic studies (aneuploids, hexaploids, uni- and multivalent formations, irregular meiotic behaviour). Hybridization among species is frequent within this group (Dujardin and Hanna, 1984b, 1985, 1989; Jauhar, 1981; Marchais and Tostain, 1997), and sequence relationships suggest that there are at least three different genomes within Pennisetum–Cenchrus: the x = 7 genome (clade B, Pennisetum species), the x = 9 genome (most Pennisetum and Cenchrus species) and the x = 17 genome (subclade Aa). The origin of the x = 17 genome could be the result of a cross between ancestors of x = 8 and x = 9, or two x = 9 taxa followed by the loss of one chromosome and, in both cases, followed by diploidization of the ancestral polyploid. Other basic chromosome numbers would be reductions from x = 9 that appeared independently in P. ramosum (x = 5), P. massaicum, P. mezianum and P. montanum (x = 8). To resolve species relationships within Pennisetum–Cenchrus, a group of plants in which reticulation and introgression are common processes, would require additional nuclear sequence loci to obtain congruent results and enhance the resolution among taxa.
At least two species of Cenchrus (C. ciliaris and C. setigerus) and 16 species of Pennisetum have been reported as facultatively or obligately apomictic (Table 1). Apomixis in P. squamulatum and C. ciliaris is linked to a single chromosome (Ozias-Akins et al., 1993) that contains a non-recombining ASGR (Ozias-Akins et al., 1998; Roche et al., 1999). The conservation of molecular markers linked to apomixis and the close relationship among species of Pennisetum and Cenchrus support the view of a single event for the evolution of this trait (Ozias-Akins et al., 2003). It has been suggested that apomixis characterized subfamily Panicoideae before its diversification into the present tribes (Brown and Emery, 1958). Our optimization of the presence of apomixis in the phylogenetic hypotheses is largely uninformative due to insufficient data. If taxa for which reproductive behaviour is not reported or unknown are considered apomictic (Table 1), then apomixis could be considered plesiomorphic, but if those taxa are sexual, then apomictic species have appeared several times during the evolution of the Pennisetum–Cenchrus clade. Because apomixis or the molecular markers linked to apomixis can be transferred to different species through hybridization, it seems more likely that apomixis is related to the interbreeding history of the group, rather than appearing independently several times. As a consequence, the acquisition of apomixis through hybridization within Pennisetum–Cenchrus would be a common mechanism that allows new genotypes to perpetuate themselves. Facultative apomixis has been shown to stabilize polyploid taxa and to permit limited gene flow in other groups of grasses. In the genera Dichanthium Willemet, Bothriochloa Kuntze and Capillipedium Stapf (tribe Andropogoneae), de Wet and Harlan (1970) documented extensive gene flow at the polyploid level, aided by apomixis. It seems likely that a similar phenomenon is occurring in Cenchrus/Pennisetum.
Taxonomic implications
The phylogenetic results presented here strongly support the unification of Cenchrus and Pennisetum, as previously suggested by Correll and Johnston (1970), and the inclusion of Odontelytrum, a monotypic genus occurring in Yemen and eastern Africa. Pennisetum as currently defined is paraphyletic, with Odontelytrum and Cenchrus embedded within it. Optimization of morphological characters within the PCO clade suggests that no morphological character constitutes a synapomorphy for any of the three genera. Due to the consistency between the present results and different phylogenetic hypotheses (including morphological, developmental and multilocus approaches), and the strong support found for the PCO clade, including the type species of the three genera, we propose unification of Pennisetum, Cenchrus and Odontelytrum. Species of Pennisetum and Odontelytrum are here transferred to Cenchrus (Appendix 4), which has priority (McNeill et al., 2006). In addition to the morphological and developmental synapomorphies of the PCO clade (= Cenchrus), Cenchrus is here characterized by having one or several spikelets accompanied by one bristle or surrounded by an involucre of multiple bristles, free or moderately to considerably fused, or having bristles fused and forming a cup-like structure (the degree of fusion varying from a small basal disc to a deep cupule), the consistency of the cupule being rigid or herbaceous.
Pennisetum, Cenchrus and Odontelytrum have been previously included under Cenchrinae by Clayton and Renvoize (1986), together with ten other genera bearing bristles. A comprehensive study including all those genera is being conducted for the tribe Paniceae (Morrone et al., 2008); however, results are too preliminary to reach any decision on the inclusion of other genera within Cenchrus as delimited here.
The complex polyploid relationships among the species shown by the nuclear gene phylogenetic analysis (Figs 3 and 4) provide another argument for combining Pennisetum and Cenchrus in a single genus. Biologically, they are clearly exchanging genes by forming allopolyploids. Their evolution is reticulate, rather than divergent, and hence is inherently difficult to incorporate into a hierarchical classification.
The present study also suggests the need for a comprehensive revision of the group to circumscribe infrageneric taxonomic categories based on monophyly, and the morphological and cytological delimitation of the new groupings. Furthermore, if the intricate pattern of interspecific relationships within the PCO clade is due to hybridization and introgression, determining the evolutionary history will require adding new species and nuclear loci into the analyses.
SUPPLEMENTARY DATA
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported by ANPCyT (Agencia Nacional de Promoción Científica y Técnica, Argentina), grants 13374, 32664 and 01286, and CONICET (Consejo Nacional de Investigaciones Científicas y Tecnológicas, Argentina), grant 5453 to O.M.; and US National Science Foundation Grant DEB-0108501 to E.A.K. Field collections were supported by the Myndel Botanica Foundation and the National Geographic Society, USA, grants 7792-05 to O.M. We thank Prof. M. Ramia, Dr H. Cota-Sánchez, Dr N. Deginani, Dr F. O. Zuloaga and genetic resources centres [Plant Gene Resources of Canada, US Department of Agriculture (USDA) and the Missouri Botanical Garden] for providing plant material. We thank one anonymous reviewer, R. J. Soreng and the Editors for their helpful comments and suggestions on the manuscript.
APPENDIX 1
Taxa studied, voucher information for the specimens sequenced here and GenBank accession numbers. ‘–’ indicates no sequence for that region. Numbers in parentheses indicate published sequences from GenBank and their reference. Material grown from seeds. Letters in bold types represent different clones.
| Taxon | Voucher | GenBank accession |
||
|---|---|---|---|---|
| ndhF | trnL-F | Kn1 | ||
| Cenchrus agrimonioides Trin. | – | AY623745(1) | – | – |
| C. brownii Roem. & Schult. | Venezuela, Ramia & Marrero 9349, SI | GU561510 | EU940005(2) | a: GU561607 |
| b: GU561608 | ||||
| c: GU561609 | ||||
| C. caliculatus Cav. | – | EF189886(3) | – | Aa: EF189760(3) |
| Ab: EF189762(3) | ||||
| Ea: EF189761(3) | ||||
| Eb: EF189763(3) | ||||
| C. ciliaris L. | – | AY029625(4) | EU940006(2) | 1Ea: EF189764(3) |
| 1Eb: EF189767(3) | ||||
| 2Da: EF189765(3) | ||||
| 2Db: EF189766(3) | ||||
| C. echinatus L. | – | AF499151(5) | EU940007(2) | Ba: EF189770(3) |
| Bb: EF189769(3) | ||||
| D: EF189768(3) | ||||
| C. incertus M.A. Curtis | Argentina, Morrone & Giussani 5166, SI | GU561514 | EU940008(2) | a: GU561598 |
| b: GU561599 | ||||
| c: GU561600 | ||||
| d: GU5616010 | ||||
| C. myosuroides Kunth | – | AF499152(5) | EU940009(2) | D: EF189772(3) |
| Ea: EF189771(3) | ||||
| Eb: EF189773(3) | ||||
| C. pilosus Kunth | – | EF189887(3) | EU940010(2) | Ea: EF189777(3) |
| Eb: EF189775(3) | ||||
| Ga: EF189774(3) | ||||
| Gb: EF189776(3) | ||||
| C. setigerus Vahl | – | AF499153(5) | EU940011(2) | Fa: EF189778(3) |
| Fb: EF189779(3) | ||||
| Ixophorus unisetus (J. Presl) Schltdl. | – | AY623749(1) | EU939980(2) | 1Ta: EF189883(3) |
| 1Tb: EF189882(3) | ||||
| 1Tc: EF189884(3) | ||||
| 1Td: EF189885(3) | ||||
| 2Ob EF189880(3) | ||||
| Odontelytrum abyssinicum Hack. | Ethiopia, Friis & al. 6699, K | GU561512 | GU561491 | – |
| Panicum miliaceum L. | – | – | – | Ia: EF189758(3) |
| Ib: EF189759(3) | ||||
| Paspalidium geminatum (Forssk.) Stapf | – | AY029662(4) | EU939981(2) | – |
| Paspalidium jubiflorum (Trin.) Hughes | – | – | – | Ea: EF189851(3) |
| Eb: EF189850(3) | ||||
| Ga: EF189798(3) | ||||
| Gb: EF189797(3) | ||||
| Pennisetum alopecuroides (L.) Spreng. | – | AY029672(4) | EU939986(2) | Ba: EF189781(3) |
| Bb: EF189780(3) | ||||
| C: EF189782(3) | ||||
| P. basedowii Summerh. & C.E. Hubb. | Australia, Pullen 10417, CANB | GU561515 | GU561495 | a: GU561550 |
| b: GU561551 | ||||
| c: GU561552 | ||||
| d: GU561553 | ||||
| P. chilense (E. Desv.) B.D. Jacks. ex R.E. Fr. | Argentina, Zuloaga & al. 8617, SI | GU561516 | EU939987(2) | a: GU561554 |
| b: GU561555 | ||||
| c: GU561556 | ||||
