Abstract
Telomerase expression, resulting from transcriptional activation of the hTERT gene, allows cells to acquire indefinite proliferative potential during cellular immortalization and tumorigenesis. However, mechanisms of hTERT gene activation in many immortal cell lines and cancer cells are poorly understood. Here, we report our studies on hTERT activation using genetically related pairs of telomerase-negative (Tel−) and -positive (Tel+) fibroblast lines. First, whereas transiently transfected plasmid reporters did not recapitulate the endogenous hTERT promoter, the promoter in chromosomally integrated bacterial artificial chromosome (BAC) reporters was activated in a subset of Tel+ cells, indicating that activation of the hTERT promoter required native chromatin context and/or distal regulatory elements. Second, the hTERT gene, located near the telomere of chromosome 5p, was translocated in all three Tel+ cell lines but not in their parental pre-crisis cells and Tel− immortal siblings. The breakage points were mapped to regions upstream of the hTERT promoter, indicating that the hTERT gene was the target of these chromosomal rearrangements. In two Tel+ cell lines, translocation of the endogenous hTERT gene appeared to be the major mechanism of its activation as the activity of hTERT promoter in many chromosomally integrated BAC reporters, with intact upstream and downstream neighboring loci, remained relatively low. Therefore, our results suggest that rearrangement of upstream sequences is an important new mechanism of hTERT promoter activation during cellular immortalization. The chromosomal rearrangements likely occurred during cellular crisis and facilitated by telomere dysfunction. Such translocations allowed the hTERT promoter to escape from the native condensed chromatin environment.
INTRODUCTION
Telomeres, consisting of TTAGGG repeats, are specialized nucleoprotein complexes that serve as protective caps of chromosomal ends and are essential for chromosomal stability and long-term cell survival. Telomeres are replenished by telomerase (Morin, 1989), a ribonucleoprotein reverse transcriptase complex containing an RNA (TER) and a protein catalytic subunit (TERT). In the absence of telomere maintenance, as in most human somatic cells, telomeres progressively shorten upon successive cell divisions. Telomere attrition in these cells leads to a permanent cell cycle arrest (known as M1 senescence) at the end of their life span (Hayflick limit). In normal cells, M1 senescence is likely to be caused by TP53- and RB1-dependent DNA damage response initiated by short telomeres. However, when these tumor-suppressing mechanisms are absent, cells fail to enter M1 senescence and continue to proliferate despite continuing telomere shortening. This leads to the loss of telomere capping activity, culminating in severe chromosomal instability and massive cell death, a process referred to as M2 crisis. During crisis, chromosomes undergo breakage-fusion-bridge cycles due to complete loss of capping in a subset of telomeres, resulting in extensive chromosomal rearrangements. Spontaneous immortal cells emerge at an extremely low frequency (~10−7) during crisis (Shay and Wright, 1989). Cells surviving crisis either have activated telomerase, as in telomerase-positive (Tel+) cell lines, or use an alternative telomere lengthening (ALT) mechanism (Cesare and Reddel, 2008), as in telomerase-negative (Tel−) cell lines, to stabilize the existing telomeres and alleviate severe chromosome instability. Although in vitro spontaneous immortalization and telomerase activation occur only after crisis, the mechanistic link between telomerase activation and short telomere induced crisis has yet to be elucidated.
hTERT transcription is the rate-limiting step in telomerase expression. The level of hTERT mRNA is generally correlated with telomerase activity (Aisner et al., 2002), whereas the RNA subunit, TERC, is expressed in all tissues (Blasco et al., 1996). It has been previously demonstrated that ectopic expression of hTERT resulted in telomere stabilization, immortalization, and tumorigenic conversion of pre-neoplastic human cells (Bodnar et al., 1998; Counter et al., 1998; Hahn et al., 1999; Zhu et al., 1999).
Whereas the hTERT gene is silenced in most somatic cells, it is activated in the majority of cancer cells (Kim et al., 1994). Multiple mechanisms of hTERT activation have been postulated in the last decade. For instances, the c-MYC protein was found to bind to the E-boxes at the hTERT core promoter and activated hTERT promoter in some cancer cells ( Wang et al., 1998; Greenberg et al., 1999). The viral oncoprotein E6 from a subset of human papillomaviruses (HPVs) also mediated hTERT transcription through the E-boxes in some epithelial cells (Klingelhutz et al., 1996; Veldman et al., 2003; Xu et al., 2008). In spite of these findings, it is known that the hTERT gene was expressed in immortalized cell lines and cancer cells that did not overexpress c-MYC or contain E6 protein. Interestingly, it was reported that the hTERT locus subjected to gene amplifications in multiple cancer cell lines. However, the copy numbers of the hTERT gene in these cells were moderate (less than 10 in the vast majority of cases). Because the hTERT gene is strongly repressed in most somatic cells, it is hardly convincing that this moderate amplification of hTERT locus alone can account for the increased level of hTERT mRNA in these cancer cells.
We have previously showed the hTERT locus was embedded in a condensed chromatin domain that was highly resistant to DNase I digestion (Wang and Zhu, 2004). Thus, such a repressive chromatin environment would explain the tight regulation of the hTERT gene in most human somatic cells (Wang et al., 2009a). In fact, telomerase activation and immortalization of human cells are extremely rare events and frequently associated with telomere-induced crisis in both in vitro culture and tumorigenesis (Maser et al., 2002; Shay and Wright, 1989; Zhu et al., 1999). Here, we report a new mechanism involved in the transcriptional activation of the hTERT gene, the chromosomal rearrangements upstream of its promoter. In all immortal fibroblast lines that we have analyzed, activation of the hTERT gene was associated with translocation of the hTERT gene to heterologous chromosomal sites. These translocations may allow the hTERT promoter to escape the repressive chromatin environment and thereby activate the telomerase expression.
MATERIALS AND METHODS
Plasmids and Bacterial Artificial Chromosomes
Plasmids pYF7 and pTRTP have been described previously (Horikawa et al., 1999; Wang and Zhu, 2003). The bacterial artificial chromosome (BAC) reporter construct 117B23-cFtRvSVP was derived from the BAC clone RPCI11-117B23 (Research Genetics), which contains a 160 kb human genomic sequence that encompasses the entire hTERT locus and its upstream and downstream neighboring loci. This reporter was constructed through a two-step recombination procedure, involving a positive and a negative selection step based on a BAC recombineering protocol from the Copeland laboratory (Lee et al., 2001; Wang et al., 2007, 2009b).
Cells and Transfection
The immortal cell lines, 3C4C, 3C104a, 3C166a, 3C167b, and pre-crisis cells 3C were derived from SV40 transformed IMR90 cells and have been described previously (Zhu et al., 1999; Wang and Zhu, 2003). GM639 and GM847 lines were purchased from Coriell Cell Repositories (Camden, NJ, USA). Cells were cultured in minimal essential medium (Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA, USA). For transient transfection experiments, 0.15 μg hTERT reporter plasmids, together with 5 ng pRL-CMV, were transfected into cells in each well of 48-well plates using FuGene 6 reagent (Roche, Indianapolis, IN, USA) and luciferase activities were determined 48 h later using the Dual Luciferase Assay system (Promega, Madison, WI, USA). Firefly luciferase (Fluc) activities from the test reporters were normalized to Renilla luciferase (Rluc) activities from the co-transfected pRL-CMV. The BAC reporter 117B23-cFtRvSVP was transfected into immortal fibroblast cells using FuGene 6 and selected with 0.5 μg/ml puromycin for 3-5 days. Isolated colonies containing both Fluc and Rluc activities were expanded for further experimental analysis.
