Abstract
The long-standing association between hosts and microbes has generated some of most intricate relationships. The studies on molecular mechanisms of host-microbe interaction have been revealing many fascinating stories. Here we zoom in on a specific topic on the interplay between bacterial effectors and plant innate immune signaling. In particular, we will summarize our recent discovery that bacterial effector proteins, AvrPto and AvrPtoB, target plant immune signaling receptor complexes to interfere with host immune responses and development.
Key words: plant innate immunity, type III effector, bacterial pathogenicity
PAMP-Triggered Immunity: The Front Line of Host Defense Responses
Disease is the exception rather than the rule for most of creatures despite that plants and animals are constantly surrounded by a diverse array of potential pathogens. Through evolution, hosts have developed the capacity to timely detect and mount effective defense responses to dampen potential infections. As an active and first line of defense, innate immunity plays pivotal roles in fending against the onslaught of outside invaders that penetrate or evade the physical barriers (such as skin and waxy epidermal cells). The invading microbes are in part detected through perception of some ‘general’ microbe-derived molecules, which were coined as pathogen-associated molecular patterns (PAMPs) or microbe-associated molecular patterns (MAMPs), by host cell-surface pattern-recognition receptors (PRRs), leading to PAMP-triggered immunity (PTI).1,2 Most identified plant PRRs are receptor-like kinases (RLKs). Two well-characterized Arabidopsis PAMP receptors are FLAGELLIN-SENSING 2 (FLS2) that perceives a conserved 22-amino acid peptide (flg22) from bacterial flagellin, and EF-TU RECEPTOR (EFR) that recognizes bacterial elongation factor EF-Tu.3,4 Affinity cross-linking assays demonstrated direct binding of flagellin to FLS2 and EF-Tu to EFR, suggesting that FLS2 and EFR are bona fide PAMP receptors.4,5 CERK1, a RLK with three LysM motifs, is involved in fungal chitin perception and signaling in Arabidopsis.6,7 However, there is no direct evidence to support that CERK1 binds to chitin. Instead, a plasma membrane glycoprotein (CEBiP) has been shown to directly bind to chitin in rice.8 CEBiP also contains two extracellular LysM motifs. Recently, it was found that the biogenesis of certain PRRs in plant innate immunity requires the endoplasmic reticulum-quality control (ER-QC) machinery. EFR, but not FLS2, cannot accumulate in the null mutant of Calreticulin 3 gene, which encodes an ER chaperon involving the proper folding of newly synthesized glycoproteins. 9 It is intriguing how the ER-QC components are specifically required for the proper accumulation of a subset of PRRs as FLS2 and EFR belong to the same family of RLKs and induce largely overlapping responses.
The intracellular signaling events downstream of PRR perception include the activation of evolutionarily conserved mitogen-activated protein kinase (MAPK) signaling cascades, transcriptional changes, and the production of antimicrobial compounds.10,11 As an early signaling event upon PAMP binding to corresponding PRR, receptor dimerization plays an essential role in triggering intracellular signaling. In plants, FLS2 rapidly dimerizes with BAK1, another RLK, upon flagellin treatment. BAK1 was originally identified as a partner of BRI1 mediating BR signaling.12–15 Interestingly, ER-QC machinery has also been implicated in the biogenesis of BRI1.16 However, BAK1 is not involved in BR binding to BRI1 and flagellin binding to FLS2.14,17 Intriguingly, BAK1 is required for multiple PAMP responses, including flagellin, EF-Tu, lipopolysachride (LPS), peptidoglycan (PGN), bacterial coldshock protein and oomycete elicitor INF1 in Arabidopsis and Nicotiana benthamiana.14,15 Thus, BAK1 appears to function in distinct receptor signaling complexes to integrate multiple PAMP perception into downstream signaling events. In addition, BAK1 is also required to constrain the spread of cell death induced by virulent bacteria Pseudomonas syringae pv. tomato DC3000.18
Bacterial Effectors Dampen Host Immunity and More: Breach of Peace