| P. clandestinum Hochst. ex Chiov. | Argentina, Morrone s.n., SI | GU561517 | EU939988(2) | GU561617 |
| P. flaccidum Griseb. | – | AF499150(5) | EU939989(2) | 1B: EF189786(3) |
| 1Ca: EF189787(3) | ||||
| 1Cb: EF189783(3) | ||||
| 2Da: EF189785(3) | ||||
| 2Db: EF189784(3) | ||||
| P. foermeranum Leeke | Namibia, Moss 2017, PRE | GU561511 | GU561496 | – |
| P. frutescens Leeke | Argentina, Deginani 1822, SI | GU561519 | EU939990(2) | a: GU561557 |
| b: GU561558 | ||||
| P. glaucocladum Stapf & C.E. Hubb. | Botswana, Smith 2403, PRE | GU561520 | GU561497 | a: GU561569 |
| b: GU561570 | ||||
| c: GU561571 | ||||
| P. glaucum (L.) R. Br. (1) | – | AF499149(5) | EU939991(2) | Ea: EF189789(3) |
| Eb: EF189788(3) | ||||
| P. glaucum (L.) R. Br. (2) | Brasil, Caxambu 375, MBM | GU561521 | GU561498 | a: GU561547 |
| b: GU561548 | ||||
| c: GU561549 | ||||
| P. hordeoides (Lam.) Steud. | Nepal, Staiton & al. 8844, K | GU561522 | GU561499 | a: GU561565 |
| b: GU561566 | ||||
| c: GU561567 | ||||
| d: GU561568 | ||||
| P. lanatum Klotzsch | India, Bohr 9207, K | GU561523 | – | D: EF189791(3) |
| India, Siddigi 1053, K | – | GU561500 | ||
| E: EF189790(3) | ||||
| P. latifolium Spreng. | Uruguay, Morrone 5231, SI | GU561524 | EU939993(2) | a: GU561559 |
| b: GU561560 | ||||
| c: GU561561 | ||||
| P. macrourum Trin. | CN 87800* | GU561525 | AY116266(7) | a: GU561562, (6) |
| b: GU561563 | ||||
| c: GU561564 | ||||
| P. massaicum Stapf | Kenya, Greenway & Kanuri 12834, K | GU561526 | GU561501 | – |
| P. mezianum Leeke | PI 214061* | GU561527 | GU561502 | a: GU561572 |
| b: GU561573 | ||||
| c: GU561574 | ||||
| P. montanum (Griseb.) Hack. | – | AY188498(6) | EU939994(2) | GU561579 |
| P. natalense Stapf | South Africa, Strey 10968, K | GU561528 | – | – |
| P. nervosum (Nees) Trin. | Argentina, Morrone 5329, SI | GU561529 | EU939996(2) | – |
| P. orientale Rich. | CN 84066* | GU561530 | GU561503 | a: GU561580 |
| b: GU561581 | ||||
| c: GU561582 | ||||
| d: GU561583 | ||||
| P. pedicellatum Trin. | CN 87902* | GU561531 | GU561504 | a: GU561584 |
| b: GU561585 | ||||
| c: GU561586 | ||||
| P. polystachion subsp. polystachion (L.) Schult. | Bolivia, Morrone & Belgrano 5060, SI | GU561533 | EU939997(2) | a: GU561587 |
| b: GU561588 | ||||
| c: GU561589 | ||||
| P. polystachion subsp. atrichum (Stapf & C.E. Hubb.) Brunken | Tanzania, Bjornstad 1704, K | GU561532 | GU561505 | a: GU561612 |
| b: GU561613 | ||||
| c: GU561614 | ||||
| d: GU561615 | ||||
| P. purpureum Schumach. | Argentina, Morrone & al. 4473, SI | GU561534 | EU939999(2) | – |
| P. ramosum (Hochst.) Schweinf. | CN84079* | GU561535 | EU929056 | a: GU561590 |
| b: GU561591 | ||||
| c: GU561592 | ||||
| P. schweinfurthii Pilg. | Ethiopia, Friis & al. 7745, K | GU561536 | GU561506 | – |
| P. setaceum (Forssk.) Chiov. | Argentina, Morrone 5373, SI | GU561537 | EU940000(2) | a: GU561593 |
| b: GU561594 | ||||
| c: GU561595 | ||||
| d: GU561597 | ||||
| e: GU561596 | ||||
| P. sieberianum (Schltdl.) Stapf & C.E. Hubb. | PI 532675* | GU561538 | EU940001(2) | GU561616 |
| P. sphacelatum (Schumach.) T. Durand & Schinz | South Africa, Smook 5934, PRE | GU561539 | – | – |
| P. squamulatum Fresen. | PI 248534* | GU561540 | EU929057 | a: GU561602 |
| b: GU561603 | ||||
| c: GU561604 | ||||
| d: GU561605 | ||||
| e: GU561606 | ||||
| P. thunbergii Kunth | CN 87791* | GU561541 | GU561507 | a: GU561575 |
| b: GU561576 | ||||
| c: GU561577 | ||||
| d: GU561578 | ||||
| P. trachypyllum Pilg. | Kenya, Bogdan 1151, K | GU561542 | GU561508 | – |
| P. tristachyum (Kunth) Spreng. | Bolivia, Morrone & al. 4234, SI | GU561543 | EU940002(2) | – |
| P. unisetum (Nees) Benth. | Sudan, Friis & Vollesen 129, K | GU561544 | EU929058 | – |
| P. villosum R. Br. ex Fresen. | – | EF189888(3) | EU940004(2) | F: EF189792(3) |
| P. violaceum (Lam.) Rich. ex Pers. | CN 88058* | GU561545 | GU561509 | a: GU561610 |
| b: GU561611 | ||||
| Pseudoraphis paradoxa Pilg. | – | – | – | Fa: EF189807(3) |
| Fb: EF189808(3) | ||||
| P. spinescens (R. Br.) Vickery | – | – | – | Fa: EF189809(3) |
| Fb: EF189810(3) | ||||
| Rupichloa acuminata (Renvoize) Salariato & Morrone | Brazil, Zuloaga & Morrone s.n., SI | AY029692(4) | GU561490 | – |
| Setaria palmifolia (J. König) Stapf | MO 801593–2 | AY029680(4) | GU561492 | B: EF189833(3) |
| Fa: EF189832(3) | ||||
| Fb: EF189834(3) | ||||
| S. parviflora (Poir.) Kerguélen | PI 316422 | AY029682(4) | GU561493 | Ga: EF189813(3) |
| Gb: EF189814(3) | ||||
| S. sphacelata (Schumach.) Stapf & C.E. Hubb. ex M.B. Moss | PI 268145 | AY029681(4) | GU561494 | Aa: EF189815(3) |
| Ab: EF189817(3) | ||||
| D: EF189816(3) | ||||
| Spinifex sericeus R. Br. | – | – | – | H: EF189822(3) |
| Ib: EF189824(3) | ||||
| Stenotaphrum secundatum (Walter) Kuntze | – | AY029684(4) | EU939985(2) | Ia: EF189854(3) |
| Ib: EF189855(3) | ||||
References: (1) Kellogg et al. (2004), (2) Donadío et al. (2009), (3) Doust et al. (2007), (4) Giussani et al. (2001), (5) Doust and Kellogg (2002), (6) Aliscioni et al. (2003), (7) Hodkinson et al. (2002).
APPENDIX 2
Cytological and morphological characters used in the cladistic analyses and coding states.
1. Chromosome basic number: x = 5 (0), x = 7 (1), x = 8 (2), x = 9 (3), x = 17 (4), x = 13 (5), x = 10 (6). 2. Ligule: membranous (0), ciliate (1), membranous–ciliate (2). 3. Contra-ligule: absent (0), present (1). 4. Leaf blade: flat (0), convolute (1). 5. Spikelets subtended by an involucre composed of bristles: absent (0), present (1). 6. Inflorescence: terminal (0), axillary (1). 7. Inflorescence-type: contracted to spiciform (0), open (1). 8. Panicle axis: scaberulous (0), glabrous (1), pubescent (2). 9. Involucre: pedicellate (0), sessile (1). 10. Pedicel of the involucre: glabrous (0), pubescent (1), scaberulous (2). 11. Spikelet: pedicellate (0), sessile (1). 12. Disarticulation at the base of the spikelet: absent (0), present (1). 13. Disarticulation at the base of the involucre: absent (0), present (1). 14. Disarticulation at the base of the upper anthecium: absent (0), present (1). 15. Disarticulation at the base of the pedicel: absent (0), present (1). This character applies to the species in which the spikelet or the involucre is pedicellate and the disarticulation point is between the pedicel and the rachis. 16. Pedicel of the spikelet: glabrous (0), pubescent (1), scaberulous (2). 17. Number of bristles: less than 20 (0), more than 21 (1). 18. Bristles: antrorsely scaberulous (0), retrorsely scaberulous (1). 19. Bristles: all the bristles plumose (0), some bristles plumose (1), without plumose bristles (2). 20. Disposition of the bristles: one whorl (0), two or more whorls (1). 21. Bristles: free (0), connate below (1), connate up to half the total length (2), connate up to two-thirds of the total length (3). 22. Length of the bristles: one conspicuously longer bristle (0), two or more conspicuously longer bristles (1), all the bristles as long as the spikelet (2), all the bristles longer than the spikelet (3). 23. Fertile spikelets per involucre: one (0), two or more (1). 24. Spikelets: isomophous (0), heteromorphous (1). 25. Lower floret: sterile (0), male (1). 26. Lower glume: absent (0), vestigial (1), complete (2). 27. Length of the lower glume: less than 1/3 of the spikelet (0), between 1/3 and half the spikelet (1), between half the spikelet and 2/3 (2), more than 2/3 of the spikelet (3), same length as the spikelet (4). 28. Consistency of the lower glume: hyaline (0), membranous (1). 29. Apex of the lower glume: acute (0), acuminate (1), rounded (2). 30. Upper glume: absent (0), vestigial (1), complete (2). 31. Length of the upper glume: less than 1/3 of the spikelet (0), between 1/3 and half the spikelet (1), between half the spikelet and 2/3 (2), more than 2/3 of the spikelet (3), same length as the spikelet (4). 32. Consistency of the upper glume: hyaline (0), membranous (1), chartaceous (2). 33. Apex of the upper glume: acute (0), acuminate (1), rounded (2). 34. Length of the lower lemma: between 1/3 and half the spikelet (0), between half the spikelet and its total length (1), same length as the spikelet (2). 35. Consistency of the lower lemma: hyaline (0), membranous (1), chartaceous (2). 36. Apex of the lower lemma: acute (0), acuminate (1), rounded (2), bidentate (3), tridentate (4). 37. Margin or apex of the lower lemma: ciliate (0), glabrous (1), scaberulous (2). 38. Consistency of the upper lemma: membranous (0), chartaceous (1), coriaceous (2). 39. Apex of the upper lemma: acute (0), acuminate (1), rounded (2), bidentate (3), tridentate (4). 40. Margin or apex of the upper lemma: ciliate (0), glabrous (1). 41. Lower palea: absent (0), vestigial (1), complete (2). 42. Apex of the lower palea: acute (0), acuminate (1), rounded (2), bidentate (3), tridentate (4). 43. Margin or apex of the lower palea: ciliate (0), glabrous (1). 44. Apex of the upper palea: acute (0), acuminate (1), rounded (2), bidentate (3), tridentate (4). 45. Margin or apex of the upper palea: ciliate (0), glabrous (1). 46. Lemma margin: flat (0), involute (1). 47. Lodicules: absent (0), present (1). 48. Styles: connate (0), free (1). 49. Anther tip: glabrous (0), penicillate (1). 50. Upper anthecium texture: smooth (0), rugose (1). 51. Consistency of the upper anthecium when mature (with caryopsis): crustaceous (0), membranous–cartilaginous (1).