Quantitative RT-PCR (Taqman) Analyses
cDNAs were synthesized from 1 μg total RNAs with an oligo(dT) primer using the SuperScript First Strand Synthesis system (Invitrogen, Carlsbad, CA, USA). PCR reactions were carried out in triplicates using StepOnePlus™ Real-Time PCR Systems (Applied Biosystems, Foster City, CA, USA). Primer and probe sequences are listed in Supplementary Table S1.
DNase I Sensitivity Assays and Southern Blot Analyses
Nuclei were isolated from proliferating cells (Wang and Zhu, 2003) and treated with 0, 2, 4, 8, 16 units per ml of DNase I (Promega, Madison, WI, USA) at 37°C for 20 min. Genomic DNA was extracted from treated nuclei and 8 μg DNA was digested with restriction enzymes, followed by Southern blot analyses and indirect labeling of genomic fragments with Rluc fragment or the probe b (Wang and Zhu, 2004). Genomic Southern blot analysis was performed as previously described (Wang and Zhu, 2003). All probes are summarized in Supplementary Table S2.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed as described previously (Wang et al., 2007). Briefly, 107 cells were cross-linked with 1% formaldehyde and stopped by 0.125 M glycine. Cell pellets were lysed and sonicated to generate DNA fragments of 200–800 base pairs, which were subsequently precipitated with antibodies against acetylated histones H3 and H4 (Millipore, Billerica, MA, USA). Isolated chromatin DNA fragments were subjected to quantitative PCR using primers and probes specific for the endogenous and transgenic hTERT promoters (Supplementary Table S1).
Fluorescent In Situ Hybridization (FISH)
Metaphase spreads and FISH analysis were performed as described (Miller et al., 1995). Briefly, proliferating cells were treated with 0.1 μg/ml Colcemid (Invitrogen, Carlsbad, CA, USA) for 1 h at 37 °C and incubated in pre-warmed hypotonic solution (0.075 M KCl) for 15 min at 37 °C. After fixation with 3:1 methanol-acetic acid mixture, cells were dropped onto pre-chilled microscopic slides to obtain chromosome spreads. FISH probes (BAC clones RPCI11-1125A11 and RPCI11-1107M2) were labeled with biotin and digoxigenin (DIG), respectively, using the Nick Translation Kit (Roche, Indianapolis, IN, USA). Following denaturation of the metaphase chromosomes, the slides were hybridized to the probes overnight at 37 °C. For the biotin-labeled probe, signals were amplified with biotinylated anti-avidin and FITC-conjugated avidin antibodies (Vector Laboratories, Burlingame, CA, USA). For the DIG –labeled probe, signal amplification was performed with Texas Red conjugated mouse anti-DIG (1:50), rat anti-mouse IgG (1:100) and Texas Red conjugated goat anti-Rat (1:50) antibodies (Jackson ImmunoResearch, West Grove, PA, USA). Samples were counter-stained with 4′, 6-diamidino-2-phenylindole (DAPI) and analyzed on a Nikon TE2000-U microscope equipped with a Roper Scientific CCD camera and NIS elements software.
RESULTS
Levels of hTERT Transcription From its Endogenous Locus and Transient Plasmid Reporter Assays
Although it has been widely reported that the hTERT gene is transcriptionally activated in most immortal cell lines, the underlying molecular mechanisms of its activation during cellular immortalization remain to be elucidated. To study this fundamental biological question of human cell immortalization, a clonal strain of SV40 transformed IMR90 cells (3C) was allowed to proliferate until a crisis induced by critically shortened telomeres (Zhu et al., 1999). Spontaneously formed colonies appeared at a frequency of less than 10−7, similar to previously reported frequencies of similar cells that escaped M2 crisis (Shay and Wright, 1989). From the initial clonal 3C strain, independent telomerase-negative (Tel−) 3C4C and 3C166a and telomerase-positive (Tel+) 3C104a and 3C167b immortalized lines were established (Wang and Zhu, 2003). These immortal lines, together with another pair of Tel+ GM639 and Tel− GM847 immortal lines (Bryan et al., 1995), were used for studying the mechanisms of telomerase activation during cellular immortalization.
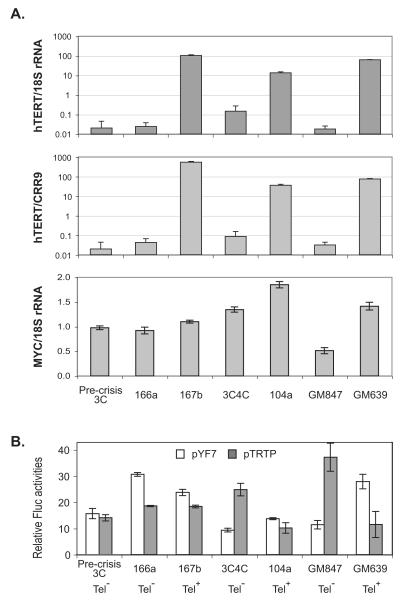
We first examined the levels of endogenous hTERT mRNA in these immortalized cells by quantitative RT-PCR analyses, in which both 18S rRNA and CRR9 mRNA were used as controls. The CRR9 gene (also called CLPTM1L), located immediately upstream of the hTERT gene, was shown to be constitutively transcribed in all cell types examined (Wang and Zhu, 2003; Wang et al., 2004). As expected from previously reported data (Ducrest et al., 2001; Wang and Zhu, 2003), Tel+ cells expressed 100-1000-fold more hTERT mRNA than pre-crisis 3C cells and Tel− immortal siblings, when normalized to either 18S rRNA or CRR9 mRNA (Fig. 1A). Among the three Tel+ immortal lines, 3C167b cells expressed the highest level of hTERT mRNA, whereas 3C104a had the lowest.
Figure 1.
Activities of the hTERT promoters and MYC expression in immortal cell lines. (A) The levels of hTERT and MYC mRNAs in pre-crisis cells and immortal cell lines. Total RNAs were reverse transcribed and subjected to quantitative PCR analysis. The hTERT mRNA levels were normalized to either 18S rRNA (upper chart) or CRR9 mRNA (middle chart). The MYC mRNA expression was normalized to 18S rRNA (lower chart). (B) Activities of transiently transfected hTERT promoters. Plasmids were transfected into fibroblast cells and luciferase activities determined 48 h later. Fluc activities from pYF7 and pTRTP were normalized to Rluc activities of co-transfected pRL-CMV and phRL-TK, respectively. The endogenous telomerase expression profile is indicated below the chart.
