Abstract
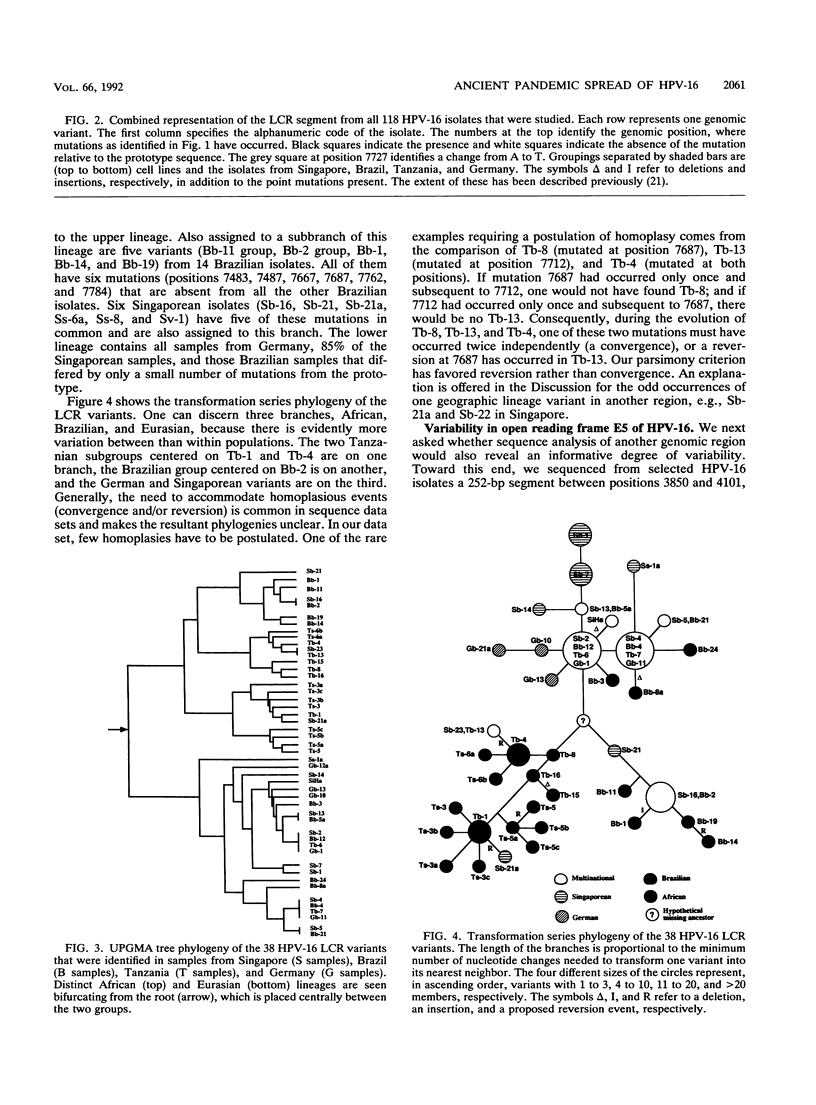
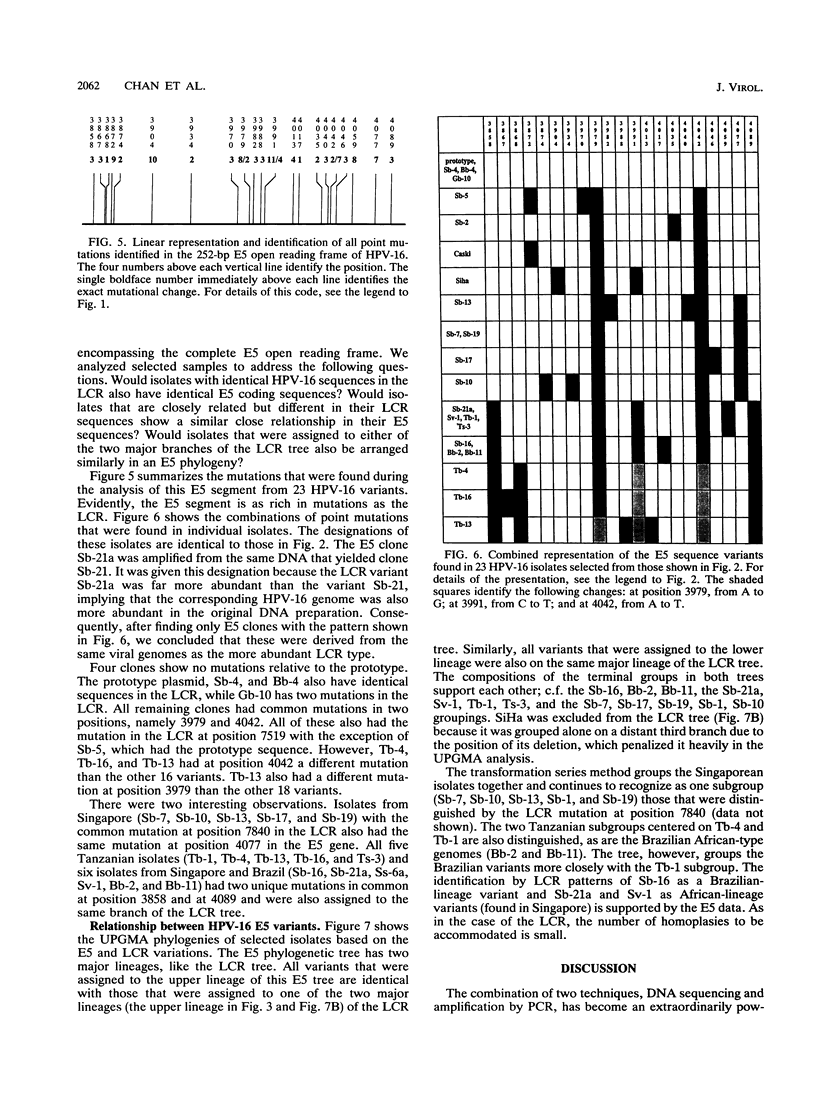
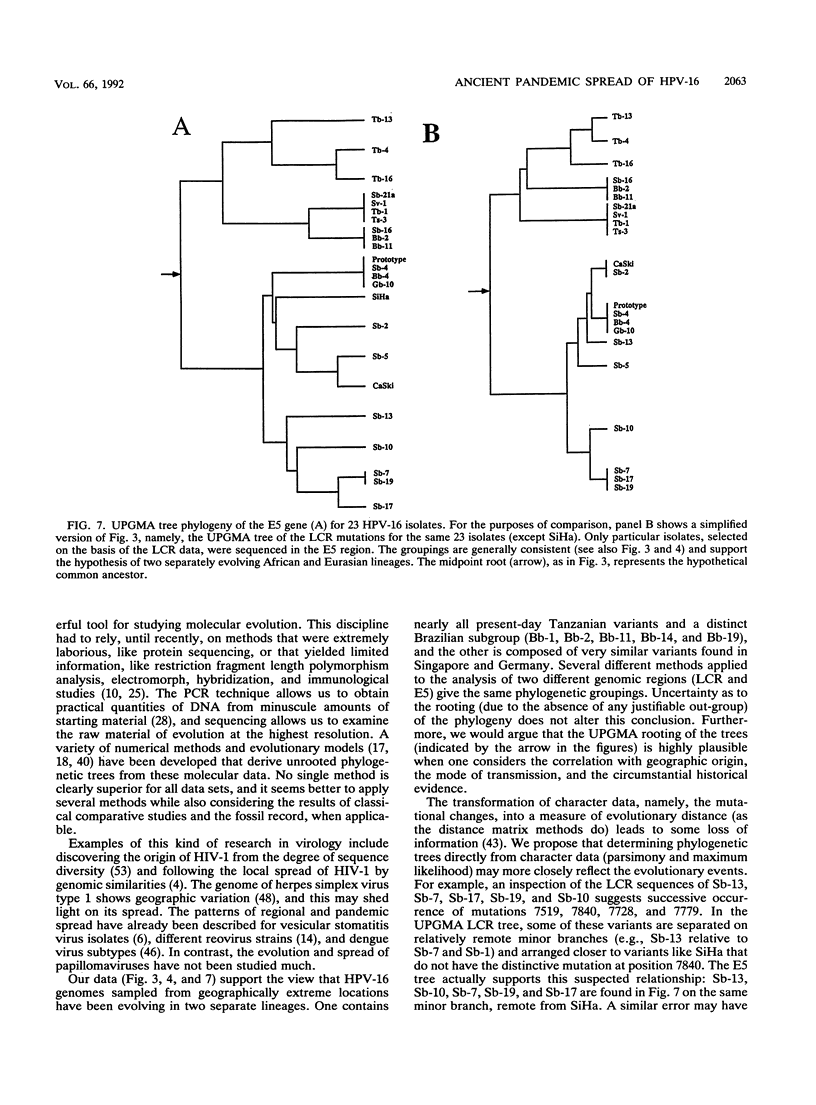
We have amplified by the polymerase chain reaction, cloned, and sequenced genomic segments of 118 human papillomavirus type 16 (HPV-16) isolates from 76 cervical biopsy, 14 cervical smear, 3 vulval biopsy, 2 penile biopsy, 2 anal biopsy, and 1 vaginal biopsy sample and two cell lines. The specimens were taken from patients in four countries--Singapore, Brazil, Tanzania, and Germany. The sequence of a 364-bp fragment of the long control region of the virus revealed 38 variants, most of which differed by one or several point mutations. Phylogenetic trees were constructed by distance matrix methods and a transformation series approach. The trees based on the long control region were supported by another set based on the complete E5 protein-coding region. Both sets had two main branches. Nearly all of the variants from Tanzania were assigned to one (African) branch, and all of the German and most of the Singaporean variants were assigned to the other (Eurasian) branch. While some German and Singaporean variants were identical, each group also contained variants that formed unique branches. In contrast to the group-internal homogeneity of the Singaporean, German, and Tanzanian variants, the Brazilian variants were clearly divided between the two branches. Exceptions to this were the seven Singaporean isolates with mutational patterns typical of the Tanzanian isolates. The data suggest that HPV-16 evolved separately for a long period in Africa and