APPENDIX 3
Taxa studied in the morphological phylogeny and voucher information.
Cenchrus brownii: Honduras, Montoya 28, SI. Mexico, Ku and Yam 410, SI. Thailand, Laegaard and Norsangsri 21870, SI. C. ciliaris: Argentina, Cabrera et al. 31024, SI; Cabrera et al. 29829, SI. Ecuador, Laegaard 53064, SI. Mexico, Lizama 1456, SI. C. echinatus: Argentina, Burkart and Gamerro 21614, SI; Hicken 12972, SI; Venturi 5509, SI. Thailand, Laegaard 21791, SI. C. incertus: Argentina, Pozner and Belgrano 173, SI. Brazil, Conrad and Dietrich 2141, SI. C. myosuroides: Argentina, Morrone and Giussani 5162, SI; Burkart 20289, SI; Burkart 22133, SI; Guaglianone and Tur 2458, SI. C. pilosus: Peru, Sánchez Vega and Guevara 6217, SI. C. setigerus: Kenya, Verdcourt 2628, SI.
Ixophorus unisetus: Bolivia, Vargas 2101, SI. Mexico, Zuloaga et al. 7360, SI.
Odontelytrum abyssinicum: Ethiopia, Ash 2595, K; Friis et al. 6699, K. Tanzania, Greenway and Kanuri 12617, K. Yemen, Bisset 281, K; Wood 1945, K.
Paspalidium geminatum: Eritrea, Pappi 6829, SI. Ethiopia, Burger 1135, SI. Tanzania, Dunipes and Jefford s.n., SI.
Pennisetum alopecuroides: Argentina, Rúgolo de Agrasar 2144, SI. P. basedowii: Australia, Pullen 10417, CANB; Wolfe and Martin 144, CANB; Paijmans 2513, CANB; Jacobs 1322, CANB; Perry 201, CANB. P. chilense: Argentina, Kiesling et al. 9467, SI; Krapovickas 3182, SI; Zuloaga and Deginani 3772, SI; Correa et al. 4477, SI; Múlgura et al. 1250, SI; Venturi 4891, SI. P. clandestinum: Argentina, Burkart 18503, SI; Nicora 9248, SI; Villar 26077, SI. P. flaccidum: Pakistan, Duthie 12666, K; Hartman 167, K; Norris 85, K; Stewart 10010, K; Winterbottom 202, K. P. foermeranum: Namibia, Ellis 1069, PRE; Moss and Jacobsen 45, PRE; Sittman 9, PRE; Smook 5182, PRE; Smook 5231, PRE. P. frutescens: Argentina, Burkart 20204, SI; Cardini 89, SI; Job 1175, SI; Jörgensen 2891, SI. Paraguay, Arenas 1745, SI. P. glaucocladum: Botswana, Gibbs Russell 2811, PRE; Smith 1694, PRE; Smith 2403, PRE. Namibia, de Winter and Marais 4761, PRE. P. glaucum: Argentina, Burkart 514, SI; Burkart 18479, SI. P. hordeoides: India, Adams 3887, K; van der Maesen 5033, K. Liberia, Baldwin 9942, K. Nepal, Stainton et al. 8844, K. P. lanatum: India, Bor 9207, K; Duthie s.n., K; Wingate s.n., BAA 13627. Pakistan, Siddigi et al. 1053, K; Stewart 8830A, K; Webster and Nasir 6491, K. P. latifolium: Argentina, Burkart and Troncoso 26263, SI; Cabrera et al. 26474, SI; Porta 209, SI; Schwarz 7647, SI; Zuluaga et al. 5054, SI. P. macrourum: Kenya, Bogdan 3514, K; Bogdan 3637, K. South Africa, Acocks 18652, PRE; Barker 588, PRE; Fugler 105, PRE; Taylor 9939, PRE; Victor 954, PRE. Tanzania, Wingfield 59, K; Wingfield 1004, K; Wingfield 914, K. P. massaicum: Kenya, Ament 799, K; Bogdan 896, K; Bogdan 3614, K; Bogdan 2409, K; Edwars 2987, K; Greenway and Kanuri 12834, K. P. mezianum: Namibia, Acocks 18046, PRE; Smook 5117, PRE. Tanzania, Greenway 9836, K; Greenway and Kanuri 11773, K; Raynal 19340, K; Richards 23694, K; Richards 25202, K. P. montanum: Argentina, Cabrera et al. 20643, SI; Cabrera et al. 34738, SI; Giardelli 998, SI; Zuloaga 3767, SI. Bolivia, Morrone and Belgrano 4931, SI. P. natalense: South Africa, Acocks 10122, PRE; Codd 1367, PRE; Edwards 2031, PRE; Strey 10968, K; Ward 4200, PRE. P. nervosum: Argentina, Burkart et al. 26853, SI; Burkart 21094, SI; Jörgensen 2406, SI; Pedersen 8316, SI; Zuloaga et al 844, SI. P. orientale: Argentina, Rúgolo de Agrasar 2188, SI. P. pedicellatum: India, Bor 9207, K. Kenya, Jeffery 538, K. Sudan, Beshir Eff. 429, K; Daws 906, K; Harrison 87, K; Simpson 7295, K. P. polystachion subsp. polystachion: Bolivia, Laegaard 22323, AAU. Costa Rica, Herrera 1544, SI. Ecuador, Laegaard 71230, AAU; Laegaard 71338, AAU; Laegaard 71517, AAU. Tanzania, Southon 224, SI. Sri Lanka, Comanor 734, SI; Cooray 69091411R, SI. P. polystachion subsp. atrichum: Kenya, Grant 878, K. Tanzania, Bjornstad 1704, K; Ngoundai 31, K. P. purpureum: Argentina, Burkart 18502a, SI. Bolivia, Zuloaga et al. 1444, SI. Costa Rica, Grayum 3433, SI. Puerto Rico, Nee 44104, SI. P. ramosum: Tanzania, Leippert 5628, SI. P. schweinfurthii: Ethiopia, Friis et al. 7745, K. Sudan, Jalen 19, K; Sherif 4028, K; Wickens 854, K. P. setaceum: Argentina, Morrone 5373, SI; Rúgolo de Agrasar 2145, SI; Rúgolo de Agrasar 2183, SI; Hurrell and Bazzano 5646, SI. Venezuela, Ramia and Grande 9341, SI. P. sieberianum: Saudi Arabia, Collenette 7909, K; Cope 166, K; Fernandez 82, K; Fernandez 1276, K. Yemen, Wood 3428, K. P. sphacelatum: Kenya, Stewart 364, K; Thulin and Tidigs 277, K; Wesche 1685, K. Tanzania, Renvoize and Abdallah 2403, K; Taylor 10316, K. South Africa, Mohle 234, PRE; Sheepers 1395, PRE; Smook 5803, SI; Smook 5934, PRE; Smook 6682, SI; Victor 1782, PRE. P. squamulatum: Kenya, Bodgan 1863, K; Bodgan 2418, K; Bodgan 3833, K; Glover and Samuel 2733, K. Tanzania, Greenway et al. 13173, K. P. thunbergii: South Africa, Drews 157, PRE; Du Toit 2491, PRE; Liebenberg 7305, PRE; Loxton 238, PRE; Pappi 241, SI; Roberts 3290, PRE; Smook 5028, PRE. P. trachypyllum: Kenya, Bogdan 1151, K; Faden and Evans 74/710, K. Uganda, Maitland 1390, K; Snowden 1445, K; Thomas 1155, K. P. tristachyum: Argentina, Schreiter 4039, SI; Venturi 1349, SI; Williulz 225, SI. Bolivia, Buchtien 457, SI. P. unisetum: Ethiopia, Friis et al. 593, K. Sudan, Friis and Vollesen 129, K. Unganda, Katende 677, K. Tanzania, Greenway and Kanuri 15170, K. P. villosum: Argentina, Crespo 33, SI; Hicken 12965, SI; Rúgolo de Agrasar 2141, SI. Eritrea, Pappi 1987, SI.
Rupichloa acuminata: Brazil, Zuloaga et al. 4843, SI; Zuloaga et al. 4766, SI.
Setaria palmifolia: Guatemala, Türckheim 1450, SI. Philippines, Fénix 117, SI. Venezuela, Zuloaga and Ortiz 4527, SI. S. parviflora: Argentina, Ragonese 2344, SI; Tivano 394, SI; Vegeti 361, SI. Bolivia, Morrone and Belgrano 4935, SI. S. sphacelata: Argentina, Morrone et al., 649, SI; Morrone et al. 5109, SI; Zuloaga and Morrone 7222, SI. Paraguay, Morrone and Pensiero, 565, SI.
Stenotaphrum secundatum: Argentina, Burkart 274, SI; Burkart 1494, SI; Lanfranchi 74, SI; Rúgolo de Agrasar 1033, SI.
APPENDIX 4.
Nomenclatural changes
Cenchrus americanus (L.) Morrone, comb. nov. Basionym: Panicum americanum L., Sp. Pl. 1: 56. 1753. LECTOTYPE. Illustration in Clusius, Rar. Pl. Hist 2: 215. 1601 (lectotype, designated by Clayton & Renvoize, in Polhill (ed.), Fl. Trop. E. Africa, Gramineae 3: 672·1982).
Panicum glaucum L., Sp. Pl.: 56. 1753, non Cenchrus glaucus Mudaliar & Sudaraj, 1957. Pennisetum glaucum (L.) R. Br., Prodr. 1: 195. 1810. LECTOTYPE: Sri Lanka, Hermann s.n. (lectotype, BM, designated as holotype by Rauschert, Feddes Repert 83(9–10): 662, 1973 ).