Previous studies showed that MYC activated hTERT transcription through binding to the E-boxes at the hTERT promoter in some cancer cells (Wang et al., 1998; Greenberg et al., 1999). To determine if activated hTERT transcription in Tel+ immortal lines resulted from elevated MYC expression, we analyzed the MYC mRNA level in these cells by quantitative RT-PCR. Compared to pre-crisis 3C cells, the levels of c-MYC mRNA varied less than two folds in all immortal cell lines and did not correlate with hTERT expression, which was more than 100-fold higher in Tel+ cells than Tel− cells. Therefore, it was unlikely that increased MYC expression was responsible for hTERT activation during immortalization of the fibroblasts used in this study.
When luciferase reporter constructs pYF7, containing a 7.5 kb genomic fragment upstream of the hTERT ATG codon (−7.4 kb to +79 bp relative to the hTERT transcription start site, or TSS) (Wang and Zhu, 2003), and pTRTP, containing a 1.6 kb genomic sequence upstream of the TSS (−1.6 kb to +28 bp) (Horikawa et al., 1999), were transiently transfected into pre-crisis cells and immortal lines, the hTERT promoter was active in both Tel+ and Tel− cells, which did not correlate with its endogenous promoter activity (Fig. 1B). In fact, the transiently transfected hTERT promoter was as active as the CMV promoter in parallel transfection experiments (Wang and Zhu, 2003). These data confirmed previously published results that transiently transfected plasmid reporters did not recapitulate the endogenous hTERT promoter (Ducrest et al., 2001; Wang and Zhu, 2003), suggesting that the endogeneous hTERT locus in these immortal cells might have been subjected to genetic or epigenetic alterations.
hTERT Promoter Activity From Chromosomally Integrated BAC Reporters
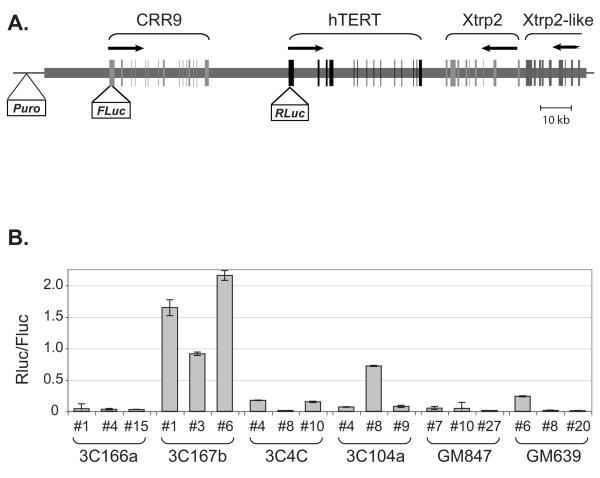
We have previously demonstrated that the endogenous hTERT core promoter was more accessible to DNase I in Tel+ cells than in Tel− cells, indicating that such an open chromatin configuration was directly responsible for the transcriptional activation of the native promoter hTERT-expressing cells (Wang and Zhu, 2003; Wang et al., 2004). Also, active transcription from transiently transfected hTERT promoters in both Tel+ and Tel− cells, in comparison with the endogenous promoter, suggested that plasmid reporters lacked distal elements required for the regulation or that epigenetic changes and/or genetic mutations were responsible for the activation of the endogenous hTERT gene. To resolve these possibilities, an hTERT BAC reporter was constructed from a BAC clone RP11CI-117B23, which contained 160 kb human genomic DNA encompassing the CRR9, hTERT, and Xtrp2 (also called SLC6A18) loci (Fig. 2A). The BAC clone was modified by recombineering, in which a Renilla luciferase (Rluc) and a Firefly luciferase (Fluc) were inserted into the initiation codons of hTERT and CRR9 genes, respectively (Wang et al, 2009b). In addition, a puromycin-resistant marker was placed in the SacB gene of the vector backbone. The modified reporter construct was transfected into immortal cell lines and stable puromycin-resistant clones were isolated. Only clones that contained equal copy numbers of Rluc and Fluc genes with no alterations within 5 kb of the hTERT core promoter, as determined by Southern blot analysis, were used for further analysis (data not shown). Because the BAC reporter was independently integrated into different chromosomal sites and each clone might have different copy numbers of the reporter, the ratio of Rluc to Fluc activities was used as a measurement of transcription from the transgenic hTERT promoter. As shown in Fig. 2B, the transgenic hTERT promoter was at least 10-fold more active in Tel+ 3C167b stable clones than that obtained from Tel−cells, suggesting that mechanisms involved in the activation of the native hTERT gene also activated the transgenic promoter in the chromosomally integrated BAC reporter. However, the activity of transgenic hTERT promoter in Tel+ 3C104a and GM639 cells was variable. In two thirds of 3C104 and GM639-derived clones, the transgenic promoter activity was not higher than that in clones from Tel− cells, suggesting that mechanisms of hTERT activation in Tel+ 3C104a and GM639 cells might lie in epigenetic and/or structural alterations at its endogenous locus. Because the chromosomal integration events were random and thus not controlled, it was likely that some of the integrated BAC reporters were not intact, thus leading to erratic transcription from the transgenic promoter due to the loss of distal elements.
Figure 2.
Activities of the hTERT promoter in chromosomally integrated BAC reporters in immortal cells. (A) A schematic diagram of the BAC reporter. The BAC reporter contains 160 kb human genomic sequence that includes three complete loci, CRR9, hTERT, and Xtrp2, and a portion of the locus for an Xtrp2-like gene. The thick horizontal line represents human genomic DNA and the thin horizontal line stands for the BAC vector sequence (pBACe3.6). Vertical lines designate the exons of genes indicated above. Horizontal arrows indicate the start sites and directions of transcription. Puro, Fluc, and Rluc denote expression cassettes of puromycin resistance gene, Firefly luciferase, and Renilla luciferase, respectively. (B) Luciferase expression from the chromosomally integrated BAC reporters in Tel+ and Tel− immortal lines. Puromycin resistance clones were selected from immortal cells transfected with the BAC reporter. Luciferase activities were measured in independent clones. The activities of transgenic hTERT promoter were shown as ratios of Rluc (from the hTERT promoter) to Fluc (from the CRR9 promoter).