To be pathogenic, many Gram-negative bacteria have evolved the ability to inject a repertoire of virulence effector proteins into host cells through type III secretion system (TTSS) to modulate diverse host cellular activities and physiology.19,20 Although the structural components of TTSS are relatively conserved, the sequences and functions of individual effectors secreted from TTSS can be highly divergent in different species of plant and animal pathogenic bacteria. Some species, such as Yersinia of animal pathogens, secrete just a few effectors, whereas Pseudomonas species of plant pathogens deliver about thirty effectors.21,22 Now, it is certain that effectors collectively determine bacterial pathogenicity by interfering with host defense responses, thus leading to effort-triggered susceptibility (ETS) on susceptible plants. However the contribution of individual effectors to virulence is trivial to observe in plants likely due to the potential functional redundancy and the limitation of diagnostic tools.23–28 In some cases, these effectors could be recognized directly or indirectly by plant disease resistance proteins (R proteins) to induce potent effector-triggered immunity (ETI) and serve as so-called avirulence (Avr) factors that turn virulent strains into avirulent ones.29–31
AvrPto was historically classified as an avirulence protein based on its ability of eliciting substantial immune responses in tomato plants carrying the immune sensors Pto and Prf.32–34 AvrPtoB, a sequence distinct effector composing of at least two functional domains (the N terminus responsible for interaction with Pto, and the C terminal E3 ligase domain) was initially identified through its interaction capacity with Pto in a yeast two-hybrid screen.35 The apparent anomaly that pathogens possess an immune elicitor was satisfactorily explained by a careful observation that AvrPto moderately promoted pathogen growth on the plants lacking Pto or Prf.36 An outstanding question was how these bacterial proteins contribute to the pathogenesis at the molecular level. The observation that type III effectors collectively suppress PTI led us to engage a versatile cellbased system to screen for individual effectors contributing to the suppression activities on susceptible hosts. AvrPto and AvrPtoB were identified from the screen as suppressors of multiple PAMP-mediated immune responses upstream of MAPK cascade at the plasma membrane. It was proposed that AvrPto and AvrPtoB may target PAMP receptors or other convergent early signaling components to suppress immunity triggered by multiple PAMPs.37
An interesting observation that transgenic plants overexpressing AvrPto resemble BR-deficient mutant led to the discovery that AvrPto and AvrPtoB directly target the signaling partner BAK1 involved in multiple PAMP-triggered immune responses and plant hormone BR-mediated signaling. 38 AvrPto interacts with BAK1 in a yeast split-ubiquitin assay designed for detecting membrane protein interaction, and AvrPto and AvrPtoB associate with BAK1 in vivo by co-immunoprecipitation assay. Importantly, extensive mutagenesis and deletion analyses suggest the biological importance of BAK1-AvrPto/AvrPtoB interaction.38 This targeting leads to the dissociation of flagellin-induced FLS2-BAK1 association, and brassinosteroidmediated BRI1-BAK1 association, thereby blocking the initiation of flagellin and BR signaling (Fig. 1). It is important to note that AvrPto and AvrPtoB do not interact with BRI1. Thus, suppression of BR signaling by AvrPto does not involve the interaction with BR receptor BRI1. Given the fact that BAK1 is involved in signaling triggered by BR, multiple PAMPs and cell death, by targeting BAK1, AvrPto and AvrPtoB efficiently suppress multiple signaling in plant immunity and development, one stone killing two birds (Fig. 2). Apparently, AvrPto also targets BAK1-independent PAMP signaling since BAK1 is essential in some, but not all PAMP-signaling responses, whereas AvrPto is equally effective in suppressing the immune responses triggered by all PAMPs tested (Fig. 2).
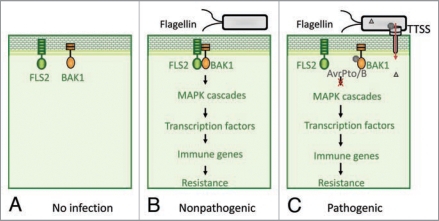
Figure 1.
Bacterial effectors suppress PAMP immunity by targeting receptor signaling complexes. (A) In the absence of infection, PAMP receptor FLS2 remains at the resting stage and there is no apparent receptor complex formation. (B) Once challenged by microbes, FLS2 detects the invading microbes by binding to the ligand-bacterial flagellin, which allows FLS2 conformational change and heterodimerization with BAK1 to form a tight receptor complex. The functional PAMP receptor complex initiates downstream signaling, including activation of MAPK cascade, transcription factors and immune genes, thereby leading to plant resistance to microbes. (C) To be pathogenic, some successful pathogens deployed virulence weapons to tear down host immune system. For instance, type III effectors AvrPto and AvrPtoB, secreted from Pst DC3000 and delivered into host cells, target BAK1-associated PAMP receptor complexes and disrupt the complex formation, thereby impeding the immune signaling and leading to disease.
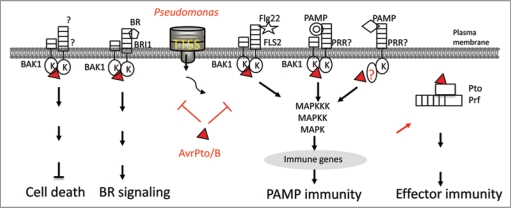
Figure 2.
