Abstract
Background and Aims
Low phosphorus (P) availability is a major constraint to soybean growth and production, especially in tropical and subtropical areas. Root traits have been shown to play critical roles in P efficiency in crops. Identification of the quantitative trait loci (QTLs) conferring superior root systems could significantly enhance genetic improvement in soybean P efficiency.
Methods
A population of 106 F9 recombinant inbred lines (RILs) derived from a cross between BD2 and BX10, which contrast in both P efficiency and root architecture, was used for mapping and QTL analysis. Twelve traits were examined in acid soils. A linkage map was constructed using 296 simple sequence repeat (SSR) markers with the Kosambi function, and the QTLs associated with these traits were detected by composite interval mapping and multiple-QTL mapping.
Key Results
The first soybean genetic map based on field data from parental genotypes contrasting both in P efficiency and root architecture was constructed. Thirty-one putative QTLs were detected on five linkage groups, with corresponding contribution ratios of 9·1–31·1 %. Thirteen putative QTLs were found for root traits, five for P content, five for biomass and five for yield traits. Three clusters of QTLs associated with the traits for root and P efficiency at low P were located on the B1 linkage group close to SSR markers Satt519 and Satt519-Sat_128, and on the D2 group close to Satt458; and one cluster was on the B1 linkage group close to Satt519 at high P.
Conclusions
Most root traits in soybean were conditioned by more than two minor QTLs. The region closer to Satt519 on the B1 linkage group might have great potential for future genetic improvement for soybean P efficiency through root selection.
Keywords: Quantitative trait loci (QTLs), soybean, Glycine max, root traits, phosphorus efficiency
INTRODUCTION
Soybean (Glycine max) is an important economic crop, providing most vegetable oils and proteins for human consumption. Because of biological nitrogen (N) fixation, soybean is also important in intercropping and/or crop rotation (Boddey et al., 1997; Abbas et al., 2001). Soybean is mostly cultivated in tropical, subtropical and temperate areas, where the soils are often deficient in phosphorus (P) due to intensive erosion, weathering, and P fixation by free Fe and Al oxides (Sample et al., 1980; Stevenson, 1986). Therefore, low P availability is often a major constraint to soybean growth and production. As P is a non-renewable resource, it is difficult to solve the problem of soil P deficiency through traditional P fertilization (Vance et al., 2003). Developing P-efficient soybean varieties with efficient P acquisition ability from both native and added P in the soils would be a sustainable and economical approach (Yan et al., 2006).
As the main organ for plants to take up nutrients, roots play important roles in P acquisition from soils. It is well documented that the ability of a plant to access P from soils depends on root physiological and morphological properties such as root length, root exudates, high-affinity Pi transporter and arbuscular mycorrhizal colonization (Bryla and Koide, 1998; Raghothama, 1999, 2000; Hinsinger, 2001; Dakora and Phillips, 2002). Root architecture, the spatial configuration of a root system in the soil, determines the soil exploration by roots, which has been shown to be important for plant P acquisition and thus could be considered as the functional prerequisite of other root traits (Lynch, 1995; Lynch and Brown, 2001; de Dorlodot et al., 2007). A study using an ‘applied core collection’ of soybean germplasm showed a close relationship between root architecture and P efficiency; P-efficient genotypes always had shallower root architecture (Zhao et al., 2004). Therefore, understanding the genetic control of root architecture in relation to P acquisition would facilitate genetic improvement for P efficiency through root modification.
Currently, a large number of studies on quantitative trait loci (QTL) analysis for root morphology and physiology as related to P nutrition in plants have been reported (Beebe et al., 2006; Li et al., 2007; Chen et al., 2009; Cichy et al., 2009; Li et al., 2009). The first description of QTLs for root traits in relation to P stress was for root weight in field-grown maize by Reiter et al. (1991), and subsequently for root architecture traits under different P conditions, such as root hair length, and lateral root branching and length (Zhu et al., 2005a, b). QTL analysis of root traits and P efficiency in legumes started later than for cereal crops. Using the recombinant inbred lines (RILs) population derived from DOR364 and G19833 with contrasting P efficiency and root architecture in common bean through paper pouch and field studies, 22 QTLs controlling basal root angle and length, as well as P uptake efficiency have been identified (Liao et al., 2004). A further study showed that the QTLs of basal root growth and development in common bean were tightly linked with the QTLs for P uptake efficiency in the field (Beebe et al., 2006). This indicated that basal root growth was vital to improve P uptake efficiency in common bean. In soybean, there are only two reports on QTL analysis of P efficiency, and few QTLs for root traits related to P efficiency have been identified (Li et al., 2005; Zhang et al., 2009). By using an RIL population derived from Kefeng1 and Nannong 1138-2, seven QTLs related to P efficiency have been detected, two of which were identified from root traits for root P content on the F linkage group (Li et al., 2005). Zhang et al. (2009) have developed a new soybean RIL population from a cross between P-efficient and P-inefficient varieties and detected 34 additive QTLs. Five QTLs were mapped for root dry weight and all of them were from the P-efficient variety. In addition, all the soybean QTLs for P efficiency have been identified under controlled conditions. QTL mapping of soybean root traits and P efficiency under natural field conditions has not been reported.
The present study analyses the QTLs controlling variations for six root traits and evaluates their relationship with QTLs for P efficiency in soybean in field trails with or without additional P supply using an F9 RIL population derived from a cross between two soybean genotypes contrasting in P efficiency and root architecture, BD2 × BX10. The results will help to better understand the genetic relationships between plant root and P efficiency.
MATERIALS AND METHODS
Plant materials
Two soybean (Glycine max) genotypes, BD2, a P-inefficient genotype with deep root architecture, and BX10, a P-efficient genotype with shallow root architecture (Zhao et al., 2004), were crossed to develop RILs in this study. Progenies were advanced by single seed descent to the F9 generation for generating 106 RILs, which were used to construct a genetic linkage map and detect QTLs associated with root traits and P efficiency.
Field trials
Two field trials were conducted in Boluo County, Guangdong Province, southern China. The native soil P at this site was generally low and changed along the soil profile, with available P content of 11·2 mg P kg−1 in the top 20 cm and almost none in the subsoil. One trial was managed at low P (no P added, LP treatment) and another at high P (160 kg P ha−2 fertilized as single super-phosphate, SSP, to the topsoil by broadcast application; HP treatment). Parental genotypes were planted with nine replications and the RILs were planted with four split plot for P levels with a randomized complete block design in each plot. Six seeds were sown per experimental plot with single rows 1·2 m in length and 0·6 m apart. Fifty days after planting, plants were extracted manually from the soil with their root systems. Shoot (SDW), root (RDW) and plant (PDW) dry weight and the ratio of root/shoot (RRS) dry weight were determined. Shoot (SPC), root (RPC) and plant (PPC) P contents were analysed colorimetrically as described by Murphy and Riley (1962) after ash digestion. At maturity, grains were harvested, and seed number (SN) as well as seed weight (SW) per plant were obtained to present yield parameters.