Eurasia. Representatives of both branches may have been transferred to Brazil via past colonial immigration. The comparable efficiencies of transfer of the African and the Eurasian variants to the New World suggest pandemic spread of HPV-16 in past centuries. Representatives of the African branch were possibly transferred to the Far East along old Arab and Indonesian sailing routes. Our data also support the view that HPV-16 is a well-defined virus type, since the variants show only a maximal genomic divergence of about 5%. The small amount of divergence in any one geographic location and the lack of marked divergence between the Tanzanian and Brazilian African genome variants two centuries after their likely introduction into the New World suggest a very slow rate of viral evolution. The phylogenetic tree therefore probably represents a minimum of several centuries of evolution, if not an age equal to that of the respective human races.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almond J. W. The attenuation of poliovirus neurovirulence. Annu Rev Microbiol. 1987;41:153–180. doi: 10.1146/annurev.mi.41.100187.001101. [DOI] [PubMed] [Google Scholar]
- Baker C. C., Phelps W. C., Lindgren V., Braun M. J., Gonda M. A., Howley P. M. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J Virol. 1987 Apr;61(4):962–971. doi: 10.1128/jvi.61.4.962-971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balfe P., Simmonds P., Ludlam C. A., Bishop J. O., Brown A. J. Concurrent evolution of human immunodeficiency virus type 1 in patients infected from the same source: rate of sequence change and low frequency of inactivating mutations. J Virol. 