Histone Acetylation and DNase I Accessibility at the Endogenous and Transgenic hTERT Promoters
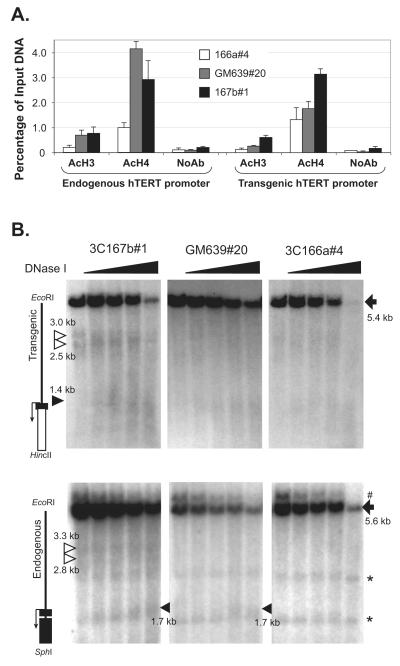
The inability of transiently transfected plasmid reporters to recapitulate the endogenous hTERT promoter was most likely due to the absence of appropriate chromatin configuration of the hTERT promoter in episomal DNAs as compared to chromosomal DNAs. Thus, the chromatin status of chromosomally integrated transgenic hTERT promoter was determined in three representative clones, 166a#4 (Tel−), GM639#20 (Tel+), and 3C167b#1 (Tel+). Both transgenic and endogenous hTERT promoters were inactive in 3C166a#4 cells and active in 3C367b#1 cells. In GM639#20 cells, however, the endogenous hTERT gene was transcribed but Rluc from the transgenic promoter was not (Fig. 2B). First, we examined the status of histone acetylation at the hTERT promoter by ChIP. Antibodies against acetylated histones H3 and H4 were used to precipitate chromatin fragments. The amounts of precipitated endogenous and transgenic promoter fragments were determined by quantitative PCR assays and normalized to input DNA. To distinguish the transgenic hTERT promoter from its endogenous counterpart, a downstream primer within the Rluc sequence was used for the PCR amplifications. As shown in Fig. 3A, acetylation of both histones H3 and H4 at the endogenous promoter was significantly more abundant in Tel+ GM639 and 3C167b cells than in Tel− 3C166a cells, consistent with their endogenous hTERT transcription profiles. However, although the histone acetylation level at the transgenic hTERT promoter in 3C167b#1 cells was higher, its level in GM639#20 cells was similar to that in Tel− 3C166a#4 cells, correlating with the Rluc expression patterns in these clones. Therefore, the level of transcription from both endogenous hTERT gene and transgenic hTERT-Rluc reporter correlated with the level of histone acetylation at their respective promoters.
Figure 3.
Chromatin status of the transgenic and endogenous hTERT promoters. (A) ChIP analyses. Antibodies used in ChIP are as follows: AcH3, acetylated histone H3; AcH4, acetylated H4; No AB, no antibody control. Precipitated DNA fragments were analyzed by quantitative PCR using primers and probes specific to the endogenous and transgenic hTERT promoters and shown as percentage of input genomic fragments. (B) DNase I accessibilities of the endogenous and transgenic hTERT promoter. Nuclei were treated with 0, 2, 4, and 8 units/ml DNase I. Genomic DNAs were isolated and digested with restriction enzymes, followed by Southern blot analysis and indirect end-labeling. The upper panels show the transgenic hTERT promoter regions, as detected by a probe specific to the Rluc sequence, and the lower panels are for the endogenous hTERT locus, as revealed by a probe specific for the second hTERT exon. Schematic diagrams of the hTERT promoter regions are shown on the left. The black rectangles are hTERT exons and the white rectangle represents the Rluc sequence. The arrows show the start site and direction of transcription. Full-length genomic bands are indicated by solid arrows on the right. The black triangles point to hypersensitive bands at the hTERT core promoters and white triangles designate the two hypersensitive bands upstream of the hTERT promoter. #, transgenic DNA band recognized by the exon 2 probe; *, non-specific bands.
Next, DNase I sensitivity assays were performed to determine the chromatin configuration of the hTERT promoter in the chromosomally integrated BAC reporters. Nuclei were isolated from 3C167b#1, GM639#20, and 3C166a#4 cells and treated with increasing concentrations of DNase I. Genomic DNAs were isolated and subjected to Southern blot analysis. As shown in Fig. 3B (upper panels), EcoRI and HincII double digestion resulted in a 5.4 kb full-length band derived from the integrated transgenic BAC construct, as it was detected by a probe specific for the Rluc sequence. In 3C167b#1 cells, the region corresponding to the transgenic hTERT promoter (~ 1.4-kb region) was more sensitive to DNase I digestion than that in GM639#20 and 3C166a#4 cells, correlating with the level of Rluc expression in these clones. Two additional DNase I hypersensitive bands (2.5 kb and 3.0 kb) were detected from the transgenic hTERT promoter in 3C167b#1 cells but not in GM639#20 and 3C166a#4 cells, corresponding to DNase I hypersensitive sites (DHSs) at 1.1 kb and 1.6 kb upstream of the hTERT TSS.
To analyze the endogenous hTERT promoter, the same genomic DNAs were digested with EcoRI (−3.9 kb, relative to TSS) and SphI (1.7 kb downstream of TSS), followed by Southern blot analysis. This blot was hybridized to a probe specific to the exon 2 sequence of the hTERT gene, resulting in a 5.6-kb full-length genomic band (Fig. 3B, lower panels). On this blot, a 1.7 kb DHS band was detected in 3C167b#1 and GM639#20 cells, but not in 3C166a#4 cells, correlating with the endogenous hTERT expression profiles in these cell types. This DHS was the same as the previously reported HS1, which appeared in all Tel+ cells (Wang and Zhu, 2003, 2004). Furthermore, the two DHSs, 1.1 kb and 1.6 kb upstream of the transgenic hTERT TSS, were also detected on the endogenous promoter in 3C167b#1 cells, but not in the other two clones. These results indicated that transcription from both the transgenic and endogenous hTERT promoters was accompanied by the opening of the hTERT promoter. The two upstream DHSs appeared on both transgenic and endogenous promoters in 3C167b cells, correlating with strong activation of both promoters in this clone.
Chromosomal Rearrangements of the hTERT Locus in Tel+ cells
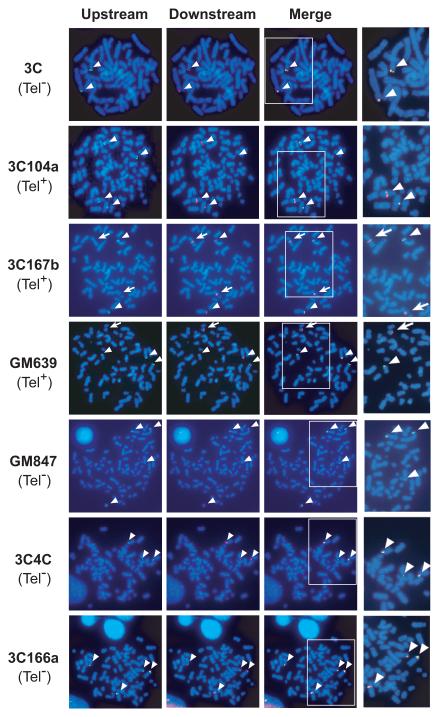
During Southern blot analyses, disproportional ratios of endogenous genomic bands were identified in Tel+ cells, suggesting the presence of chromosomal rearrangements within or near the hTERT locus. These observations prompted us to examine the endogenous hTERT locus by FISH. Two BAC constructs were used as probes: BAC RPCI11-1125A11, containing a 157 kb human genomic sequence 58 kb upstream of the hTERT TSS, and RPCI11-1107M2, with a 197 kb human sequence starting at 88 kb downstream of the hTERT TSS. In pre-crisis 3C cells, both probes hybridized to the tips of the short arms of two chromosomes, consistent with the localization of hTERT locus near the telomere of chromosome 5p (Fig. 4). In all immortal cell lines tested, there were three or four hybridization signals with either probe, indicating that these immortal cells were tetraploid or near-tetraploid. Notably, the upstream probe generated fewer hybridization signals than the downstream probe in Tel+ 3C167b and GM639 cells. The overlapping signals generated by both probes were located at the chromosomal tips (arrowheads in Fig. 4), whereas regions recognized only by the downstream probe resided in the internal portions of chromosomes in these cells (arrows in Fig. 4), indicating that these internal loci resulted from chromosomal rearrangements. In Tel+ 3C104a cells, both probes identified four loci: two located at chromosomal tips and the other two in the middle of different chromosomes. These results suggested that a subset of hTERT alleles in Tel+ cells were rearranged and translocated to heterologous chromosomal sites. In Tel− 3C4C, 3C166a, and GM847 cells, both upstream and downstream probes hybridized to the same three or four signals in each cell line and these signals were all at chromosomal tips (Fig. 4).