Cenchrus abyssinicus (Hack.) Morrone, comb. nov. Basionym: Odontelytrum abyssinicum Hack., Oesterr. Bot. Z. 48: 86. 1898. TYPE: Ethiopia. Gaffat to Debra Tabor, 2700 m, 1863, Shimper 1121 (holotype, B; isotype, K).
Cenchrus advena (Wipff & Veldkamp) Morrone, comb. nov. Basionym: Pennisetum advena Wipff & Veldkamp, Sida 18(4): 1033, f. 1. 1999. TYPE: United States. Texas, Brazos Co., cultivated at Texas A&M University, 18 Sep 1990, J.K. Wipff 1723 (holotype, L; isotypes, K, MO, US, UTC).
Cenchrus annuus (Mez) Morrone, comb. nov. Basionym: Pennisetum annuum Mez, Bot. Jahrb. Syst. 56 (Beibl. 125): 7. 1921. TYPE: Peru. Lima-Oroya, 17 Apr 1910, A. Weberbauer 5354 (holotype, B; isotype, US).
Cenchrus arnhemicus (F. Muell.) Morrone, comb. nov. Basionym: Pennisetum arnhemicum F. Muell., Fragm. 7: 109. 1873. TYPE: Australia. Upper river Victoria River, F. Mueller s.n. (holotype, MEL).
Cenchrus bambusiformis (E. Fourn.) Morrone, comb. nov. Basionym: Gymnotrix bambusiformis E. Fourn., Mexic. Pl. 2: 48. 1886. Pennisetum bambusiforme (E. Fourn.) Hemsl. ex B.D. Jacks., Index Kew. 2: 458. 1895. TYPE: Mexico. Mirador, Mar 1842, J.G. Schaffner338 (holotype, P; isotypes, P, US-207605).
Cenchrus basedowii (Summerh. & C.E. Hubb.) Morrone, comb. nov. Basionym: Pennisetum basedowii Summerh. & C.E. Hubb., Bull. Misc. Inform. Kew 1926: 440. 1926. TYPE: Australia. King Sound, May River, Basedow 13 (holotype, K).
Cenchrus caninus (Reinw. ex Blume) Morrone, comb. nov. Basionym: Saccharum caninum Reinw. ex Blume, Catal. Hort. Bogor.: 38. 1823. Pennisetum caninum (Reinw. ex Blume) Koord., Exkurs.-Fl. Java 1: 140. 1911. SYNTYPES. Indonesia. Java, Reinwardt s.n., Junghuhn s.n. and Zollinger s.n. (types not located).
Gymnotrix macrostachys Brongn., Voy. Monde 2(2): 104, t. 11. 1830, non Cenchrus macrostchyus Hochst. ex Steud., 1854. Pennisetum macrostachys (Brongn.) Trin., Mém. Acad. Imp. Sci. Saint-Pétersbourg, Sér. 6, Sci. Math., Seconde Pt. Sci. Nat. 3, 1(2–3): 177. 1834. TYPE: ‘Moluccas’ (type not located).
Cenchrus chilensis (E. Desv.) Morrone, comb. nov. Basionym: Gymnotrix chilensis E. Desv., Fl. Chile 6: 251, t. 74. 1853. Pennisetum chilense (E. Desv.) B.D. Jacks. ex R.E. Fr., Nova Acta Regiae Soc. Sci. Upsal. 1: 172. 1905. TYPE: Chile, C. Gay s.n. (holotype, P; isotypes, K, W).
Cenchrus clandestinus (Hochst. ex Chiov.) Morrone, comb. nov. Basionym: Pennisetum clandestinum Hochst. ex Chiov., Annuario Reale Ist. Bot. Roma 8: 41, pl. 5, fig. 2. 1903. TYPE: Ethiopia, Schimper 2084 (holotype, FI; isotypes, G, K, TUB).
Cenchrus complanatus (Nees) Morrone, comb. nov. Basionym: Gymnotrix complanata Nees, Bonplandia (Hanover) 3: 83. 1855. Pennisetum complanatum (Nees) Hemsl., Biol. Cent.-Amer., Bot. 3(19): 507. 1885. TYPE: Panama, Seemann 1560 (holotype, BM; isotype, US-0093598).
Cenchrus compressus (R. Br.) Morrone, comb. nov. Basionym: Pennisetum compressum R. Br., Prodr.: 195. 1810. TYPE. Australia. New South Wales, R. Brown 6139 (holotype, K).
Panicum alopecuroides L., Sp. Pl. 1: 55. 1753, non Cenchrus alopecuroides Thunb., 1794. Pennisetum alopecuroides (L.) Spreng., Syst. Veg. 1: 303. 1825. LECTOTYPE: China, without collector (lectotype, LINN-80·1, designated by Veldkamp in Cafferty, Jarvis & Turland, Taxon 49(2): 253, 2000).
Alopecurus hordeiformis L., Sp. Pl. 1: 60. 1753. Pennisetum hordeiforme (L.) Spreng., Syst. Veg 1: 302. 1825, non Cenchrus hordeiformis Thunb., 1794. LECTOTYPE: India, Hudson 29 (lectotype, LINN-82·2, designated by Cope in Cafferty, Jarvis & Turland, Taxon 49(2): 245, 2000).
Cenchrus crinitus (Kunth) Morrone, comb. nov. Basionym: Gymnotrix crinita Kunth, Nov. Gen. Sp. (quarto ed.) 1: 112. 1815(1816). Pennisetum crinitum (Kunth) Spreng., Syst. Veg. 1: 302. 1825. TYPE: Mexico. Michoacán, F.W.H.A. von Humboldt & A.J.A. Bonpland, s.n. (holotype, P).
Cenchrus distachyus (E. Fourn.) Morrone, comb. nov. Basionym: Gymnotrix distachya E. Fourn., Mexic. Pl. 2: 48. 1886. Pennisetum distachyum (E. Fourn.) Rupr. ex Chase, Contr. U.S. Natl. Herb. 22(4): 229. 1921. LECTOTYPE: Mexico. Barranca de San Martin prope Zacuapan, Galeotti 5680 (lectotype, BR, designated by Chase, Contr. U.S. Natl. Herb. 22(4): 230, 1921).
Cenchrus domingensis (Spreng. ex Schult.) Morrone, comb. nov. Basionym: Gymnotrix domingensis Spreng. ex Schult., Mant. 2: 284. 1824. Pennisetum domingense (Spreng. ex Schult.) Spreng., Syst. Veg. 1: 302. 1825. TYPE: Santo Domingo, Bertero s.n. (type not located).
Cenchrus dowsonii (Stapf & C.E. Hubb.) Morrone, comb. nov. Basionym: Pennisetum dowsonii Stapf & C.E. Hubb., Bull. Misc. Inform. Kew 1933: 279. 1933. TYPE: Kenya. Naivasha District, Aberdares Range, plain of Lake OĺBolossat, 2100 m, W.J. Dowson 562 (holotype, K; isotype, EA).
Cenchrus durus (Beal) Morrone, comb. nov. Basionym: Pennisetum durum Beal, Grass. N. Amer. 2: 163. 1896. LECTOTYPE: Mexico. Chihuahua, Potrero Mts., 12 Oct 1886, C.G. Pringle 817 (lectotype MSC, designated by Chase, Contr. U.S. Natl. Herb. 22(4): 229, 1921; isolectotypes, CM, MO-2977366, MO-3727999, US-691229).
Cenchrus flaccidus (Griseb.) Morrone, comb. nov. Basionym: Pennisetum flaccidum Griseb., Gött. Nach. 1868: 86. 1868. TYPE: India. Kashmir, Ladak, 1900–1300, Nubra s.n. [Thomson] (holotype, GOET?).
Cenchrus flexilis (Mez) Morrone, comb. nov. Basionym: Pennisetum flexile Mez, Notizbl. Bot. Gard. Berlin-Dahlem 7: 51. 1917. TYPE: India. Kaschmir, Scinujpur, Clarke 29026 (holotype, B).
Cenchrus foermeranus (Leeke) Morrone, comb. nov. Basionym: Pennisetum foermeranum Leeke, Z. Naturwiss. 79: 26·1907. SINTYPE: Namibia. Herero-land, Windhoek, 1897, I. Fischer 77; Windhoek, R. Foermer 46 (syntypes, B; isosyntypes, K).
Cenchrus glaucocladus (Stapf & C.E. Hubb.) Morrone, comb. nov. Basionym: Pennisetum glaucocladum Stapf & C.E. Hubb., Bull. Misc. Inform. Kew 1933: 276. 1933. TYPE: Zimbabwe. Hunyani River, 1410 m, F. Eyles 4903 (holotype, K).
Cenchrus hohenackeri (Hochst. ex Steud.) Morrone, comb. nov. Basionym: Pennisetum hohenackeri Hochst. ex Steud., Syn. Pl. glum. 1: 103. 1854. TYPE: India. Nilgiri Hills, R.F. Hohenacker 930 (holotype, K; isotypes, L, M, US-978422, US-1127293, US-3243707).
Cenchrus hordeoides (Lam.) Morrone, comb. nov. Basionym: Panicum hordeoides Lam., Tabl. Encycl. 1: 170. 1791. Pennisetum hordeoides (Lam.) Steud., Syn. Pl. Glumac. 1: 103. 1854. TYPE: Sierra Leone, Smeathman s.n. (holotype, P).
Cenchrus intectus (Chase) Morrone, comb. nov. Basionym: Pennisetum intectum Chase, Contr. U.S. Natl. Herb. 24(8): 485. 1927. TYPE: Ecuador: Loja: between Loja and San Lucas, ca. 2500 m, 6 Sep 1923, A.S. Hitchcock 21477 (holotype, US-1163845).
Cenchrus lanatus (Klotzsch) Morrone, comb. nov. Basionym: Pennisetum lanatum Klotzsch, Bot. Ergebn. Reise Waldemar: 65, fig. 99. 1862. TYPE: India, Hoffmeister s.n. (holotype, C?).
Cenchrus latifolius (Spreng.) Morrone, comb. nov. Basionym: Pennisetum latifolium Spreng., Syst. Veg. 1: 302. 1825. TYPE: Uruguay. Montevideo, F. Sellow s.n. (holotype, B?).