Root sampling and measurements
A representative plant was selected from each repetition in the field to measure root traits. A square block of soil (40 × 40 cm) with the plant base at the centre was dug to reach the end of the tap root to get the complete plant root system (1 m was the maximum depth for the soil block if the depth of the tap root was more than 1 m). In the field, root width (RW) was measured as the widest width between two basal roots in natural situations and used as the index for root architecture.
Roots were carefully cleaned before being scanned into the computer and the digital images were quantified with computer image analysis software (Win-Rhizo Pro, Régent Instruments, Québec, Canada) for total root length (RL) and total root surface area (RSA).
Construction of a genetic linkage map
The parental genotypes, BD2 and BX10, were first surveyed for 621 simple sequence repeat (SSR) markers selected from an integrated soybean genetic linkage map (Cregan et al., 1999; Song et al., 2004). The primer sequences were obtained from the SoyBase website of the USDA, ARS Soybean Genome Database (http://soybase.agron.iastate.edu/). DNA was extracted from fresh leaf tissues at flowering stage according to the protocol described by Wang and Fang (2003) with slight modifications. All DNA samples were diluted to 20 ng μL−1 with Tris-EDTA buffer (pH 8·0) prior to amplification in polymerase chain reactions (PCRs). The reactions were performed on a Biometra Thermocycler PCR system with 33 cycles. Following PCR amplification, 4 µL of 10× loading buffer was added to the amplified products, and the amplified products were then loaded on 7·2 % denatured polyacrylamide gels in 0·5× TBE buffer. After electrophoresis, the gels were stained with silver to detect polymorphisms.
Of the SSR markers tested, 296 polymorphic and informative markers showing 47·67 % polymorphisms between the parents were chosen as anchors to construct the linkage groups covering all of the 20 linkage groups. Marker order and distance were determined by the Map Manager program QTXb17 (http://www.mapmanager.org/) using the Kosambi function. JoinMap 3.0 software (Stam, 1993; Van Ooijen and Voorrips, 2001) was used for the calculation of genetic map and 277 markers were assigned to the 20 linkage groups as expected from the integrated map (Cregan et al., 1999; Song et al., 2004).
QTL mapping and analysis
QTL detections were performed by using MapQTL 4.0 software (Van Ooijen, 2004) at 5·0 cM intervals to map QTL to the SSR map of the RIL population. QTLs were detected successively by interval mapping (IM) (Lander and Botstein, 1989; Jansen, 1993) and multiple-QTL mapping (MQM) (Jansen, 1994). The IM method was used first to determine the putative QTL involved in the variation of the considered traits. MQM was then performed to eliminate the interference from background or the other QTLs. Background markers were selected to reduce the residual genetic variance. The closest marker to each local logarithm-of-odds (LOD) score peak was considered as a co-factor while testing the presence of a novel QTL elsewhere in the genome. A threshold LOD value of 2·5 was used to justify the presence of a QTL. The LOD significant threshold was estimated from test permutation analysis. The additive effects of the detected QTL were estimated from MQM results. An additive effect represents the mean effect of the replacement of the BD2 allele by the BX10 allele at a particular locus. The additive effect was estimated for each trait by using the following equation: Additive effect = (mean of the ‘BD2’ allele – mean of the ‘BX10’ allele)/2 (Van Ooijen, 2004). The contribution of each detected QTL to the total variance (R2) was estimated by variance component analysis.
Statistical analysis
In total, there were 12 traits, including six root and six P efficiency traits, used for normality, variance and QTL analysis. As the parental genotypes were planted with more repetitions than the RILs in the field, they were analysed separately from the RILs in the two field trials. The frequency distribution of the RILs for all the tested traits was performed using the univariate procedure of SAS (SAS Institute Inc., 1990) and normal distributions were determined using the Shapiro–Wilk test. Broad sense heritability (h2b) was estimated for each trait by using the following equation:
where VG is the variance between RILs and VE is the variance within RILs (Zhu, 2002).
RESULTS
Genetic linkage map construction
Most of the markers used segregated in an expected 1 : 1 ratio in the RIL population. After a χ2 test, 237 of 296 markers (80·07 %) segregated in a 1 : 1 ratio and were revealed as co-dominant markers. Fifty-nine markers (19·93 %) on 14 linkage groups showed distorted segregation with 16 and 43 distorted to BD2 and BX10, respectively. Among the distorted markers, 37 produced a significant (P < 0·05) segregation, and most of them were on the A1, B1, C2 and N linkage groups.
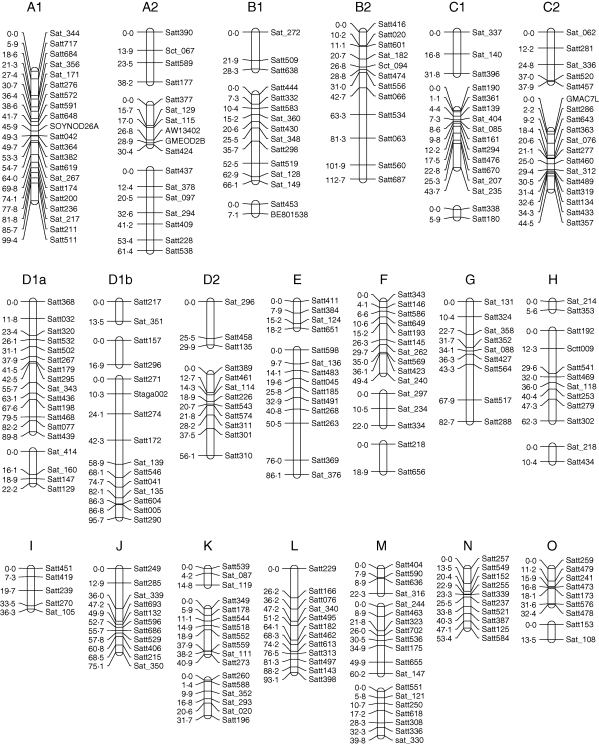
The constructed map contained 20 linkage groups whose total length was 1802·91 cM and had 277 SSR markers (Fig. 1). The average distance between the markers was 6·51 cM. The number of markers on each group ranged from five to 21, and the length of each group ranged from 5·93 to 99·43 cM. In general, SSR markers spread relatively evenly in each linkage group. There were three gaps greater than 25 cM on the D2, E and L linkage groups, due to the lack of appropriate SSR markers. Most of the discordant marker orders occurred within a region smaller than 5 cM. The sequences and distances of the markers were consistent with the soybean public integrated genetic map except for a few SSR markers (Cregan et al., 1999; Song et al., 2004).
Fig. 1.
Soybean genetic linkage map based on an RIL population derived from BD2 and BX10 genotypes using SSR markers.