Figure 4.
FISH detection of the hTERT locus in immortal cell lines. Pre-crisis and immortal cells were hybridized to an upstream BAC RP11-1125A11 (a 157-kb BAC clone located 58 kb upstream to the hTERT TSS, green) and a downstream BAC RP11-1107M2 (a 197-kb BAC beginning 88 kb downstream of the hTERT TSS, red). The chromosomes were counterstained with Hoechst 33342. Signals recognized by both upstream and downstream probes are indicated by arrowheads and those detected only by the downstream probe are labeled by arrows.
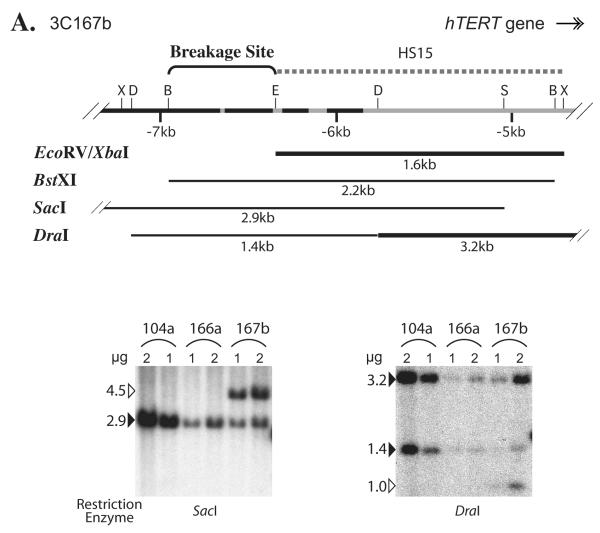
To map the chromosomal breakage sites, various genomic regions at or near the hTERT locus in immortal cell lines were examined by Southern blot analysis. Genomic DNAs were digested with different restriction enzymes, followed by Southern blot analysis using probes specific to various regions near the hTERT locus. The sites of rearrangements were mapped by comparing the intensities of different genomic bands on the same blots that were hybridized to distinct probes. Fig. 5A shows an example of such an analysis of a region upstream of the hTERT TSS with probe HS15. First, the HS15 probe recognized two SacI fragments in 3C167b cells, a predicted 2.9 kb fragment and a 4.5 kb larger-than-expected fragment, suggesting that the rearrangement was within this SacI fragment. Second, the 1.6 kb EcoRV-XbaI band was stronger in 3C167b cells than in 166a cells (data not shown), indicating that the breakage point was upstream of the EcoRV site. Third, the 2.2 kb BstXI band recognized by HS15 had the same intensity in 3C167b and 3C166a cells, suggesting that the breakage point was within this BstXI fragment (data not shown). Finally, whereas the 1.4 kb DraI band intensity was equal in 3C166a and 3C167b cells, the 3.2 kb DraI fragment was much stronger in 3C167b cells and an extra 1.0 kb band was also present in 3C167b cells, confirming that the breakage site was within the 1.4 kb region (Fig. 5A). Taken together, these data mapped the chromosomal breakage point in 3C167b cells to a 608 bp genomic region between the BstXI (−7.0 kb, relative to the hTERT TSS) and EcoRV (−6.4 kb) sites (Fig. 5A).
Figure 5.
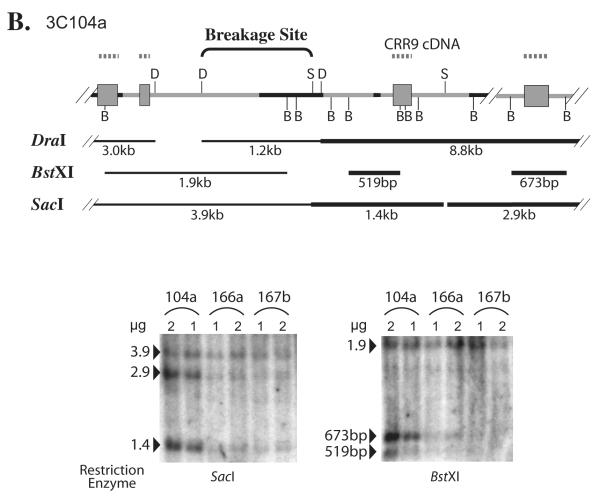
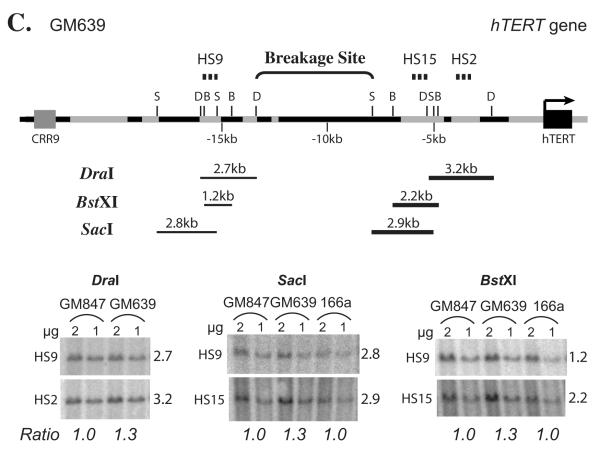
Breakage of genomic regions 5′ to the hTERT gene in immortal cell lines, 3C167b (A), 3C104a (B), and GM639 (C), by Southern blot analyses. Diagrams above each pair of representative Southern blots are schematic interpretations of chromosomal rearrangements. Thin lines represent introns and intergenic regions, and black portions denote short repetitive elements. Gray rectangles correspond to CRR9 exons, whereas the black rectangle in (C) symbolizes the first two exons of hTERT. B, BstXI; D, DraI; S, SacI; X, XbaI. Dashed lines above each diagram show the probes used in Southern blot analyses: (A) HS15, (B) CRR9 cDNA, (C) HS2, HS9, and HS15. Lines below the genomic sequence diagrams are restriction fragments that were examined by Southern blot analyses. Thick restriction fragment lines indicate the bands that were more intense in Tel+ lines than in Tel− cells, whereas thin restriction fragment lines show that their intensities were the same in Tel+ and Tel− cells. For Southern blot analysis, one and two micrograms of genomic DNAs, as indicated in each figure, were digested with restriction enzymes. In (A) and (B), numbers on the left of each blot are sizes of genomic bands in kb. In (C), the probes are shown on the left of each blot, whereas sizes of genomic bands are on the right. The relative intensity ratios of lower bands to the corresponding upper bands are shown at the bottom, as determined by phosphoimaging quantification.