Multiple functions of AvrPto and AvrPtoB as virulence and avirulence factors. BAK1 is a signaling partner of BR receptor BRI 1, multiple PAMP receptors PRRs, and unknown receptor for cell death. By targeting BAK1, AvrPto and AvrPtoB efficiently suppress multiple signaling in plant immunity, development and cell death control, one stone killing two birds. Apparently, AvrPto also targets BAK1-independent PAMP signaling. In the presence of Pto and Prf, AvrPto triggers potent effector-mediated immunity in tomato.
Conclusions and Challenges
Great strides are being made in understanding the biological and enzymatic functions of effectors in the past few years with the development of new technologies and more sensitive and quantitative assays. The newly identified effector targets in hosts have been revealed to be often novel and essential components in innate immunity. For instance, tandem mass spectrometry analysis revealed that OspF from Shigella encodes a novel enzyme, phosphothreonine lyase, in the suppression of MAPK activity.39 HopAI1, an OspF family member in plant pathogen P. syringae, also dephosphorylates MAPKs probably through the same biochemical activity.40 P. syringae effector HopU1, a mono-ADP-ribosyltransferase (ADP-RT), modifies several Arabidopsis RNA-binding proteins, which represent a novel class of ADP-RT substrates. Importantly, the Arabidopsis mutant lacking one of these RNA binding proteins is immunocompromised to pathogen infection, suggesting an essential role of this protein in innate immunity.41 HopM1, another P. syringae effector, targets and degrades Arabidopsis MIN7 protein, a member of the ARF family of guanine nucleotide exchange factors involved in vesicle trafficking, which was also found t be an important player in plant innate immunity.42 Currently, profound knowledge on mechanistic bases of effector hijacking host cellular signaling is mostly derived from overexpression of individual effector genes in host cells. In nature, minute amount of effectors is likely delivered into hosts by pathogens. Thus, the interpretation of data needs to be cautious with the particular system deployed. Another complication is whether the secretion of effectors into hosts possesses the host specificity and secretion hierarchy among the repertoire of effectors on different hosts. Multiple dynamic host targets of effectors have also been proven to commonly exist.43 With the rapid development of innovative technologies and in-depth understanding of host cellular signaling pathways, it is brightly believed that biologists will continue to unravel the dynamic and intimate interaction between microbes and hosts.
Abbreviations
- BAK1
BRI1-associated kinase
- PAMP
pathogen-associated molecular pattern
- PRR
pattern recognition receptor
- TTSS
type III secretion system
- ETI
effector-triggered immunity
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/10301
References
- 1.Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 2.He P, Shan L, Sheen J. Elicitation and suppression of microbe-associated molecular pattern-triggered immunity in plant-microbe interactions. Cell Microbiol. 2007;9:1385–1396. doi: 10.1111/j.1462-5822.2007.00944.x. [DOI] [PubMed] [Google Scholar]
- 3.Gomez-Gomez L, Boller T. FLS2: an LRR receptorlike kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 4.Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 5.Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell. 2006;18:465–476. doi: 10.1105/tpc.105.036574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, et al. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:19613–19618. doi: 10.1073/pnas.0705147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, et al. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell. 2008;20:471–481. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyama C, Dohmae N, Takio K, et al. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci USA. 2006;103:11086–10891. doi: 10.1073/pnas.0508882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Zhao-Hui C, Batoux M, Nekrasov V, Roux M, Chinchilla D, et al. Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0905532106. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nat Immunol. 2005;6:973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- 11.Nurnberger T, Brunner F, Kemmerling B, Piater L. Innate immunity in plants and animals: striking similarities and obvious differences. Immunol Rev. 2004;198:249–266. doi: 10.1111/j.0105-2896.2004.0119.x. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 13.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 14.Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nurnberger T, Jones JD, et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448:497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 15.Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, et al. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA. 2007;104:12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin H, Yan Z, Nam KH, Li J. Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol Cell. 2007;26:821–830. doi: 10.1016/j.molcel.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinoshita T, Cano-Delgado A, Seto H, Hiranuma S, Fujioka S, Yoshida S, et al. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature. 2005;433:167–171. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- 18.Kemmerling B, Schwedt A, Rodriguez P, Mazzotta S, Frank M, Qamar SA, et al. The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr Biol. 2007;17:1116–1122. doi: 10.1016/j.cub.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 19.Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 20.Galan JE, Collmer A. Type III secretion machines: bacterial devices for protein delivery into host cells. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 21.Lindeberg M, Cartinhour S, Myers CR, Schechter LM, Schneider DJ, Collmer A. Closing the circle on the discovery of genes encoding Hrp regulon members and type III secretion system effectors in the genomes of three model Pseudomonas syringae strains. Mol Plant Microbe Interact. 2006;19:1151–1158. doi: 10.1094/MPMI-19-1151. [DOI] [PubMed] [Google Scholar]
- 22.Navarro L, Alto NM, Dixon JE. Functions of the Yersinia effector proteins in inhibiting host immune responses. Curr Opin Microbiol. 2005;8:21–27. doi: 10.1016/j.mib.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 23.Shan L, He P, Sheen J. Intercepting host MAPK signaling cascades by bacterial type III effectors. Cell Host Microbe. 2007;1:167–174. doi: 10.1016/j.chom.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 24.Abramovitch RB, Anderson JC, Martin GB. Bacterial elicitation and evasion of plant innate immunity. Nat Rev Mol Cell Biol. 2006;7:601–611. doi: 10.1038/nrm1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grant SR, Fisher EJ, Chang JH, Mole BM, Dangl JL. Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu Rev Microbiol. 2006;60:425–449. doi: 10.1146/annurev.micro.60.080805.142251. [DOI] [PubMed] [Google Scholar]
- 26.Mudgett MB. New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu Rev Plant Biol. 2005;56:509–531. doi: 10.1146/annurev.arplant.56.032604.144218. [DOI] [PubMed] [Google Scholar]
- 27.Block A, Li G, Fu ZQ, Alfano JR. Phytopathogen type III effector weaponry and their plant targets. Curr Opin Plant Biol. 2008;11:396–403. doi: 10.1016/j.jbi.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Speth EB, Lee YN, He SY. Pathogen virulence factors as molecular probes of basic plant cellular functions. Curr Opin Plant Biol. 2007;10:580–586. doi: 10.1016/j.pbi.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Hostmicrobe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 30.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 31.Bent AF, Mackey D. Elicitors, effectors and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol. 2007;45:399–436. doi: 10.1146/annurev.phyto.45.062806.094427. [DOI] [PubMed] [Google Scholar]
- 32.Ronald PC, Salmeron JM, Carland FM, Staskawicz BJ. The cloned avirulence gene avrPto induces disease resistance in tomato cultivars containing the Pto resistance gene. J Bacteriol. 1992;174:1604–1611. doi: 10.1128/jb.174.5.1604-1611.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin GB, Brommonschenkel SH, Chunwongse J, Frary A, Ganal MW, Spivey R, et al. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- 34.Salmeron JM, Oldroyd GE, Rommens CM, Scofield SR, Kim HS, Lavelle DT, et al. Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell. 1996;86:123–133. doi: 10.1016/s0092-8674(00)80083-5. [DOI] [PubMed] [Google Scholar]
- 35.Kim YJ, Lin NC, Martin GB. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell. 2002;109:589–598. doi: 10.1016/s0092-8674(02)00743-2. [DOI] [PubMed] [Google Scholar]
- 36.Shan L, He P, Zhou JM, Tang X. A cluster of mutations disrupt the avirulence but not the virulence function of AvrPto. Mol Plant Microbe Interact. 2000;13:592–598. doi: 10.1094/MPMI.2000.13.6.592. [DOI] [PubMed] [Google Scholar]
- 37.He P, Shan L, Lin NC, Martin GB, Kemmerling B, Nurnberger T, et al. Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell. 2006;125:563–575. doi: 10.1016/j.cell.2006.02.047. [DOI] [PubMed] [Google Scholar]
- 38.Shan L, He P, Li J, Heese A, Peck SC, Nurnberger T, et al. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptorsignaling complexes and impede plant immunity. Cell Host Microbe. 2008;4:17–27. doi: 10.1016/j.chom.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Xu H, Zhou Y, Zhang J, Long C, Li S, et al. The phosphothreonine lyase activity of a bacterial type III effector family. Science. 2007;315:1000–1003. doi: 10.1126/science.1138960. [DOI] [PubMed] [Google Scholar]
- 40.Zhang J, Shao F, Li Y, Cui H, Chen L, Li H, et al. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe. 2007;1:175–185. doi: 10.1016/j.chom.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 41.Fu ZQ, Guo M, Jeong BR, Tian F, Elthon TE, Cerny RL, et al. A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature. 2007;447:284–288. doi: 10.1038/nature05737. [DOI] [PubMed] [Google Scholar]
- 42.Nomura K, Debroy S, Lee YH, Pumplin N, Jones J, He SY. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science. 2006;313:220–223. doi: 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
- 43.Luo Y, Caldwell KS, Wroblewski T, Wright ME, Michelmore RW. Proteolysis of a negative regulator of innate immunity is dependent on resistance genes in tomato and Nicotiana benthamiana and induced by multiple bacterial effectors. Plant Cell. 2009;21:2458–2472. doi: 10.1105/tpc.107.056044. [DOI] [PMC free article] [PubMed] [Google Scholar]