In order to examine the fidelity of the genetic map constructed in this study, mapping of the gene controlling flower colour was carried out. The location of the gene encoding flower colour was found to be on the linkage group F tightly associated with SSR marker Satt423 at a distance of 0·046 cM (LOD 30·88). This result was comparable with the public genetic map (Cregan et al., 1999) and previous research (Liu et al., 2000).
Phenotypic variation and correlation
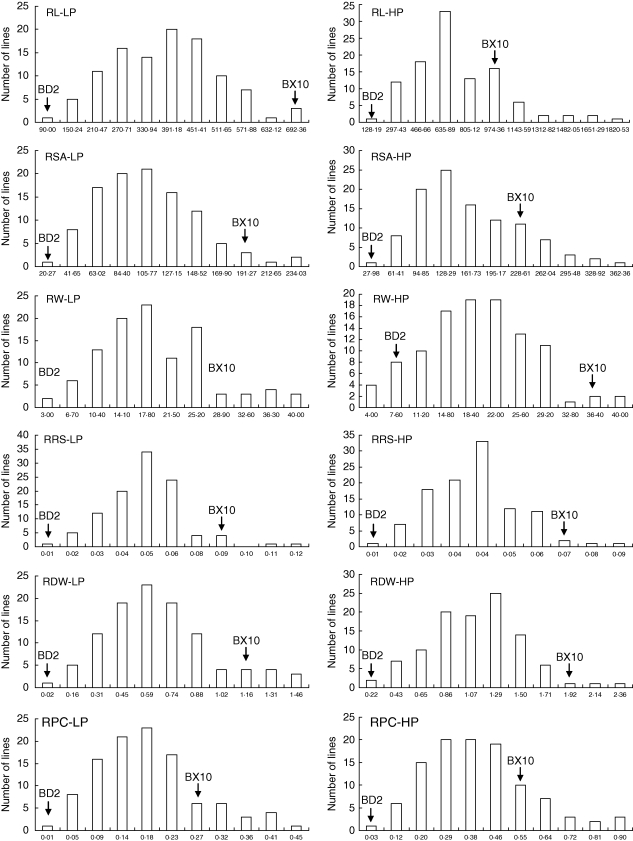
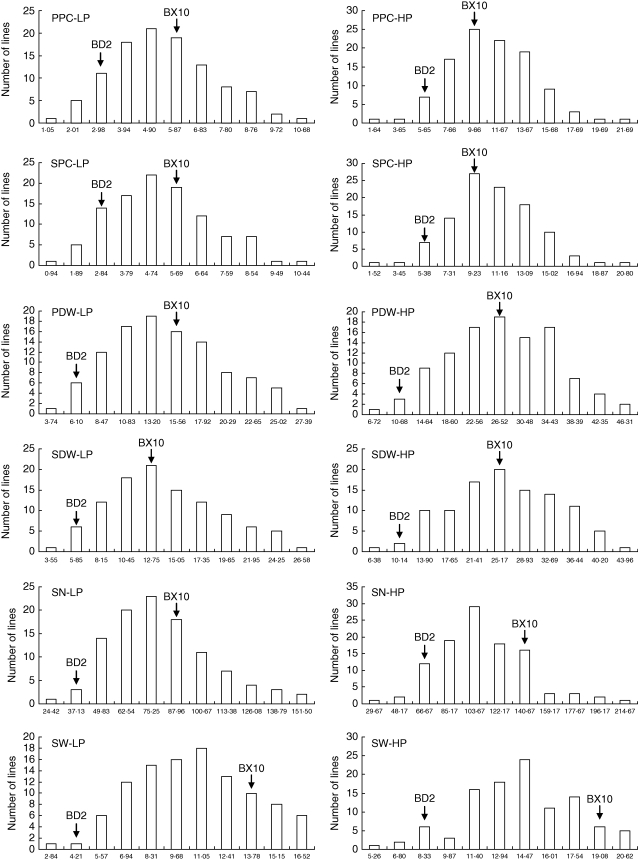
There were 12 traits tested in the present study, including six root and six P efficiency traits. The P efficiency traits consisted of two P content, two biomass and two yield traits. The frequency distribution of the 106 RILs and two parents for 12 traits was normal as determined by Shapiro–Wilks test under both low and high P conditions (Figs 2 and 3). Values of the mean and range for 12 traits are summarized in Table 1. The values of all the tested traits of the P-inefficient parental genotype, BD2, were smaller than those of the P-efficient genotype, BX10, especially at low P level (Table 1). The RILs exhibited a wide range of variations for the tested traits. In general, most traits had higher broad-range heritability (h2b) and lower coefficient of variations (CV%) at low P than those at high P. The yield trait (i.e. seed weight) had higher h2b and lower CV% than P content and biomass traits, indicating a more stable heritability and less environmental effects on yield.
Fig. 2.
Frequency distribution of root traits under low and high P conditions in 106 RILs derived from BD2 and BX10 genotypes.
Fig. 3.
Frequency distribution of P content, biomass and yield traits under low and high P conditions in 106 RILs derived from BD2 and BX10 genotypes.
Table 1.
Means, range, variation and broad-sense heritability (h2b) of root traits from two soybean parental genotypes and their 106 RILs in the field under different P levels.