Through similar analysis, the chromosomal breaking point in 3C104a was mapped to a 1.1-kb region within the second intron of the CRR9 gene (Fig. 5B). For GM639 cells, the difference of bands between these cells and control Tel− cells was smaller. As shown in Fig. 5C, the bands recognized by the downstream probes HS15 and HS2 were about 30% stronger in GM639 cells than those in Tel− GM847 and 3C166a cells, when normalized to the bands detected by the upstream HS9 probe. These results mapped the breakage point in GM639 cells to the 5.3 kb region between DraI (−13.3 kb) and SacI (−7.9 kb) sites upstream of the hTERT core promoter (Fig. 5C). Because nearly all the sequences between the DraI and SacI sites were short repetitive elements, it was technically very difficult to further map the chromosomal breakage point in GM639 cells. By similar Southern blot analyses, we did not detect any chromosomal rearrangement in pre-crisis 3C cells and Tel− 3C4C, 3C166a, and GM847 cells (Fig. 5 and data not shown).
Based on FISH experiments (Fig. 4) and Southern blot analyses (Fig. 5 and data not shown), there were two and four hTERT alleles in pre-crisis 3C cells and 3C166a cells, respectively. Using these numbers as references, we determined the copy numbers of normal and rearranged hTERT alleles in immortal cell lines by quantifying the intensities of genomic bands on Southern blots (Table 1). Evidently, there was no correlation between the endogenous hTERT expression and copy number of the hTERT gene. However, higher hTERT expression was observed in cells in which chromosomal rearrangements were identified near the hTERT promoter, such as 3C167b cells. Therefore, these results suggested that chromosomal translocations played an important role in transcriptional activation of the hTERT gene in immortal cells.
TABLE 1.
Estimated Copy Numbers of Non-Rearranged and Rearranged hTERT Alleles and the Endogenous hTERT Expression
| Cells | 3C | 3C104a | 3C167b | GM639 | GM847 | 3C4C | 3C166a |
|---|---|---|---|---|---|---|---|
| Total | 2 | 3 12 | 8 | 4 | 4 | 3 | 4 |
| Non-rearranged a | 2 | 4 | 4 | 3 | 4 | 3 | 4 |
| Rearranged a | 0 | 3 8 | 4 | 1 | 0 | 0 | 0 |
| Chromosomal breakage sites (distance to the hTERT TSS) |
~ | ~ 40 kb | 6.5-7 kb | 8-13 kb | ~ | ~ | ~ |
|
| |||||||
| Fold of elevated hTERT b mRNA expression |
- | 42x | 325x | 192x | - | - | - |
Copy numbers of non-rearranged and rearranged alleles were determined by quantifying the intensities of genomic bands in a number of Southern blots, such as the ones shown in panels A and D of Fig. 5, based on the assumption that pre-crisis 3C and 3C166a cells had two and four copies of normal hTERT alleles, respectively.
Fold increase of hTERT mRNA expression in Tel+ cells is relative to the average level of hTERT expression in all Tel− cells, normalized to 18S rRNA, as shown in the upper panel of Fig. 1A.
DISCUSSION
Telomerase expression is critical for cellular immortalization and cancer development, and its expression requires transcriptional activation of the hTERT gene. By detailed studies of the hTERT promoter in the SV40 transformed pre-crisis fibroblasts and three pairs of spontaneously-derived Tel+ and Tel− immortal lines, we revealed two mechanisms of hTERT activation in human immortal fibroblasts, the chromatin-dependent activation of hTERT promoter and the rearrangements of its upstream sequences. First, the hTERT promoter from a chromosomally integrated BAC reporter was active in Tel+ 3C167b cells, regardless of its integration sites. However, the activation of hTERT promoter was not recapitulated in transiently transfected small plasmid reporters. The transcriptional activation was associated with increased histone acetylation and DNase I accessibility at both endogenous and transgenic hTERT promoters. Therefore, the activation of hTERT promoter in 3C167b cells depended on its chromatin context. Second, by FISH and Southern blot experiments, genomic DNA rearrangements were detected upstream of the hTERT promoter in all three Tel+ lines, whereas none were found in pre-crisis cells and Tel− immortal lines. In all cases of rearrangements, extra copies of the hTERT gene were translocated to heterologous chromosomal sites. These results indicated that genomic rearrangements near the 5′ end of the hTERT gene played a major role in hTERT transcription in immortal fibroblasts. To the best of our knowledge, this is the first evidence that chromosomal translocation of the hTERT gene is a mechanism of telomerase activation and cellular immortalization.
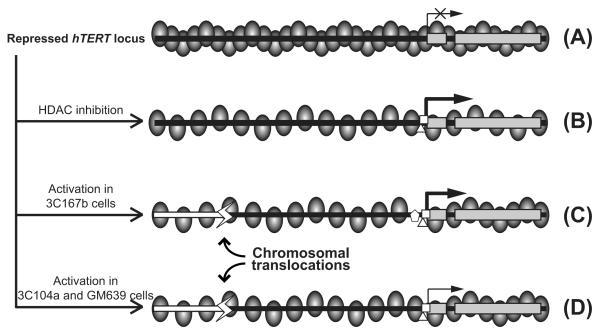
The native hTERT locus is embedded in a large nuclease-resistant chromatin domain in normal human fibroblasts and many other cell lines (Wang and Zhu, 2004) (Fig. 6A). As a result, the hTERT promoter is stringently repressed in these cells. This repression can be relieved by inhibition of HDACs (Cong and Bacchetti, 2000; Wang and Zhu, 2003), as shown in Fig. 6B. It has been reported that the hTERT gene was amplified in cancer cell lines and primary cancers, including those of brain, breast, cervix, liver, and lung origins (Zhang et al., 2000, 2002; Takuma et al., 2004), as well as primary and metastatic melanomas (Pirker et al., 2003). In most cases, the amplification ratio, the ratio of hTERT gene copy number to number of chromosome 5p, was less than two; and only in very rare cases, the amplification ratio was over ten (Zhang et al., 2000; Pirker et al., 2003; Cao et al., 2008). This suggested that modest amplification of the hTERT gene occurred in most cancers. Given that the expression level of the endogenous hTERT gene is low in most human somatic tissues ((Wright et al., 1996; Horikawa et al., 2005) and data not shown), this modest amplification might not be sufficient to account for the large increases of hTERT mRNA expression in cancer cells. In fact, no statistical association was found between the copy number of hTERT gene and its expression level or telomerase activity in a large set of colorectal carcinomas (Palmqvist et al., 2005). Thus, additional mechanisms are likely involved in hTERT activation in cancer cells. Indeed, the transgenic hTERT promoter in chromosomally integrated BAC reporter was highly active in 3C167b cells, consistent with the presence of transacting factors being involved in the activation of the endogenous hTERT promoter (Fig. 6C). Apparently, this activation was dependent on the chromatin context because transiently transfected plasmid reporters did not recapitulate the endogenous hTERT promoter activity in these Tel+ cells (Fig. 1B and (Wang and Zhu, 2003, 2004)).