Cenchrus longissimus (S.L. Chen & Y.X. Jin) Morrone, comb. nov. Basionym: Pennisetum longissimum S.L. Chen & Y.X. Jin, Bull. Bot. Res., Harbin 4(1) 65, fig. 2. 1984. TYPE: China. Guizhou, Duyun Xian, 23 Aug 1930, Y. Tsiang 6040 (holotype, JSBI).
Cenchrus macrourus (Trin.) Morrone, comb. nov. Basionym: Pennisetum macrourum Trin., Gram. Panic.: 64. 1826. SINTYPE: South Africa, Link s.n.; Cape of Good Hope, Swartz s.n. (sintype of Swartz s.n., LE).
Cenchrus massaicus (Stapf) Morrone, comb. nov. Basionym: Pennisetum massaicum Stapf, Bull. Misc. Inform. Kew 1906: 82. 1906. LECTOTYPE: Kenya. Machakos District, Makindu, Linton 72 (lectotype, K, designated by Stapf & Hubbard, Bull. Misc. Inform. Kew 1933: 273, 1933).
Cenchrus mezianus (Leeke) Morrone, comb. nov. Basionym: Pennisetum mezianum Leeke, Z. Naturwiss. 79: 39. 1907. SYNTYPES: Tanzania, Arusha-Moshi, Uhlig 1076, Tanzania/Kenya, Burraberge, Uhlig 35; Kenya, Makinde River, Hässner 584 (isosyntype of Uhlig 1076, K; sintype of Uhlig 35, B, isosintype, EA; sintype of Hässner 584, B).
Cenchrus mildbraedii (Mez) Morrone, comb. nov. Basionym: Pennisetum mildbraedii Mez, Notizbl. Bot. Gart. Berlin-Dahlem 7: 52. 1917. TYPE: Rwanda. Sabinio to Mgahinga, Mildbraed 1763 (holotype, B).
Cenchrus monostigma (Pilg.) Morrone, comb. nov. Basionym: Pennisetum monostigma Pilg., Bot. Jahrb. Syst. 30(1): 120. 1901. SYNTYPES: Cameroon. Zwischen Manus-Quelle und Kamewrum-Pic, 2800 m, Feb 1891, Preuss 822; Cameroon. Manus-Quelle, 1891, Preuss 984 (syntypes, B).
Cenchrus occidentalis (Chase) Morrone, comb. nov. Basionym: Pennisetum occidentale Chase, Contr. U.S. Natl. Herb. 24(8): 483. 1927. TYPE: Ecuador: Guayas, west of Guayaquil, 20 Jun 1923, A.S. Hitchcock 19953 (holotype, US-1163831).
Cenchrus orientalis (Rich.) Morrone, comb. nov. Basionym: Pennisetum orientale Rich., in Persoon, Syn. Pl. 1: 72. 1805. TYPE: ‘Cenchrus orientalis Willd. (ined.) Hab. in Oriente.’ (type not located).
Cenchrus pauperus (Nees ex Steud.) Morrone, comb. nov. Basionym: Pennisetum pauperum Nees ex Steud., Syn. Pl. Glumac. 1: 102. 1854. TYPE: Ecuador. Galapagos Islands. Anon. s.n. (holotype, P; isotype, K).
Cenchrus pedicellatus (Trin.) Morrone, comb. nov. Basionym: Pennisetum pedicellatum Trin., Mem. Acad. Imp. Sci. Saint-Pétersbourg, Sér. 6, Sci. Math., Seconde Pt. Sci. Nat. 3,1(2–3): 184. 1834. TYPE: Cape Verde Islands, St. Iago, D. Peters s.n. (holotype, LE-TRIN-1101·01).
Cenchrus pedicellatus subsp. unispiculus (Brunken) Morrone, comb. nov. Basionym: Pennisetum pedicellatum Trin. subsp. unispiculum Brunken, Bot. J. Linn. Soc. 79(1): 62. 1979. TYPE: Ghana. Cape Verde Islands, R. Innes 30227 (holotype, PRE; isotype, K).
Cenchrus peruvianus (Trin.) Morrone, comb. nov. Basionym: Pennisetum peruvianum Trin., Linnaea 10(3): 295. 1836. TYPE: Peru: Andes Peruviae, 1834, E.F. Poeppig (holotype, LE; isotypes, BM, US).
Cenchrus petiolaris (Hochst.) Morrone, comb. nov. Basionym: Gymnotrix petiolaris Hochst., Flora 27: 250. 1844. Pennisetum petiolare (Hochst.) Chiov., Annuario Reale Ist. Bot. Roma 8(3): 324. 1908. TYPE: Ethiopia. Mt. Scholoda, Schimper 136 (isotype, K).
Cenchrus pilcomayensis (Mez) Morrone, comb. nov. Basionym: Pennisetum pilcomayense Mez, Bot. Jahrb. Syst. 56(Beibl. 125): 7. 1921. TYPE. Paraguay. In regione cursus inferioris fluminis Pilcomayo, May 1906, T. Rojas 61 (holoype, B; isotypes, P, US-978374).
Pennisetum frutescens Leeke, Z. Naturwiss. 79: 35. 1907, non Cenchrus frutescens L., 1753. TYPE: Argentina. Chaco, Fuerte Sarmiento, Dragones, P.G. Lorentz & G.H.E.W. Hieronymus 584 (holoype, B; isotype, GOET).
Cenchrus polystachios (L.) Morrone, comb. nov. Basionym: Panicum polystachion L., Syst. Nat. (ed. 10) 2: 870. 1759. Pennisetum polystachion (L.) Schult., Mant. 2: 146. 1824. LECTOTYPE: India, without collector (lectotype, LINN-80·4, designated by van der Zon, Wageningen Agric. Univ. Pap. 92–1: 335, 1992).
Cenchrus polystachios subsp. atrichus (Stapf & C.E. Hubb.) Morrone, comb. nov. Basionym: Pennisetum atrichum Stapf & C.E. Hubb., Bull. Misc. Inform. Kew 1933: 282. 1933. Pennisetum polystachion subsp. atrichum (Stapf & C.E. Hubb.) Brunken, Bot. J. Linn. Soc. 79(1): 63. 1979. TYPE: Malawi. Zomba, 1170 m, Manning 4 (holotype, K).
Cenchrus procerus (Stapf) Morrone, comb. nov. Basionym: Beckeropsis procera Stapf, Bull. Misc. Inform. Kew 1933: 272. 1933. Pennisetum procerum (Stapf). W.D. Clayton, Kew Bull. 32(3): 580. 1978. TYPE: Kenya. Nakuru, Hitchcock 25117 (holotype, K; isotype, BR).
Cenchrus prolificus (Chase), Morrone, comb. nov. Basionym: Pennisetum prolificum Chase, Contr. U.S. Natl. Herb. 22(4): 231, fig. 75. 1921. TYPE: Mexico. Veracruz, Barranca of Metlac, ca. 900 m, 29 Jan 1895, C. G. Pringle 6075 (holotype, US-250836; isotype, MO-2977369).
Cenchrus purpureus (Schumach.) Morrone, comb. nov. Basionym: Pennisetum purpureum Schumach., Beskr. Guin. Pl.: 44. 1827. TYPE: Ghana, Thonning 355 (holotype, C; isotype, BM).
Cenchrus quianningensis (S.L. Zhong) Morrone, comb. nov. Basionym: Pennisetum quianningense S.L. Zhong, J.S. SouthW. Agricv. Coll. 1982(4): 75, pl. 1. 1982. TYPE: China. Sichuan, Qian'ning, 12 Aug 1974, West Sichuan Veget. Exped. 05820 (holotype, SWAU).
Cenchrus ramosus (Hochst.) Morrone, comb. nov. Basionym: Gymnotrix ramosa Hochst., Flora 27(16): 252. 1844. Pennisetum ramosum (Hochst.) Schweinf., Beitr. Fl. Aethiop.: 301. 1867. TYPE: Sudan. Sennaar, T. Kotschy 199 (isotypes, BM, G, K, L, MO).
Cenchrus rigidus (Griseb.) Morrone, comb. nov. Basionym: Gymnotrix rigida Griseb., Abh. Königl. Ges. Wiss. Göttingen 19: 263. 1874. Pennisetum rigidum (Griseb.) Hack., Anales Mus. Nac. Buenos Aires 11: 84. 1904. TYPE: Argentina. Córdoba: Ascochinga, Apr 1871, P.G. Lorentz 47 (holotype, GOET; isotypes, CORD, W).
Cenchrus riparius (Hochst. ex A. Rich.) Morrone, comb. nov. Basionym: Pennisetum riparium Hochst. ex A. Rich., Tent. Fl. Abyss. 2: 381. 1851. TYPE: Ethiopia. Adua (Adoa), G.H.W. Schimper 84 (holotype, P; isotypes, B, BR, G, K, M, US-1061597).
Cenchrus rupestris (Chase) Morrone, comb. nov. Basionym: Pennisetum rupestre Chase, Contr. U.S. Natl. Herb. 24(8): 484. 1927. TYPE: Peru: Matucana, alt. 2400 m, 12 Apr-3 May 1922, J.F. MacBride & W. Featherstone 453 (holotype, US-1161395).
Cenchrus sagittatus (Henrard) Morrone, comb. nov. Basionym: Pennisetum sagittatum Henrard, Blumea Suppl. 1: 229, tab. 16, fig. 26. 1937. TYPE: Bolivia. Sur Yungas, La Florida, 1700 m, 4 Feb 1932, L.R. Parodi 10069 (holotype, L; isotypes, BAA, K, US-1539315).
Cenchrus setaceus (Forssk.) Morrone, comb. nov. Basionym: Phalaris setacea Forssk., Fl. Aegypt.-Arab.: 17. 1775. Pennisetum setaceum (Forssk.) Chiov., Boll. Soc. Bot. Ital. 1923: 113. 1923. TYPE: Egypt, P. Forsskål s.n. (isotype, BM).
Cenchrus shaanxiensis (S.L. Chen & Y.X. Jin) Morrone, comb. nov. Basionym: Pennisetum shaanxiense S.L. Chen & Y.X. Jin, Bull. Bot. Res., Harbin 4(1): 68, fig. 3. 1984. TYPE: China. Shaanxi, Luoyang Xian, 870 m, 2 Nov 1958, C.L. Tang 957 (holotype, JSBI).