| P level | Trait | Parents |
RIL |
h2b | ||||
|---|---|---|---|---|---|---|---|---|
| BD2 | BX10 | Mean | Mean | Range | CV (%) | |||
| Low (LP) | Root | |||||||
| RL | 104·36 ± 5·35 | 713·34 ± 27·38 | 408·85 ± 41·95 | 372·07 ± 16·91 | 90·00–692·35 | 41·00 | 0·54 | |
| RSA | 21·05 ± 0·77 | 192·98 ± 4·96 | 107·01 ± 11·70 | 89·01 ± 4·21 | 20·27–234·02 | 46·17 | 0·60 | |
| RW | 3·57 ± 0·18 | 29·14 ± 1·32 | 16·36 ± 1·79 | 16·48 ± 0·83 | 3·00–40·00 | 52·85 | 0·59 | |
| RRS | 0·0092 ± 0·0002 | 0·0863 ± 0·0027 | 0·0477 ± 0·0053 | 0·0488 ± 0·0017 | 0·01–0·12 | 45·75 | 0·51 | |
| RDW | 0·0669 ± 0·0045 | 1·1466 ± 0·050 | 0·6068 ± 0·0745 | 0·5820 ± 0·0290 | 0·02–1·45 | 52·52 | 0·47 | |
| RPC | 0·0132 ± 0·0005 | 0·2527 ± 0·0100 | 0·1330 ± 0·0163 | 0·1645 ± 0·0034 | 0·01–0·45 | 54·80 | 0·54 | |
| P content | ||||||||
| PPC | 2·87 ± 0·23 | 5·73 ± 0·40 | 4·30 ± 0·41 | 4·76 ± 0·19 | 1·00–10·69 | 42·31 | 0·76 | |
| SPC | 2·86 ± 0·23 | 5·48 ± 0·40 | 4·47 ± 0·41 | 4·59 ± 0·18 | 0·94–10·44 | 42·47 | 0·75 | |
| Biomass | ||||||||
| PDW | 7·34 ± 0·43 | 14·44 ± 0·74 | 10·88 ± 0·96 | 12·9656 ± 1·10 | 3·74–27·38 | 42·44 | 0·77 | |
| SDW | 7·27 ± 0·43 | 13·29 ± 0·73 | 10·28 ± 0·95 | 12·3836 ± 0·99 | 3·55–26·57 | 42·63 | 0·75 | |
| Yield | ||||||||
| SN | 40·68 ± 3·92 | 87·83 ± 5·51 | 64·26 ± 6·67 | 73·46 ± 2·52 | 24·42–151·50 | 35·372 | 0·64 | |
| SW | 4·45 ± 0·32 | 13·21 ± 0·55 | 8·83 ± 1·10 | 9·78 ± 0·29 | 2·84–16·52 | 31·55 | 0·81 | |
| High (HP) | Root | |||||||
| RL | 156·99 ± 6·86 | 985·66 ± 44·57 | 571·33 ± 58·55 | 622·47 ± 35·36 | 128·19–1820·52 | 55·00 | 0·68 | |
| RSA | 40·49 ± 1·41 | 233·98 ± 5·36 | 137·24 ± 13·00 | 140·59 ± 6·80 | 27·98–362·36 | 50·00 | 0·72 | |
| RW | 5·61 ± 0·34 | 35·91 ± 0·78 | 20·76 ± 2·01 | 16·41 ± 0·78 | 4·00–40·00 | 46·81 | 0·68 | |
| RRS | 0·0080 ± 0·0002 | 0·0756 ± 0·0024 | 0·0418 ± 0·0045 | 0·0408 ± 0·0010 | 0·01–0·09 | 27·31 | 0·63 | |
| RDW | 0·0925 ± 0·005 | 1·9293 ± 0·051 | 1·0109 ± 0·1222 | 0·9034 ± 0·0390 | 0·22–2·35 | 44·55 | 0·58 | |
| RPC | 0·0276 ± 0·0007 | 0·5010 ± 0·0209 | 0·2643 ± 0·0325 | 0·3519 ± 0·0060 | 0·03–0·90 | 50·43 | 0·65 | |
| P content | ||||||||
| PPC | 5·59 ± 0·27 | 9·64 ± 0·32 | 7·62 ± 0·43 | 10·03 ± 0·32 | 1·64–21·70 | 33·13 | 0·85 | |
| SPC | 5·57 ± 0·27 | 9·14 ± .032 | 7·35 ± 0·42 | 9·67 ± 0·31 | 1·52–20·80 | 33·07 | 0·84 | |
| Biomass | ||||||||
| PDW | 11·67 ± 0·53 | 27·46 ± 0·87 | 19·57 ± 1·68 | 22·82 ± 2·33 | 6·72–46·31 | 32·96 | 0·86 | |
| SDW | 11·58 ± 0·53 | 25·53 ± 0·86 | 18·56 ± 1·65 | 21·912 ± 2·30 | 6·38–43·96 | 32·88 | 0·85 | |
| Yield | ||||||||
| SN | 72·94 ± 6·03 | 137·04 ± 5·58 | 105·00 ± 9·16 | 100·02 ± 3·43 | 29·67–214·67 | 35·32 | 0·79 | |
| SW | 8·87 ± 0·98 | 20·04 ± 0·95 | 14·46 ± 1·50 | 12·89 ± 0·37 | 5·26–20·61 | 22·99 | 0·93 | |
RL, total root length; RSA, total root surface area; RW, root width; RRS, ratio of root/shoot; RDW, root dry weight; RPC, root P content; PPC, plant P content; SPC, shoot P content; PDW, plant dry weight; SDW, shoot dry weight; SN, seed number per plant; SW, seed weight per plant.
The correlation coefficients between root traits and P efficiency parameters at both low P and high P levels are shown in Table 2. Under low P conditions, there were negative correlations between the ratio of root/shoot (RRS) and P content (PPC and SPC), as well as biomass (PDW and SDW), whereas significant positive correlations existed among the other five root traits and P efficiency parameters. Under high P conditions, positive correlations among all the root traits and P efficiency parameters were found, most of which reached significance. Also at high P, the relationships of root traits with P content and biomass parameters were much closer than with yield traits, suggesting that root traits were more closely correlated with P uptake and biomass formation. Furthermore, the correlations among root width and P content, biomass and yield traits at low P were stronger than at high P, indicating that root width might play an important role in response to P stress in soybean.
Table 2.
Phenotypic correlation coefficients among root traits and P content, biomass and yield traits of 106 RILs in the field under different P levels
| P level | Trait | PPC | SPC | PDW | SDW | SN | SW |
|---|---|---|---|---|---|---|---|
| LP | RL | 0·43** | 0·56** | 0·47** | 0·55** | 0·44** | 0·46** |
| RSA | 0·59** | 0·70** | 0·65** | 0·70** | 0·47** | 0·47** | |
| RW | 0·56** | 0·62** | 0·62** | 0·63** | 0·46** | 0·43** | |
| RRS | −0·21** | −0·03 | −0·17 | −0·04 | 0·32** | 0·32** | |
| RDW | 0·67** | 0·80** | 0·74** | 0·81** | 0·53** | 0·51** | |
| RPC | 0·70** | 0·87** | 0·76** | 0·82** | 0·51** | 0·49** | |
| HP | RL | 0·38** | 0·55** | 0·44** | 0·56** | 0·22* | 0·25* |
| RSA | 0·59** | 0·73** | 0·65** | 0·73** | 0·37** | 0·29** | |
| RW | 0·43** | 0·43** | 0·44** | 0·43** | 0·08 | 0·21* | |
| RRS | 0·09 | 0·28** | 0·14 | 0·24* | 0·21** | 0·11 | |
| RDW | 0·74** | 0·83** | 0·80** | 0·82** | 0·40** | 0·34** | |
| RPC | 0·71** | 0·90** | 0·73** | 0·80** | 0·32** | 0·26** |
Asterisks indicate significant at *5 % level of probability, **1 % level of probability. RL, total root length; RSA, total root surface area; RW, root width; RRS, ratio of root/shoot; RDW, root dry weight; RPC, root P content; PPC, plant P content; SPC, shoot P content; PDW, plant dry weight; SDW, shoot dry weight; SN, seed number per plant; SW, seed weight per plant.
Mapping QTLs for root traits
In total, 31 putative QTLs were found to be associated with the 12 tested traits (Tables 3 and 4). Among them, 18 and 13 QTLs were identified at low P and high P level, respectively. Furthermore, 13 putative QTLs were found for root traits (Table 3), five for P content, five for biomass and eight for yield traits (Table 4).
Table 3.