Figure 6.
Models of hTERT rearrangement and activation in human immortal fibroblasts. (A) The hTERT locus is embedded in a condensed chromatin domain in normal human fibroblasts and pre-crisis cells. As a result, the promoter is inaccessible to transcription activators and thereby silenced. (B) Inhibition of HDACs results in chromatin de-condensation, opening of the hTERT promoter, and hTERT transcription. (C) Transcriptional activation of the hTERT promoter and chromosomal translocation of the hTERT locus result in higher hTERT transcription in 3C167b cells. (D) In Tel+ GM639 and 3C104a cells, chromosomal rearrangements upstream of the hTERT promoter lead to partial chromatin de-condensation of the translocated hTERT alleles. Ovals represent nucleosomes. Gray rectangles denote the first two exons of hTERT. Black horizontal bars are genomic DNA upstream of the hTERT promoter and white bars are heterologous genomic sequence fused upstream to the translocated hTERT alleles. Horizontal arrows indicate the TSS and direction of transcription, and thickness of the arrows represents relative levels of transcription. White polygons at the hTERT promoter symbolize various transcription activators.
Our studies presented here indicated that genetic rearrangements might contribute to the activation of hTERT expression in Tel+ cells. Indeed, the levels of hTERT mRNA correlated better with the distance between chromosomal breakage points and the hTERT promoter than the hTERT gene copy numbers (Table 1). These results suggested that the chromosomal rearrangements disrupted the condensed chromatin domain that strongly repressed the hTERT promoter. In this scenario, the fusion of heterologous DNA sequences with the translocated hTERT gene may generate a more permissible environment for its transcription (Figs. 6C and D).
All immortal fibroblast lines used in this report were derived from pre-crisis fibroblast cells, indicating that translocation of the hTERT gene might have been caused by telomere-induced crisis (Shay and Wright 1989; Wang and Zhu 2003). During crisis, short dysfunctional telomeres led to end-to-end chromosome fusions, resulting in breakage-fusion-bridge cycles and consequently a mitotic catastrophe (Counter et al., 1994; Zhu et al., 1999). The chromosomal breakage at the hTERT locus and subsequent ligation to heterologous sequences by non-homologous end joining (NHEJ) mechanisms resulted in nonreciprocal translocations, such as those in Tel+ immortal lines. The extremely rare cells in which the hTERT gene was rearranged from its upstream region thus survived this crisis and were ultimately established as immortal cells. This might explain the observation that in vitro immortalized Tel+ fibroblast cell lines are very rare and always arise following short telomere-induced crisis.
Telomere-induced crisis occurs during tumorigenesis (Maser et al.; 2002) and has been proposed to be an important barrier for cancer progression (Greenberg, 2005; Hornsby, 2007). Activation of the hTERT gene by telomere-crisis may also occur during tumor development. Our results supported the notion that telomere-induced crisis provides a mutational mechanism that can generate the diverse genetic alterations required for cancer initiation or progression (Greenberg, 2005; Hornsby, 2007). We propose that one of these genetic alterations is telomerase activation via rearrangements of the hTERT gene.
While we and others found that the hTERT promoter in transiently transfected plasmids was similarly active in Tel+ and Tel− cells (Ducrest et al., 2001; Wang and Zhu, 2003, and Fig. 1B), a few other studies showed that the transiently transfected hTERT promoter was more active in Tel+ cells than in Tel− cells (Cong et al., 1999; Takakura et al., 1999). Although the cause for such discrepancies is currently unknown, it is likely that mechanisms for hTERT activation are cell-specific. Cellular immortalization might be caused by de-repression of hTERT promoter through chromosomal rearrangements in the fibroblast models used in this study. Telomerase expression in other cell types, however, might involve activation of the hTERT promoter by transcriptional regulators, such as MYC. In the latter case, it is conceivable that transiently transfected hTERT promoter is more active in those Tel+ cells than Tel− cells.
In summary, our experiments revealed chromosomal rearrangement as a novel mechanism in transcriptional activation of hTERT gene during cellular immortalization. In particular, rearrangements at the upstream sequence of hTERT gene are crucial for telomerase activation in immortal cells, which should help us further understand telomerase activation during cancer development.
Supplementary Material
ACKNOWLEDGEMENT
We thank Nan Li and Laura Carrel for technical assistance on Fluorescent In-Situ Hybridization. We also thank Anne Stanley, David Stanford, and Joe Bednarczyk of the Macromolecular and Molecular Genetics Core Facilities at Penn State College of Medicine for their excellent services. JZ is a Research Scholar of American Cancer Society.
REFERENCES
- Aisner DL, Wright WE, Shay JW. Telomerase regulation: not just flipping the switch. Curr Opin Genet Dev. 2002;12:80–85. doi: 10.1016/s0959-437x(01)00268-4. [DOI] [PubMed] [Google Scholar]
- Blasco MA, Rizen M, Greider CW, Hanahan D. Differential regulation of telomerase activity and telomerase RNA during multi-stage tumorigenesis. Nature Genetics. 1996;12:200–204. doi: 10.1038/ng0296-200. [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Bryan TM, Reddel RR. Increased copy number of the TERT and TERC telomerase subunit genes in cancer cells. Cancer Sci. 2008;99:1092–1099. doi: 10.1111/j.1349-7006.2008.00815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ, Reddel RR. Telomere uncapping and alternative lengthening of telomeres. Mech Ageing Dev. 2008;129:99–108. doi: 10.1016/j.mad.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Cong YS, Bacchetti S. Histone deacetylation is involved in the transcriptional repression of hTERT in normal human cells. J Biol Chem. 2000;275:35665–35668. doi: 10.1074/jbc.C000637200. [DOI] [PubMed] [Google Scholar]
- Cong YS, Wen J, Bacchetti S. The human telomerase catalytic subunit hTERT: organization of the gene and characterization of the promoter. Hum Mol Genet. 1999;8:137–142. doi: 10.1093/hmg/8.1.137. [DOI] [PubMed] [Google Scholar]
- Counter CM, Botelho FM, Wang P, Harley CB, Bacchetti S. Stabilization of short telomeres and telomerase activity accompany immortalization of Epstein-Barr virus-transformed human B lymphocytes. J Virol. 1994;68:3410–3414. doi: 10.1128/jvi.68.5.3410-3414.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counter CM, Hahn WC, Wei W, Caddle SD, Beijersbergen RL, Lansdorp PM, Sedivy JM, Weinberg RA. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc Natl Acad Sci USA. 1998;95:14723–14728. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducrest AL, Amacker M, Mathieu YD, Cuthbert AP, Trott DA, Newbold RF, Nabholz M, Lingner J. Regulation of human telomerase activity: repression by normal chromosome 3 abolishes nuclear telomerase reverse transcriptase transcripts but does not affect c-Myc activity. Cancer Res. 2001;61:7594–7602. [PubMed] [Google Scholar]