Cenchrus sichuanensis (S.L. Chen & Y.X. Jin) Morrone, comb. nov. Basionym: Pennisetum sichuanense S.L. Chen & Y.X. Jin, Bull. Nanjing Bot. Gard. 1988–1989: 5. 1990. TYPE: China. Sichuan, Derong Xian, 2000–3000 m, c.i. 3366 (type not located).
Cenchrus sphacelatus (Nees) Morrone, comb. nov. Basionym: Gymnotrix sphacelata Nees, Fl. Afr. Austral. Ill.: 68. 1841. Pennisetum sphacelatum (Nees.) T. Durand & Schinz, Consp. Fl. Afr. 5: 784. 1894. SYNTYPES: South Africa. Stormberg, J.F. Drège s.n.; Gekau to Mbashe (Basche), J.F. Drège s.n; Gekau, J.F. Drège s.n. (isosyntypes, K).
Cenchrus squamulatus (Fresen.) Morrone, comb. nov. Basionym: Pennisetum squamulatum Fresen., Mus. Senckenberg. 2: 137. 1837. TYPE: Ethiopia. Semien (Simen), Rüppell s.n. (holotype, FR).
Cenchrus stramineus (Peter) Morrone, comb. nov. Basionym: Pennisetum stramineum Peter, Repert. Spec. Nov. Regni Veg. Beih. 40(1): 71, fig. 37. 1930. TYPE: Tanzania. Masai District, Ngorongoro, Peter 43215 (holotype, B).
Cenchrus subangustus (Schumach.) Morrone, comb. nov. Basionym: Panicum subangustum Schumach., Beskr. Guin. Pl.: 59. 1827. Pennisetum subangustum (Schumach.) Stapf & C.E. Hubb., Bull. Misc. Inform. Kew 1933: 271. 1933. TYPE: Ghana. Thonning s.n. (holotype, C).
Cenchrus tempisquensis (R.W. Pohl) Morrone, comb. nov. Basionym: Pennisetum tempisquense R.W. Pohl, Fieldiana, Bot. 38(2): 6, fig. 2. 1976. TYPE: Costa Rica. Guanacaste, 8 km N of Hacienda Palo Verde, 14 km WSW of Bagaces, 10 m, 20 Feb 1969, R.W. Pohl & G. Davidse 11725 (holotype, ISC; isotypes, CR-47189, F, K, UC, US-3055850).
Cenchrus thulinii (S.M. Phillips) Morrone, comb. nov. Basionym: Pennisetum thulinii S.M. Phillips, Kew Bull. 46(3): 535. 1991. TYPE: Ethiopia. Arussi Prov., Chilalo awraja, Katar river, ca. 20 km SW of Asella, 2200 m, M. Thulin 1541 (holotype, K; isotypes, EA, UPS).
Cenchrus thunbergii (Kunth) Morrone, comb. nov. Basionym: Pennisetum thunbergii Kunth, Révis. Gramin. 1: 50. 1829. TYPE: South Africa, Thunberg s.n. (holotype, UPS).
Cenchrus trachyphyllus (Pilg.) Morrone, comb. nov. Basionym: Pennisetum trachyphyllum Pilg., Bot. Jahrb. Syst. 30(1): 122. 1901. SINTYPES: Tanzania. Usambara, Lutindi, Jul 1893, C. Holst 3253; Bulua, Sep 12893, C. Holst 5003; Usambara, Kwai, Oct 1899, 4600 m, Albers 170; Wegen, Sep 1899, Albers 363; Usagara, W.-Uluguru, 4700 m, Stuhlmann 9087 (isosintype of C. Holst 3253, K, M; sintype of Albers 170, B).
Cenchrus trisetus (Leeke) Morrone, comb. nov. Basionym: Pennisetum trisetum Leeke, Z. Naturwiss. 79: 30. 1907. TYPE: Ethiopia,. Begemeder, Efak, Schimper 1411 (holotype, B; isotype, K).
Cenchrus unisetus (Nees) Morrone, comb. nov. Basionym: Gymnotrix uniseta Nees, Fl. Afr. Austral. Ill.: 66. 1841. Pennisetum unisetum (Nees) Benth., J. Linn. Soc., Bot. 19: 47. 1881. TYPE: South Africa, Durban (Port Natal), J.F. Drège s.n. (isotypes, K, L, S).
Cenchrus violaceus (Lam.) Morrone, comb. nov. Basionym: Panicum violaceum Lam., Tabl. Encycl. 1: 169. 1791. Pennisetum violaceum (Lam.) Rich. ex Pers., Syn. Pl. 1: 72. 1805. TYPE: Senegal, D. Rousillon s.n. (holotype, P).
Cenchrus weberbaueri (Mez) Morrone, comb. nov. Basionym: Pennisetum weberbaueri Mez, Notizbl. Bot. Gart. Berlin-Dahlem 7: 50. 1917. TYPE: Peru: Dept. Junin, Tarma, 3000–3300 m, 10 Feb 1903, A. Weberbauer 2393 (holotype, B).
LITERATURE CITED
- Ahsan SMN, Vahidy AA, Ali SI. Chromosome numbers and incidence of polyploidy in Panicoideae (Poaceae) from Pakistan. Annals of the Missouri Botanical Garden. 1994;81:775–783. [Google Scholar]
- Akenova ME, Chehheda HR. Morphology, cytology and forage potential of Pennisetum americanum (L.) K. Schum.×P. purpureum Schum. amphidiploids. Euphytica. 1981;30:397–404. [Google Scholar]
- Akiyama Y, Goel S, Chen Z, Hanna WW, Ozias-Akins P. Pennisetum squamulatum: is the predominant cytotype hexaploid or octaploid? Journal of Heredity. 2006;97:521–524. doi: 10.1093/jhered/esl005. [DOI] [PubMed] [Google Scholar]
- Aliscioni SS, Giussani LM, Zuloaga FO, Kellogg EA. A molecular phylogeny of Panicum (Poaceae: Paniceae): tests of monophyly and phylogenetic placement within the Panicoideae. American Journal of Botany. 2003;90:796–821. doi: 10.3732/ajb.90.5.796. [DOI] [PubMed] [Google Scholar]
- Avdulov NP. Karyo-sistematischen Untersuchungen der Familie Gramineen. Trudy po Prikladnoi Botanike, Genetike i Selektsii. 1931;44:1–428. [Google Scholar]
- Bess EC, Doust AN, Kellogg EA. A naked grass in the ‘Bristle Clade’: a phylogenetic and developmental study of Panicum section Bulbosa (Paniceae: Poaceae) International Journal of Plant Sciences. 2005;166:371–381. [Google Scholar]
- Bir SS, Sahni M. SOCGI plant chromosome number reports – IV. Journal of Cytology and Genetics. 1986;1:152–154. [Google Scholar]
- Bir SS, Sahni M. Chromosomal and morphological variations in grasses of Punjab. Journal of Cytology and Genetics. 1987;22:12–22. [Google Scholar]
- Brown WV. A cytological study of some Texas Gramineae. Bulletin of the Torrey Botanical Club. 1950;77:63–76. [Google Scholar]
- Brown WV, Emery WH. Apomixis in the Gramineae: Panicoideae. American Journal of Botany. 1958;45:253–263. [Google Scholar]
- Brunken JN. A systematic study of Pennisetum sect. Pennisetum (Gramineae) American Journal of Botany. 1977;2:161–167. [Google Scholar]
- Brunken JN. Cytotaxonomy and evolution in Pennisetum section Brevivalvula (Gramineae) in tropical Africa. Botanical Journal of the Linnean Society. 1979;79:37–49. [Google Scholar]
- Burton GW. A cytological study of some species in the tribe Paniceae. American Journal of Botany. 1942;29:355–361. [Google Scholar]
- Chase A. The Linnaean concept of pearl millet. American Journal of Botany. 1921;8:41–49. [Google Scholar]
- Chatterji AK, Timothy DH. Microsporogenesis and embryogenesis in Pennisetum flaccidum Griseb. Crop Science. 1969;9:219–222. [Google Scholar]
- Chen SL, Phillips SM. Cenchrus. In: Wu ZY, Raven PH, editors. Flora of China, Poaceae, vol. 22. Beijing/St. Louis, MO: Science Press/Missouri Botanical Garden Press; 2006. pp. 552–553. [Google Scholar]
- Chopanov P, Yurtsev BN. Chromosome numbers of some grasses of Turkmenia II. Botanicheskii Zhurnal SSSR. 1976;61:1240–1244. [Google Scholar]
- Christopher J, Abraham A. Studies on the cytology and phylogeny of South Indian grasses. III. Subfamily VI: Panicoideae, tribe Paniceae. Cytologia. 1976;41:621–637. [Google Scholar]
- Clark LG, Zhang WP, Wendel JF. A phylogeny of the grass family (Poaceae) based on ndhF sequence data. Systematic Botany. 1995;20:436–460. [Google Scholar]
- Clayton WD. Gramineae. 101. Pennisetum. In: Hepper FN, editor. Flora of West Tropical Africa (III) London: Crown Agents; 1972. pp. 459–462. [Google Scholar]
- Clayton WD. Gramineae (Paniceae, Isachneae and Arundinelleae) In: Launert E, Pope GV, editors. Flora Zambesiaca. 3. Vol. 10. Whitstable: Whitstable Litho Printers Ltd; 1989. pp. 1–231. [Google Scholar]
- Clayton WD, Renvoize SA. Pennisetum. In: Polhill RM, editor. Flora of tropical East Africa. Rotterdam: A.A. Balkema; 1982. pp. 672–690. [Google Scholar]
- Clayton WD, Renvoize SA. Genera Graminum. Grasses of the world. London: Her Majesty's Stationery Office; 1986. [Google Scholar]
- Clayton WD, Harman KT, Williamson H. GrassBase – The Online World Grass Flora. (2006 onwards) http://www.kew.org/data/grasses-db.html . Accessed 20 April 2009. [Google Scholar]
- Connor HE. Breeding systems in New Zealand grasses IX: sex ratios in dioecious Spinifex sericeus. New Zealand Journal of Botany. 1984;22:569–574. [Google Scholar]
- Correll DS, Johnston MC. Manual of the vascular plants of Texas. Texas: Texas Research Foundation, Renner; 1970. [Google Scholar]
- Crins WJ. The genera of Paniceae (Gramineae: Panicoideae) in the southeastern United States. Journal of the Arnold Arboretum, Supplementary Series. 1991;1:171–312. [Google Scholar]
- Davidse G, Pohl RW. Chromosome numbers and notes on some Central American grasses. Canadian Journal of Botany. 1972;50:273–283. [Google Scholar]
- Davidse G, Pohl RW. Chromosome numbers, meiotic behavior, and notes on tropical American grasses. Canadian Journal of Botany. 1974;52:317–328. [Google Scholar]
- Davidse G, Pohl RW. Chromosome numbers of Tropical American grasses (Gramineae) Annals of the Missouri Botanical Garden. 1978;65:637–649. [Google Scholar]