Putative QTLs detected for six root traits by MQM using 106 RILs derived from BD2 and BX10 genotypes in the field under different P levels
| P level | Trait | Locus name | Interval | LG* | Position (cM)† | LOD‡ | Variance (%)§ | Additive effect¶ |
|---|---|---|---|---|---|---|---|---|
| LP | RL | qRL-b1_2-A | Satt519-Sat_128 | B1 | 4·997 | 3·81 | 14·4 | −80·54 |
| RL | qRL-d1a_1-B | Sat_343 | D1a | 3·205 | 3·36 | 16·8 | −91·52 | |
| RSA | qRSA-b1_2-A | Satt519-Sat_128 | B1 | 4·997 | 4·35 | 18·7 | −20·53 | |
| RSA | qRSA-d1a_1-B | Sat_343 | D1a | 3·205 | 2·72 | 14·4 | −19·21 | |
| RW | qRW-b1_2-A | Sat_348-Satt298 | B1 | 5·01 | 2·80 | 14·9 | −3·39 | |
| RRS | qRRS-b1_2-A | Satt519 | B1 | 0·003 | 2·85 | 11·8 | −0·0064 | |
| RDW | qRDW-b1_2-A | Satt519-Sat_128 | B1 | 4·997 | 3·02 | 15·2 | −0·1196 | |
| RDW | qRDW-d2_1-B | Satt458 | D2 | 0·021 | 2·98 | 10·9 | −0·0990 | |
| RPC | qRPC-d2_1-A | Satt458 | D2 | 0·021 | 2·95 | 12·0 | −0·0312 | |
| HP | RL | qRL-b1_2-C | Satt519 | B1 | 0·003 | 3·35 | 13·7 | −136·71 |
| RSA | qRSA-b1_2-C | Satt519 | B1 | 0·003 | 3·92 | 15·8 | −28·25 | |
| RDW | qRDW-b1_2-C | Satt519 | B1 | 0·003 | 3·86 | 15·5 | −0·1604 | |
| RPC | qRPC-b1_2-B | Satt519 | B1 | 0·003 | 3·33 | 13·5 | −0·0660 |
* Linkage group.
† QTL position from the first marker.
‡ Maximum likelihood LOD score for the individual QTL.
§ Phenotypic variation explained by each QTL.
¶ The mean effect of the replacement of the BD2 allele by the BX10 allele at a particular locus.
Table 4.
Putative QTLs detected for P content, biomass and yield traits by MQM using 106 RILs derived from BD2 and BX10 genotypes in the field under different P levels
| P level | Trait | Locus name | Interval | LG* | Position (cM)† | LOD‡ | Variance (%)§ | Additive effect¶ |
|---|---|---|---|---|---|---|---|---|
| PPC | qPPC-d2_1-A | Sat_296-Satt458 | D2 | 20·000 | 2·53 | 18·2 | −0·8578 | |
| SPC | qSPC-d2_1-A | Satt458 | D2 | 0·021 | 2·79 | 11·4 | −0·2602 | |
| PDW | qPDW-d2_1-A | Sat_296-Satt458 | D2 | 20·000 | 2·78 | 20·0 | −2·44 | |
| SDW | qSDW-d2_1-B | Satt458 | D2 | 0·021 | 2·61 | 10·7 | −0·88 | |
| SDW | qSDW-b1_2-A | Satt519-Sat_128 | B1 | 4·997 | 3·09 | 22·3 | −1·19 | |
| SN | qSN-b1_2-A | Satt519 | B1 | 0·003 | 5·04 | 19·7 | −11·68 | |
| SN | qSN-c1_2-B | Sat_404 | C1 | 0·032 | 2·78 | 11·4 | −8·85 | |
| SW | qSW-b1_2-A | Satt519-Sat_128 | B1 | 4·997 | 6·71 | 31·1 | −1·77 | |
| SW | qSW-c1_2-B | Sat_404 | C1 | 0·032 | 2·76 | 11·3 | −1·05 | |
| HP | PPC | qPPC-b1_2-B | Satt519 | B1 | 0·003 | 2·52 | 10·4 | −1·0844 |
| SPC | qSPC-b1_2-B | Satt519 | B1 | 0·003 | 4·66 | 18·3 | −0·6521 | |
| SPC | qSPC-f_1-C | Satt423 | F | 0·049 | 2·73 | 9·1 | 0·4544 | |
| PDW | qPDW-b1_2-B | Satt519 | B1 | 0·003 | 3·79 | 15·2 | −2·97 | |
| SDW | qSDW-b1_2-C | Satt519 | B1 | 0·003 | 5·28 | 20·5 | −1·78 | |
| SN | qSN-b1_2-C | Satt519 | B1 | 0·003 | 3·55 | 14·3 | −13·54 | |
| SN | qSN-c1_2-D | Sat_404 | C1 | 0·032 | 2·78 | 11·4 | −12·03 | |
| SW | qSW-b1_2-C | Satt519 | B1 | 0·003 | 3·85 | 15·4 | −1·18 | |
| SW | qSW-b1_2-D | Satt519-Sat_128 | B1 | 4·997 | 3·90 | 16·7 | −1·29 |
*Linkage group.
† QTL position from the first marker.
‡ Maximum likelihood LOD score for the individual QTL.
§ Phenotypic variation explained by each QTL.
¶ The mean effect of the replacement of the BD2 allele by the BX10 allele at a particular locus.
All the QTLs for root traits could explain more than 10 % of phenotypic variation, ranging from 10·9 to 18·7 % individually (Table 3). All of the favourable alleles were derived from the parent BX10 as indicated by negative values of additive effects. Nine QTLs were found on the B1, D1a and D2 linkage groups at low P, which were involved in six root traits. There were only four QTLs involved in four root traits found on the B1 linkage group under high P. Three putative QTLs for total root length (RL) were detected, two of which were mapped at low P and one at high P. Three QTLs for total root surface area (RSA) were identified, and were located on the same marker intervals and linked to the same SSR markers as the three QTLs controlling RL at low P and high P levels, respectively. This indicated that RL and RSA were genetically closer. There was one QTL for root width (RW) and one for the RRS at low P but not at high P (Table 3). Three putative QTLs for root dry weight (RDW) were detected with two at low P and one at high P. Two QTLs controlling root P content (RPC) were identified at low P and high P, respectively.
Mapping QTLs for P content, biomass and yield
Eighteen QTLs associated with six traits for P content, biomass and yield were found, and were located on the B1, C1, D2 and F linkage groups (Table 4). Among them, five QTLs controlled for P content, five for biomass and eight for yield traits. Each QTL accounted for 9·1–31·1 % of the phenotypic variation, and 17 of them could explain more than 10 % of the phenotypic variance individually. Most of the favourable alleles were derived from the parent BX10, which had superior root traits, especially under P stress.
Two QTL controlling plant P content (PPC) were found, one at low P and one at high P (Table 4). One QTL related to shoot P content (SPC) at low P and two at high P were found. Two QTLs for plant dry weight (PDW) were detected, one at low P in the interval Sat_296–Satt458 on the D2 linkage group, and the other at high P on the B1 linkage group linking to Satt519. Three QTLs for shoot dry weight (SDW) were found, two of them at low P.