- Greenberg RA. Telomeres, crisis and cancer. Curr Mol Med. 2005;5:213–218. doi: 10.2174/1566524053586590. [DOI] [PubMed] [Google Scholar]
- Greenberg RA, O’Hagan RC, Deng H, Xiao Q, Hann SR, Adams RR, Lichtsteiner S, Chin L, Morin GB, DePinho RA. Telomerase reverse transcriptase gene is a direct target of c-Myc but is not functionally equivalent in cellular transformation. Oncogene. 1999;18:1219–1226. doi: 10.1038/sj.onc.1202669. [DOI] [PubMed] [Google Scholar]
- Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- Horikawa I, Cable PL, Afshari C, Barrett JC. Cloning and characterization of the promoter region of human telomerase reverse transcriptase gene. Cancer Res. 1999;59:826–830. [PubMed] [Google Scholar]
- Horikawa I, Chiang YJ, Patterson T, Feigenbaum L, Leem SH, Michishita E, Larionov V, Hodes RJ, Barrett JC. Differential cis-regulation of human versus mouse TERT gene expression in vivo: identification of a human-specific repressive element. Proc Natl Acad Sci USA. 2005;102:18437–18442. doi: 10.1073/pnas.0508964102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornsby PJ. Senescence as an anticancer mechanism. J Clin Oncol. 2007;25(14):1852–7. doi: 10.1200/JCO.2006.10.3101. [DOI] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Klingelhutz AJ, Foster SA, McDougall JK. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature. 1996;380:79–82. doi: 10.1038/380079a0. [DOI] [PubMed] [Google Scholar]
- Lee EC, Yu D, de Velasco J Martinez, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Maser RS, DePinho RA, Masera G. Connecting chromosomes, crisis, and cancer. Science. 2002;297:565–569. doi: 10.1126/science.297.5581.565. [DOI] [PubMed] [Google Scholar]
- Miller AP, Gustashaw K, Wolff DJ, Rider SH, Monaco AP, Eble B, Schlessinger D, Gorski JL, van Ommen GJ, Weissenbach J, Willard HF. Three genes that escape X chromosome inactivation are clustered within a 6 Mb YAC contig and STS map in Xp11.21-p11.22. Hum Mol Genet. 1995;4:731–739. doi: 10.1093/hmg/4.4.731. [DOI] [PubMed] [Google Scholar]
- Morin GB. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- Palmqvist R, Zhang A, Xu D, Golovleva I, Norrback KF, Gruber A, Oberg A, Stenling R, Roos G. hTERT gene copy number is not associated with hTERT RNA expression or telomerase activity in colorectal cancer. Int J Cancer. 2005;116:395–400. doi: 10.1002/ijc.21020. [DOI] [PubMed] [Google Scholar]
- Pirker C, Holzmann K, Spiegl-Kreinecker S, Elbling L, Thallinger C, Pehamberger H, Micksche M, Berger W. Chromosomal imbalances in primary and metastatic melanomas: over-representation of essential telomerase genes. Melanoma Res. 2003;13:483–492. doi: 10.1097/00008390-200310000-00007. [DOI] [PubMed] [Google Scholar]
- Shay JW, Wright WE. Quantitation of the frequency of immortalization of normal human diploid fibroblasts by SV40 large T-antigen. Exp Cell Res. 1989;184:109–118. doi: 10.1016/0014-4827(89)90369-8. [DOI] [PubMed] [Google Scholar]
- Takakura M, Kyo S, Kanaya T, Hirano H, Takeda J, Yutsudo M, Inoue M. Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res. 1999;59:551–557. [PubMed] [Google Scholar]
- Takuma Y, Nouso K, Kobayashi Y, Nakamura S, Tanaka H, Matsumoto E, Fujikawa T, Suzuki M, Hanafusa T, Shiratori Y. Telomerase reverse transcriptase gene amplification in hepatocellular carcinoma. J Gastroenterol Hepatol. 2004;19:1300–1304. doi: 10.1111/j.1440-1746.2004.03447.x. [DOI] [PubMed] [Google Scholar]
- Veldman T, Liu X, Yuan H, Schlegel R. Human papillomavirus E6 and Myc proteins associate in vivo and bind to and cooperatively activate the telomerase reverse transcriptase promoter. Proc Natl Acad Sci U S A. 2003;100:8211–8216. doi: 10.1073/pnas.1435900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Xie LY, Allan S, Beach D, Hannon GJ. Myc activates telomerase. Genes Dev. 1998;12:1769–1774. doi: 10.1101/gad.12.12.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Hu C, Zhu J. Transcriptional silencing of a novel hTERT reporter locus during in vitro differentiation of mouse embryonic stem cells. Mol Biol Cell. 2007;18:669–677. doi: 10.1091/mbc.E06-09-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Robertson GP, Zhu J. A novel human homologue of Drosophila polycomblike gene is up-regulated in multiple cancers. Gene. 2004;343:69–78. doi: 10.1016/j.gene.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Wang S, Zhao Y, Hu C, Zhu J. Differential repression of human and mouse TERT genes during cell differentiation. Nucleic Acids Res. 2009a;37:2618–2629. doi: 10.1093/nar/gkp125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhao Y, Leiby M, Zhu J. A new positive/negative selection scheme for precise BAC recombineering. Mol Biotechnol. 2009b;42:110–116. doi: 10.1007/s12033-009-9142-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhu J. Evidence for a relief of repression mechanism for activation of the human telomerase reverse transcriptase promoter. J Biol Chem. 2003;278:18842–18850. doi: 10.1074/jbc.M209544200. [DOI] [PubMed] [Google Scholar]
- Wang S, Zhu J. The hTERT gene is embedded in a nuclease-resistant chromatin domain. J Biol Chem. 2004;279:55401–55410. doi: 10.1074/jbc.M411352200. [DOI] [PubMed] [Google Scholar]
- Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW. Telomerase activity in human germline and embryonic tissues and cells. Dev Gen. 1996;18:173–179. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Xu M, Luo W, Elzi DJ, Grandori C, Galloway DA. NFX1 interacts with mSin3A/histone deacetylase to repress hTERT transcription in keratinocytes. Mol Cell Biol. 2008;28:4819–4828. doi: 10.1128/MCB.01969-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Zheng C, Hou M, Lindvall C, Wallin KL, Angstrom T, Yang X, Hellstrom AC, Blennow E, Bjorkholm M, Zetterberg A, Gruber A, Xu D. Amplification of the telomerase reverse transcriptase (hTERT) gene in cervical carcinomas. Genes Chromosomes Cancer. 2002;34:269–275. doi: 10.1002/gcc.10071. [DOI] [PubMed] [Google Scholar]
- Zhang A, Zheng C, Lindvall C, Hou M, Ekedahl J, Lewensohn R, Yan Z, Yang X, Henriksson M, Blennow E, Nordenskjold M, Zetterberg A, Bjorkholm M, Gruber A, Xu D. Frequent amplification of the telomerase reverse transcriptase gene in human tumors. Cancer Res. 2000;60:6230–6235. [PubMed] [Google Scholar]
- Zhu J, Wang H, Bishop JM, Blackburn EH. Telomerase extends the lifespan of virus-transformed human cells without net telomere lengthening. Proc Natl Acad Sci U S A. 1999;96:3723–3728. doi: 10.1073/pnas.96.7.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.