- DeLisle D. Taxonomy and distribution of the genus Cenchrus. Iowa State Journal of Science. 1963;37:259–351. [Google Scholar]
- DeLisle D. Chromosome numbers in Cenchrus (Gramineae) American Journal of Botany. 1964;51:1133–1134. [Google Scholar]
- Donadío S, Giussani LM, Kellogg EA, Zuloaga FO, Morrone O. A preliminary molecular phylogeny of Pennisetum and Cenchrus (Poaceae-Paniceae) based on the trnL-F, rpl16 chloroplast markers. Taxon. 2009;58:392–404. [Google Scholar]
- Doust AN, Kellogg EA. Inflorescence diversification in the panicoid ‘Bristle grass’ clade (Paniceae, Poaceae): evidence from molecular phylogenies and developmental morphology. American Journal of Botany. 2002;88:1203–1222. doi: 10.3732/ajb.89.8.1203. [DOI] [PubMed] [Google Scholar]
- Doust AN, Penly AM, Jacobs SWL, Kellogg EA. Congruence, conflict and polyploidization shown by nuclear and chloroplast markers in the monophyletic ‘bristle clade’ (Paniceae, Panicoideae, Poaceae) Systematic Botany. 2007;32:531–544. [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987;19:11–15. [Google Scholar]
- Dujardin M. Additional chromosome numbers and meiotic behavior in tropical African grasses from western Zaire. Canadian Journal of Botany. 1979;57:864–876. [Google Scholar]
- Dujardin M, Hanna W. Cytogenetics of double cross hybrids between Pennisetum americanum – P. purpureum amphiploids and P. americanum × Pennisetum squamulatum interspecific hybrids. Theoretical and Applied Genetics. 1984a;69:97–100. doi: 10.1007/BF00262551. [DOI] [PubMed] [Google Scholar]
- Dujardin M, Hanna W. Microsporogenesis, reproductive behavior, and fertility in five Pennisetum species. Theoretical and Applied Genetics. 1984b;67:197–201. doi: 10.1007/BF00317033. [DOI] [PubMed] [Google Scholar]
- Dujardin M, Hanna W. Cytology and reproduction of reciprocal backcrosses between pearl millet and sexual and apomictic hybrids of pearl millet × Pennisetum squamulatum. Crop Science. 1985;25:59–62. [Google Scholar]
- Dujardin M, Hanna W. Crossability of pearl millet with wild Pennisetum species. Crop Science. 1989;29:77–80. [Google Scholar]
- Duvall MR, Noll JD, Minn AH. Phylogenetics of Paniceae (Poaceae) American Journal of Botany. 2001;88:1988–1992. [PubMed] [Google Scholar]
- Emery WHP. A study of reproduction in Setaria macrostachya and its relatives in the Southwestern United States and Northern Mexico. Bulletin of the Torrey Botanical Club. 1957;84:106–121. [Google Scholar]
- Farris JS, Albert VA, Källersjö M, Lipscomb D, Kluge AG. Parsimony jackknifing outperforms neighbor-joining. Cladistics. 1996;12:99–124. doi: 10.1111/j.1096-0031.1996.tb00196.x. [DOI] [PubMed] [Google Scholar]
- Filgueiras TS. O gênero Cenchrus L. no Brasil (Gramineae: Panicoideae) Acta Amazonica. 1984;14:95–127. [Google Scholar]
- Giussani LM, Cota-Sánchez H, Zuloaga FO, Kellogg EA. A molecular phylogeny of the grass subfamily Panicoideae (Poaceae) shows multiple origins of C4 photosynthesis. American Journal of Botany. 2001;88:1993–2012. [PubMed] [Google Scholar]
- Giussani LM, Zuloaga FO, Quarín CL, Cota-Sánchez JH, Ubayasena K, Morrone O. Phylogenetic relationships in the genus Paspalum (Poaceae: Panicoideae: Paniceae): an assessment of the Quadrifaria and Virgata informal Groups. Systematic Botany. 2009;34:32–43. [Google Scholar]
- Goel S, Chen Z, Conner JA, Akiyama Y, Hanna WW, Ozias-Akins P. Delineation by fluorescence in situ hybridization of a single hemizygous chromosomal region associated with aposporous embryo sac formation in Pennisetum squamulatum and Cenchrus ciliaris. Genetics. 2003;163:1069–1082. doi: 10.1093/genetics/163.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloboff PA, Farris JS, Nixon KC. TNT, a free program for phylogenetic analysis. Cladistics. 2008;24:774–786. [Google Scholar]
- Gómez-Martínez R, Culham A. Phylogeny of the subfamily Panicoideae with emphasis on the tribe Paniceae: evidence from the trnLF cpDNA region. In: Jacobs SWL, Everett JE, editors. Grasses: systematics and evolution. Collingwood, Vic: CSIRO Publishing; 2000. pp. 136–140. [Google Scholar]
- Gould FW. Chromosome numbers in southwestern grasses. American Journal of Botany. 1958;45:757–767. [Google Scholar]
- Gould FW. Chromosome numbers in some Mexican grasses. Boletín de la Sociedad Botánica de Mexico. 1965;29:49–62. [Google Scholar]
- Gould FW. Chromosome numbers of Texas grasses. Canadian Journal of Botany. 1968;46:1315–1325. [Google Scholar]
- Gould FW, Soderstom TR. Chromosome numbers of tropical American grasses. American Journal of Botany. 1967;54:676–683. [Google Scholar]
- Gould FW, Soderstrom TR. Chromosome numbers of some Mexican and Colombian grasses. Canadian Journal of Botany. 1970;48:1633–1639. [Google Scholar]
- Gould FW, Soderstrom TR. Chromosome numbers of some Ceylon grasses. Canadian Journal of Botany. 1974;52:1075–1090. [Google Scholar]
- Gupta PK, Singh RV. Variations in chromosomes and flavonoids in Setaria Beauv. Nucleus. 1977;20:167–171. [Google Scholar]
- Gustafsson Å. Apomixis in higher plants. I. The mechanism of apomixis. 1946:3–67. Lunds Universitets Årsskrift. N. F. Avd. 2, Bd 42, Nr. 3. [Google Scholar]
- Harlan JR, de Wet JMJ. Toward a rational classification of cultivated plants. Taxon. 1971;20:509–517. [Google Scholar]
- Hodkinson TR, Chase MW, Lledo MD, Salamin N, Renvoize SA. Phylogenetics of Miscanthus, Saccharum and related genera (Saccharinae, Andropogoneae, Poaceae) based on DNA sequences from ITS nuclear ribosomal DNA and plastid trnL intron and trnL-F intergenic spacers. Journal of Plant Research. 2002;115:381–392. doi: 10.1007/s10265-002-0049-3. [DOI] [PubMed] [Google Scholar]
- Hrishi NJ. Studies on the cytogenetics of six species of Pennisetum and their comparative morphology and anatomy. Genetica. 1952;26:280–356. doi: 10.1007/BF01690619. [DOI] [PubMed] [Google Scholar]
- Hsu CC. Preliminary chromosome studies on the vascular plants of Taiwan (V). Cytotaxonomy on some monocotyledons. Taiwania. 1972;17:48–65. [Google Scholar]
- Hunziker JH, Zuloaga FO, Morrone O, Escobar A. Estudios cromosómicos en Paniceae sudamericanas (Poaceae: Panicoideae) Darwiniana. 1998;35:29–36. [Google Scholar]
- Jauhar PP. Cytogenetics and breeding of pearl millet and related species. New York: Alan. R Liss, Inc; 1981. [Google Scholar]
- Jensen KB, Highnight K, Wipff KJ. IOPB chromosome data 1. International Organization of Plant Biosystematists Newsletter. 1989;13:20–21. [Google Scholar]
- Judziewicz EJ. 187. Poaceae (Gramineae) In: Görts-Van Rijn ARA, editor. Flora of the Guianas. Series A, Phanaerogams. Vol. 8. Königstein: Koeltz Scientific Books; 1990. pp. 1–127. [Google Scholar]
- Kammacher P, Anoma G, Adjanohoun E, Aké Assi L. Nombres chromosomiques de Graminées de Côte-d́Ivoire. Candollea. 1973;28:191–217. [Google Scholar]
- Kellogg EA, Hiser KM, Doust AN. Taxonomy, phylogeny, and inflorescence development of the genus Ixophorus (Panicoideae: Poaceae) International Journal of Plant Sciences. 2004;165:1089–1105. [Google Scholar]
- Kellogg EA, Aliscioni SS, Morrone O, Pensiero J, Zuloaga F. A phylogeny of Setaria (Poaceae, Panicoideae, Paniceae) and related genera based on the chloroplast gene ndhF. International Journal of Plant Sciences. 2009;170:117–131. [Google Scholar]
- Khosla PK, Mhera PN. In IOPB chromosome number reports XLII. Taxon. 1973;22:647–654. [Google Scholar]
- Khosla PK, Sharma ML. Cytological observations on some species of Setaria. Nucleus. 1973;16:38–41. [Google Scholar]
- Marchais L, Tostain S. Analysis of reproductive isolation between pearl millet (Pennisetum glaucum (L.) R.Br.) and P. ramosum, P. schweinfurthii, P. squamulatum, Cenchrus ciliaris. Euphytica. 1997;93:97–105. [Google Scholar]
- Martel E, De Nay D, Siljak-Yakovlev S, Brown S, Sarr A. Genome size variation and basic chromosome number in pearl millet and fourteen related Pennisetum species. Journal of Heredity. 1997;88:139–143. [Google Scholar]
- Martel E, Poncet V, Lamy F, Siljak-Yakolev S, Lejune B, Sarr A. Chromosome evolution of Pennisetum species (Poaceae): implications of ITS phylogeny. Plant Systematics and Evolution. 2004;249:139–149. [Google Scholar]
- McNeill J, Barrie FR, Burdet HM, et al. International Code of Botanical Nomenclature (Vienna) Regnum Vegetabile. 2006;146:1–568. [Google Scholar]
- Mehra PN. Cytology of East Indian grasses. India: PN Mehra Chandigarh; 1982. [Google Scholar]