Four QTLs for seed number per plant (SN) were identified (Table 4). Two were at low P on the B1 linkage group linked to Satt519 and the C1 linkage group to Sat_404, respectively. The other two were detected at high P on the B1 linkage group. Four QTLs for seed weight per plant (SW) were found, with one major QTL in the interval Satt519–Sat_128 on the B1 linkage group, explaining 31·1 % of the variance at low P (Table 4).
Linkage analysis of root traits with P efficiency parameters
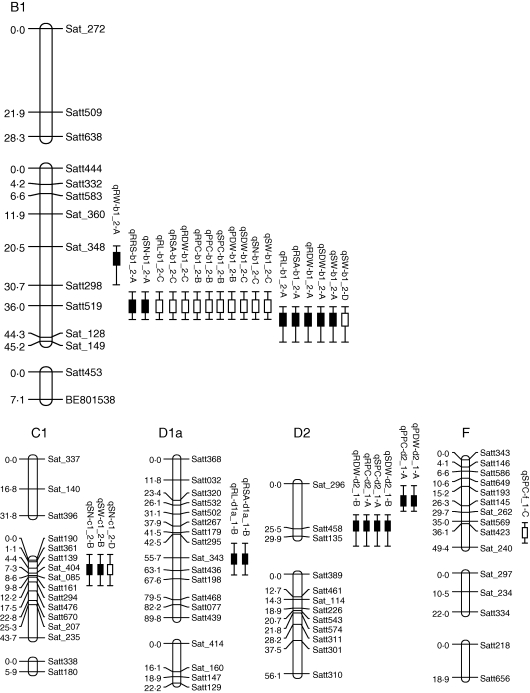
Further QTL linkage analysis found that there were four clusters of QTLs conferring root traits and P efficiency parameters simultaneously (Fig.4). Among these, three clusters of QTLs were identified from low P and one from high P. The QTL cluster for RRS and SN was on the B1 linkage group close to Satt519. The QTL cluster conferring RL, RSA, RDW, SDW and SW were on the B1 linkage group in the interval Satt519–Sat_128. The QTL cluster associated with RDW, RPC, SDW and SPC was found on the D2 linkage group close to Satt458. Interestingly, the only QTL cluster from high P controlling RL, RSA, RDW, RPC, PDW, PPC, SDW, SPC, SN and SW was on the same region as the QTL for RRS and SN at low P, which was on the B1 linkage group close to Satt519. The QTL for SW at high P was also located on the B1 linkage group in the interval Satt519–Sat_128, which conferred RL, RSA, RDW, SDW and SW at low P.
In addition, there were some QTLs associated with two or more traits presenting relative characters (Fig. 4). The QTL on the C1 linkage group close to Sat_404 controlled two seed formation parameters, SN and SW at low P, and SN at high P. The QTL on the D1a group close to Sat_343 conferred two root traits, RSA and RL, at low P.
Fig. 4.
Five linkage groups harbouring 31 putative QTLs for root traits as related to P efficiency in an RIL population under low and high P conditions in field trails. The designation on the right is the marker name, and that on the left is the Kosambi function. The box delineates the position of the QTL having the highest LOD value, and the whiskers of each box delineate the QTL support interval. The solid and open boxes represent the QTLs identified at low and high P, respectively.
DISCUSSION
Mapping QTL is becoming increasingly important in modern breeding programmes through marker-assisted selection (MAS) and gene discovery (Price, 2006). There are many successful examples of using QTLs to facilitate crop cultivar development, especially for disease resistance (Su et al., 2002; Zuo et al., 2007; Beaver and Osorno 2009; Swarbrick et al., 2009) and quality improvement (Blair et al., 2009; Sabouri 2009). Slow progress has been made in the improvement of traits such as adaptation to abiotic stresses (e.g. drought, low P availability) with better root morphology and architecture using conventional breeding methods. The difficulty in directly measuring root traits and the significance of genotype × environment interactions make the selection of these traits more complicated. Therefore, using the genomic approach to examine complex traits such as root morphology and those of architecture should help breeders to devise more effective selection strategies. For instance, the QTLs influencing root morphology and other drought-related traits have been successfully used in MAS to improve upland rice varieties (Champoux et al., 1995; Price and Tomos, 1997; Yadav et al., 1997; Steele et al., 2006). Until now, there has been a lack of successful examples of using QTLs to facilitate genetic improvement in crop P efficiency. In the present study, QTL analysis was used to further define root traits associated with P efficiency parameters, which might facilitate the genetic improvement of P efficiency in soybean, obviating the need for the direct evaluation of the root system.
The selection of suitable parental genotypes is critical in identifying important QTLs. For example, most important QTLs associated with root traits and P efficiency in common bean were detected using a cross between DOR364 and G19833, which not only contrast for P efficiency but also for root architecture (Liao et al., 2004; Beebe et al., 2006). The two parental genotypes used in the present study also contrast for both P efficiency and root architecture (Zhao et al., 2004). The distinct evolutionary background of these two germplasms, with BX10 derived from Brazil and BD2 from southern China, may have contributed to the physiological differences that were observed between them (Table 1). Therefore, the genetic linkage map reported here is the first soybean genetic map constructed based on field data from the parental genotypes contrasting both in P efficiency and root architecture, as well as their F9 RILs (Fig. 1), which should prove useful for mapping QTLs for root and P efficiency traits.
As the hidden part of plants, roots are difficult to quantify. Therefore, studies on roots are lagging behind those on shoots (Epstein, 2004). Although some QTLs controlling root traits have been reported, most are based on root weight and/or some root morphological traits such as root length and specific root length (Reiter et al., 1991; Beebe et al., 2006). Along with root morphological traits, root architecture plays an important role in P efficiency due to the low mobility of P in soils. This has been shown by the results from a common bean study, which found that three QTLs for root architecture traits in growth pouch were associated with QTLs for P acquisition in the field (Liao et al., 2004). Thus far, however, no QTLs conferring root morphological and/or architectural traits as related to P efficiency in soybean have been reported. The present study detected 13 putative QTLs for six root traits (Table 3). Interestingly, all the QTLs for root traits were from BX10, the shallower root genotype, especially for RL with additive effects of 80·54 and 136·71 at low P and high P, respectively. This suggests these regions of the soybean genome could be used for improvement of the root system under both low P and high P conditions.
Phosphorus efficiency is defined as the ability of plants to produce higher biomass or yield, and/or take up more P, under inadequate P conditions (Yan et al., 2006). Interestingly, none of the QTLs reported here is close to the soybean QTLs controlling SDW, RDW, PDW, PUE and PAE reported by Zhang et al. (2009) (Table 4). This is probably because the QTLs detected by Zhang et al. (2009) were from a pot experiment with limited soil volume, which might inhibit root growth. In turn, this condition could inhibit overall plant growth and P uptake, and thus not reflect P efficiency in agricultural fields. The present study analysed QTLs based on data from the field, and therefore the results should be more representative than previous reports. In addition, although this and Zhang et al.'s (2009) study used the same soil type, acid soils, other environmental conditions such as temperature, photoperiod, light intensity and water regime could also affect soybean growth, and the RIL genotypes developed for the different objectives of these two studies could also contribute to the differences observed.
In order to study the relationship among root traits and P efficiency, phenotypic correlations were calculated and QTL linkage analysis was conducted. Significant correlations exist among root traits and most P efficiency parameters (Table 2), indicating that these traits might be closely related to each other. Some of the putative QTLs detected could control root traits and P efficiency parameters simultaneously (Fig. 4). This genetically demonstrates that root traits play critical roles in soybean P efficiency, like the functions of roots in common bean (Liao et al., 2004; Beebe et al., 2006), rice (Zhu et al., 2005b, 2006; Shimizu et al., 2008; Li et al., 2009) and maize (Zhu et al., 2005a, b, 2006). More interestingly, three of four QTLs conferring root traits together with P efficiency parameters were from the low P treatment, and two of them were on the B1 linkage group close to the marker Satt519, indicating that root traits contributed more to P efficiency under P stress. The only QTL associated with both root traits and P efficiency parameters identified under high P conditions is also located in the same region, suggesting that this is an important QTL which might have great commercial potential for future genetic improvement of soybean P efficiency.
Conclusions
A soybean genetic linkage map was constructed based on field data using the F9 RILs from a cross between BD2 and BX10. In total, 31 QTLs controlling root traits and P efficiency parameters were identified. Among them, 13 QTLs were found for root traits, and 18 for P efficiency parameters. At least four QTLs conferred root traits and P efficiency parameters simultaneously, suggesting critical roles of roots in soybean P efficiency. These findings could have great potential for improving soybean genetic breeding on acid soils, where P deficiency is predominant.
ACKNOWLEDGEMENTS
We thank Dr Shaowei Huang for helping with QTL analysis and Larry York with the English text. This research is in part supported by the 7th international symposium on plant–soil interactions at low pH, the National Key Basic Research Special Funds of China (Grant No. 2005CB120902) and the National Natural Science Foundation of China (Grant No. 30890131).
LITERATURE CITED
- Abbas M, Monib M, Rammah A, Fayez M, Hegazi N. Intercropping of sesbania (Sesbania sesban) and leucaena (Leucaena leucocephala) with five annual grasses under semi-arid conditions as affected by inoculation with specific rhizobia and associative diazotrophs. Agronomy. 2001;21:517–525. [Google Scholar]
- Beaver JS, Osorno JM. Achievements and limitations of contemporary common bean breeding using conventional and molecular approaches. Euphytica. 2009;168:145–175. [Google Scholar]
- Beebe SE, Rojas-Pierce M, Yan XL, et al. Quantitative trait loci for root architecture traits correlated with phosphorus acquisition in common bean. Crop Science. 2006;46:413–423. [Google Scholar]
- Blair MW, Sandoval TA, Caldas GV, Beebe SE, Paez MI. Quantitative trait locus analysis of seed phosphorus and seed phytate content in a recombinant inbred line population of common bean. Crop Science. 2009;49:237–246. [Google Scholar]
- Boddey RM, Sa JCD, Alves BJR, Urquiaga S. The contribution of biological nitrogen fixation for sustainable agricultural systems in the tropics. Soil Biology and Biochemistry. 1997;29:787–799. [Google Scholar]
- Bryla D, Koide RT. Mycorrhizal response of two tomato genotypes relates to their ability to acquire and utilize phosphorus. Annals of Botany. 1998;82:849–857. [Google Scholar]
- Chen JY, Xu L, Cai YL, Xu J. Identification of QTLs for phosphorus utilization efficiency in maize (Zea mays L.) across P levels. Euphytica. 2009;167:245–252. [Google Scholar]
- Cichy KA, Blair MW, Mendoza CHG, Snapp SS, Kelly JD. QTL analysis of root architecture traits and low phosphorus tolerance in an Andean bean population. Crop Science. 2009;49:59–68. [Google Scholar]
- Champoux MC, Wang G, Sarkarung S, et al. Locating genes associated with root morphology and drought avoidance in rice via linkage to molecular markers. Theoretical and Applied Genetics. 1995;90:969–981. doi: 10.1007/BF00222910. [DOI] [PubMed] [Google Scholar]
- Cregan PB, Jarvik T, Bush AL, et al. An integrated genetic linkage map of the soybean genome. Crop Science. 1999;39:1464–1490. [Google Scholar]
- Dakora FD, Phillips DA. Root exudates as mediators of mineral acquisition in low-nutrient environments. Plant and Soil. 2002;245:35–47. [Google Scholar]
- de Dorlodot S, Forster B, Pages L, Price A, Tuberosa R, Draye X. Root system architecture: opportunities and constraints for genetic improvement of crops. Trends in Plant Science. 2007;12:474–481. doi: 10.1016/j.tplants.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Epstein E. Plant biologists need to get back to their roots. Nature. 2004;430:804–805. doi: 10.1038/430829c. [DOI] [PubMed] [Google Scholar]
- Hinsinger P. Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: a review. Plant and Soil. 2001;237:173–195. [Google Scholar]
- Jansen RC. Interval mapping of multiple quantitative trait loci. Genetics. 1993;135:205–211. doi: 10.1093/genetics/135.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RC. Controlling the Type I and Type II errors in mapping quantitative trait loci. Genetics. 1994;138:871–881. doi: 10.1093/genetics/138.3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Xie Y, Dai AY, Liu LF, Li ZC. Root and shoot traits responses to phosphorus deficiency and QTL analysis at seedling stage using introgression lines of rice. Journal of Genetics and Genomics. 2009;36:173–183. doi: 10.1016/S1673-8527(08)60104-6. [DOI] [PubMed] [Google Scholar]
- Li YD, Wang YJ, Tong YP, Gao JG, Zhang JS, Chen SY. QTL mapping of phosphorus deficiency tolerance in soybean (Glycine max L. Merr.) Euphytica. 2005;142:137–142. [Google Scholar]
- Li ZX, Ni ZF, Peng HR, et al. Molecular mapping of QTLs for root response to phosphorus deficiency at seedling stage in wheat (Triticum aestivum L.) Proceedings of the National Academy of Sciences. 2007;17:1177–1184. [Google Scholar]
- Liao H, Yan XL, Rubio G, Beebe SE, Blair MW, Lynch JP. Genetic mapping of basal root gravitropism and phosphorus acquisition efficiency in common bean. Functional Plant Biology. 2004;31:959–970. doi: 10.1071/FP03255. [DOI] [PubMed] [Google Scholar]