- Mehra PN, Rememanandan P. Cytological investigation in West Himalayan Panicoideae. Cytologia. 1973;38:259–270. [Google Scholar]
- Mehra PN, Sharma ML. In IOPB chromosome number reports XXXIX. Taxon. 1973;22:115–118. [Google Scholar]
- Mehra PN, Sharma ML. Cytological studies in some central and eastern Himalayan grasses. II. The Paniceae. Cytologia. 1975;40:75–89. [Google Scholar]
- Miège J. Quatrième liste de nombres chromosomiques d‘especies d́Afrique occidentale. Revue de Cytologie et de Biologie Vegetales. 1962;24:149–164. [Google Scholar]
- Morgenstern B. DIALIGN: multiple DNA and protein sequence alignment at BiBiServ. Nucleic Acids Research. 2004;32:W33–W36. doi: 10.1093/nar/gkh373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern B, Frech K, Dress A, Werner T. DIALIGN: finding local similarities by multiple sequence alignment. Bioinformatics. 1998;14:290–294. doi: 10.1093/bioinformatics/14.3.290. [DOI] [PubMed] [Google Scholar]
- Morrone O, Hunziker JH, Zuloaga FO, Escobar A. Números cromosómicos en Paniceae sudamericanas (Poaceae: Panicoideae) Darwiniana. 1995;33:53–60. [Google Scholar]
- Morrone O, Aegesen L, Scataglini A, et al. The Fourth International Conference on the Comparative Biology of the Monocotyledons; the Fifth International Symposium on Grass Systematics and Evolution. Copenhagen: University of Copenhagen; 2008. Phylogeny of the Paniceae (Poaceae: Panicoideae) integrating chloroplast DNA sequences and morphology; p. 43. [Google Scholar]
- Naithani SP, Sisodia KP. Preliminary meiotic study in Pennisetum pedicellatum Trin. Current Science. 1966;35:343–344. [Google Scholar]
- Norrmann GA, Quarín CL, Killeen TJ. Chromosome numbers in Bolivian grasses (Gramineae) Annals of the Missouri Botanical Garden. 1994;81:768–774. [Google Scholar]
- Núñez O. Investigaciones cariosistemáticas en las Gramíneas Argentinas de la tribu Paniceae. Revista de la Facultad de Agronomía de La Plata. 1952;28:229–255. [Google Scholar]
- Olmstead RG, Sweere JA. Combining data in phylogenetic systematics: an empirical approach using three molecular data sets in the Solanaceae. Systematic Biology. 1994;43:467–481. [Google Scholar]
- Ozias-Akins P. Apomixis: developmental characteristics and genetics. Critical Reviews in Plant Sciences. 2006;25:199–214. [Google Scholar]
- Ozias-Akins P, Lubbers EL, Hanna WW, McNay JW. Transmission of the apomictic mode of reproduction in Pennisetum: co-inheritance of the trait and molecular markers. Theoretical and Applied Genetics. 1993;85:632–638. doi: 10.1007/BF00220923. [DOI] [PubMed] [Google Scholar]
- Ozias-Akins P, Roche D, Hanna WW. Tight clustering and hemizygosity of apomixis-linked molecular markers in Pennisetum squamulatum implies genetic control of apospory by a divergent locus which may have no allelic form in sexual genotypes. Proceedings of the National Academy of Sciences USA. 1998;95:5127–5132. doi: 10.1073/pnas.95.9.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozias-Akins P, Akiyama Y, Hanna WW. Molecular characterization of the genomic region linked with apomixis in Pennisetum/Cenchrus. Functional and Integrative Genomics. 2003;3:94–104. doi: 10.1007/s10142-003-0084-8. [DOI] [PubMed] [Google Scholar]
- Pilger R. Gramineae. III: Unterfamilie Panicoideae. In: Engler A, Prantl K, editors. Die natürlichen pflanzenfamilien. 2nd edn. Leipzig: Engelmann; 1940. pp. 1–208. [Google Scholar]
- Pohl RW. Burger WC, editor. Family No. 15, Gramineae. Fieldiana: Botany, New Series. 1980:1–608. Flora Costaricensis. [Google Scholar]
- Pohl RW, Davidse G. Chromosome numbers of Costa Rica grasses. Brittonia. 1971;23:293–324. [Google Scholar]
- Pohl RW, Davidse G. Cenchrus 5. In: Davidse G, Sousa MS, Chater AO, editors. Flora Mesoamerica. Vol. 6. Mexico: Universidad Nacional Autónoma de Mexico; 1994. pp. 374–327. Sutton and MJ Hufp. [Google Scholar]
- Rao YS, Rao SA, Mengesha MH. New evidence on the phylogeny of basic chromosome number in Pennisetum. Current Science. 1989;58:869–871. [Google Scholar]
- Reeder JR. In IOPB chromosome number reports XI. Taxon. 1967;16:215–222. [Google Scholar]
- Reeder JR. Notes on Mexican grasses VIII. Miscellaneous chromosome numbers-2. Bulletin of the Torrey Botanical Club. 1968;95:69–86. [Google Scholar]
- Renno J-F, Schmelzer GH, de Jong JH. Variation and geographical distribution of ploidy levels in Pennisetum section Brevivalvula (Poaceae) in Burkina Faso, Benin and southern Niger. Plant Systematics and Evolution. 1995;198:89–100. [Google Scholar]
- Roche D, Cong P, Chen ZB, et al. An apospory-specific genomic region is conserved between buffelgrass (Cenchrus ciliaris L.) and Pennisetum squamulatum Fresen. Plant Journal. 1999;19:203–208. doi: 10.1046/j.1365-313x.1999.00514.x. [DOI] [PubMed] [Google Scholar]
- Rúgolo de Agrasar ZE, Puglia ML. Gramíneas ornamentales. In: Hurrel JA, editor. Plantas de la Argentina. Silvestres y cultivadas. Buenos Aires: Editorial LOLA; 2004. pp. 1–336. [Google Scholar]
- Schmelzer GH. Review of Pennisetum section Brevivalvula (Poaceae) Euphytica. 1997;1:1–20. [Google Scholar]
- Scholz H. Kikuyochloa, genus novum (Poaceae: Paniceae) Feddes Repertorium. 2006;117:512–518. [Google Scholar]
- Shanthamma C. Reproductive behavior of Pennisetum macrostachyum Benth., and a new basic chromosome number in the genus Pennisetum. Bulletin of the Torrey Botanical Club. 1979;106:73–78. [Google Scholar]
- Singh M, Burson BL, Finlayson SA. Isolation of candidate genes for apomictic development in buffelgrass (Pennisetum ciliare) Plant Molecular Biology. 2007;64:673–682. doi: 10.1007/s11103-007-9188-1. [DOI] [PubMed] [Google Scholar]
- Sinha R, Bhardwaj PP, Singh RK. SOCGI plant chromosome number report-IX. Journal of Cytology and Genetics. 1990;25:140–143. [Google Scholar]
- Sisodia KPS. Cytological studies on some species in genus Pennisetum. Theoretical and Applied Genetics. 1970;40:26–31. doi: 10.1007/BF00280983. [DOI] [PubMed] [Google Scholar]
- Stapf O, Hubbard CE. Pennisetum. In: Prain D, editor. Flora of Tropical Africa. Ashford: L. Reeve & Co. Ltd; 1934. pp. 954–1070. [Google Scholar]
- Swaminathan MS, Nath J. Two new basic chromosome numbers in the genus Pennisetum. Nature. 1956;178:1241–1242. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Tateoka T. Chromosome numbers of some east African grasses. American Journal of Botany. 1965;52:864–869. [Google Scholar]
- Türpe AM. Las especies sudamericanas del género Pennisetum L.C. Richard (Gramineae) Lilloa. 1983;36:105–129. [Google Scholar]
- Watson L, Dallwitz MJ. The grass genera of the world. Cambridge: C.A.B: International, University Press; 1992. [Google Scholar]
- Webster RD. Genera of the North American Paniceae (Poaceae: Panicoideae) Systematic Botany. 1988;13:576–609. [Google Scholar]
- Wen XS, Ye XL, Li YQ, Chen ZL, Xu SX. Embryological studies on apomixis in Pennisetum squamulatum. Acta Botanica Sinica. 1998;40:598–604. [Google Scholar]
- de Wet JMJ. Chromosome numbers of a few South African grasses. Cytologia. 1954;19:97–103. [Google Scholar]
- de Wet JMJ. Chromosome numbers and some morphological attributes of various South African grasses. American Journal of Botany. 1960;47:44–49. [Google Scholar]
- de Wet JMJ, Harlan JR. Apomixis, polyploidy, and speciation in Dichanthium. Evolution. 1970;24:270–277. doi: 10.1111/j.1558-5646.1970.tb01760.x. [DOI] [PubMed] [Google Scholar]
- Wipff JK. Nomenclature changes in Pennisetum (Poaceae: Paniceae) Sida. 2001;19:523–530. [Google Scholar]
- Wipff JK. Pennisetum Rich. In: Barkworth ME, Capel KM, Long S, Piep MB, editors. Flora of North America. North of Mexico, vol. 25. Magnoliophyta: Commelinidae (in part): Poaceae, part 2. New York: Oxford University Press; 2003. pp. 515–529. [Google Scholar]
- Zuloaga FO, Morrone O. Zulaoga FO, Morrone O, Davidse G, et al., editors. Cenchrus. Catalogue of New World Grasses (Poaceae): III. Subfamilies Panicoideae, Aristidoideae, Arundinoideae, and Danthonioideae. Contributions from the US National Herbarium. 2003;46:144–150. [Google Scholar]
- Zuloaga FO, Morrone O, Giussani LM. A cladistic analysis of the Paniceae: a preliminary approach. In: Jacobs SWL, Everett J, editors. Grasses: Systematics and Evolution. Melbourne: CSIRO; 2000. pp. 123–134. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.