- Liu F, Zhuang BC, Zhang JS, Chen SY. Construction and analysis of soybean genetic map. Acta Genetica Sinica. 2000;27:1018–1026. [PubMed] [Google Scholar]
- Lynch JP. Root architecture and plant productivity. Plant Physiology. 1995;109:7–13. doi: 10.1104/pp.109.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP, Brown KM. Topsoil foraging – an architectural adaptation of plants to low phosphorus availability. Plant and Soil. 2001;237:225–237. [Google Scholar]
- Murphy J, Riley JP. A modified single solution method for the determination of phosphorus in natural waters. Analytica Chimica Acta. 1962;27:31–36. [Google Scholar]
- Price AH. Believe it or not, QTLs are accurate! Trends in Plant Science. 2006;11:213–216. doi: 10.1016/j.tplants.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Price AH, Tomos AD. Genetic dissection of root growth in rice (Oryza sativa L.). Mapping quantitative trait loci using molecular markers. Theoretical and Applied Genetics. 1997;95:143–152. [Google Scholar]
- Raghothama KG. Phosphate acquisition. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50:665–693. doi: 10.1146/annurev.arplant.50.1.665. [DOI] [PubMed] [Google Scholar]
- Raghothama KG. Phosphorus acquisition: plant in the driver's seat! Trends in Plant Science. 2000;5:412–413. doi: 10.1016/s1360-1385(00)01746-5. [DOI] [PubMed] [Google Scholar]
- Reiter RS, Coor JG, Sussman MR, Gabelman WH. Genetic analysis of tolerance to low-phosphorus stress in maize using restriction fragment length polymorphisms. Theoretical and Applied Genetics. 1991;82:561–568. doi: 10.1007/BF00226791. [DOI] [PubMed] [Google Scholar]
- Sabouri H. QTL detection of rice grain quality traits by microsatellite markers using an indica rice (Oryza sativa L.) combination. Journal of Genetics. 2009;88:81–85. doi: 10.1007/s12041-009-0011-4. [DOI] [PubMed] [Google Scholar]
- Sample EC, Soper RJ, Racz GJ. Reaction of phosphate fertilizers in soils. In: Khasawneh FE, Sample EC, Kamprath EJ, editors. The role of phosphorus in agriculture. Madison, WI: American Society of Agronomy; 1980. pp. 263–310. [Google Scholar]
- Shimizu A, Kato K, Komatsu A, Motomura K, Ikehashi H. Genetic analysis of root elongation induced by phosphorus deficiency in rice (Oryza sativa L.): fine QTL mapping and multivariate analysis of related traits. Theoretical and Applied Genetics. 2008;117:987–996. doi: 10.1007/s00122-008-0838-8. [DOI] [PubMed] [Google Scholar]
- Song QJ, Marek LF, Shoemaker RC, et al. A new integrated genetic linkage map of the soybean. Theoretical and Applied Genetics. 2004;109:122–128. doi: 10.1007/s00122-004-1602-3. [DOI] [PubMed] [Google Scholar]
- Stam P. Construction of integrated genetic linkage maps by means of a new computer package: JoinMap. The Plant Journal. 1993;3:739–744. [Google Scholar]
- Steele KA, Price AH, Shashidhar HE, Witcombe JR. Marker-assisted selection to introgress rice QTLs controlling root traits into an Indian upland rice variety. Theoretical and Applied Genetics. 2006;112:208–221. doi: 10.1007/s00122-005-0110-4. [DOI] [PubMed] [Google Scholar]
- Stevenson FJ. Cycles of soil: carbon, nitrogen, phosphorus, sulfur, micronutrients. New York: John Wiley & Sons Inc; 1986. [Google Scholar]
- Su CC, Cheng XN, Zhai HQ, Wan JM. Detection and analysis of QTL for resistance to the brown planthopper, Nilaparvata lugens (Stal), in rice (Oryza sativa L.), using backcross inbred lines. Acta Genetica Sinica. 2002;29:332–338. [PubMed] [Google Scholar]
- Swarbrick PJ, Scholes JD, Press MC, Slate J. A major QTL for resistance of rice to the parasitic plant Striga hermonthica is not dependent on genetic background. Pest Management Science. 2009;65:528–532. doi: 10.1002/ps.1719. [DOI] [PubMed] [Google Scholar]
- Van Ooijen JW. MapQTL 4.0, software for the mapping of quantitative trait loci in experimental populations. Wageningen, The Netherlands: Kyazma BV; 2004. [Google Scholar]
- Van Ooijen JW, Voorrips RE. JoinMap3.0, software for the calculation of genetic maps. Wageningen, The Netherlands: Kyazma BV; 2001. [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL. Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytologist. 2003;157:423–447. doi: 10.1046/j.1469-8137.2003.00695.x. [DOI] [PubMed] [Google Scholar]
- Wang J, Fang XJ. Isolation of plant DNA. Molecular Plant Breeding. 2003;1:281–288. [Google Scholar]
- Yadav R, Courtois B, Huang N, McLaren G. Mapping genes controlling root morphology and root distribution in a doubled-haploid population of rice. Theoretical and Applied Genetics. 1997;94:619–632. [Google Scholar]
- Yan XL, Wu P, Ling HQ, Xu GH, Xu FS, Zhang QF. Plant nutriomics in China: an overview. Annals of Botany. 2006;98:473–482. doi: 10.1093/aob/mcl116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Cheng H, Geng LY, et al. Detection of quantitative trait loci for phosphorus deficiency tolerance at soybean seedling stage. Euphytica. 2009;167:313–322. [Google Scholar]
- Zhao J, Fu JB, Liao H, et al. Characterization of root architecture in an applied core collection for phosphorus efficiency of soybean germplasm. Chinese Science Bulletin. 2004;49:1611–1620. [Google Scholar]
- Zhu J. Genetics. 3rd edn. Beijing: Chinese Agricultural Press; 2002. [Google Scholar]
- Zhu JM, Kaeppler SM, Lynch JP. Mapping of QTL controlling root hair length in maize (Zea mays L.) under phosphorus deficiency. Plant and Soil. 2005a;270:299–310. [Google Scholar]
- Zhu JM, Kaeppler SM, Lynch JP. Mapping of QTLs for lateral root branching and length in maize (Zea mays L.) under differential phosphorus supply. Theoretical and Applied Genetics. 2005b;111:688–695. doi: 10.1007/s00122-005-2051-3. [DOI] [PubMed] [Google Scholar]
- Zhu JM, Mickelson SM, Kaeppler SM, Lynch JP. Detection of quantitative trait loci for seminal root traits in maize (Zea mays L.) seedlings grown under differential phosphorus levels. Theoretical and Applied Genetics. 2006;113:1–10. doi: 10.1007/s00122-006-0260-z. [DOI] [PubMed] [Google Scholar]
- Zuo SM, Yin YJ, Zhang L, Zhang YF, Chen ZX, Pan XB. Breeding value and further mapping of a QTL qSB-11 conferring the rice sheath blight resistance. Zhongguo Shuidao Kexue. 2007;21:136–142. [Google Scholar]