1990 Dec;64(12):6221–6233. doi: 10.1128/jvi.64.12.6221-6233.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilsel P. A., Nichol S. T. Polymerase errors accumulating during natural evolution of the glycoprotein gene of vesicular stomatitis virus Indiana serotype isolates. J Virol. 1990 Oct;64(10):4873–4883. doi: 10.1128/jvi.64.10.4873-4883.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. M., George M., Jr, Wilson A. C. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. M., Prager E. M., Wang A., Wilson A. C. Mitochondrial DNA sequences of primates: tempo and mode of evolution. J Mol Evol. 1982;18(4):225–239. doi: 10.1007/BF01734101. [DOI] [PubMed] [Google Scholar]
- Bubb V., McCance D. J., Schlegel R. DNA sequence of the HPV-16 E5 ORF and the structural conservation of its encoded protein. Virology. 1988 Mar;163(1):243–246. doi: 10.1016/0042-6822(88)90259-0. [DOI] [PubMed] [Google Scholar]
- Cann R. L., Stoneking M., Wilson A. C. Mitochondrial DNA and human evolution. Nature. 1987 Jan 1;325(6099):31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- Chong T., Chan W. K., Bernard H. U. Transcriptional activation of human papillomavirus 16 by nuclear factor I, AP1, steroid receptors and a possibly novel transcription factor, PVF: a model for the composition of genital papillomavirus enhancers. Nucleic Acids Res. 1990 Feb 11;18(3):465–470. doi: 10.1093/nar/18.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow V. T., Tham K. M., Bernard H. U. Thermus aquaticus DNA polymerase-catalysed chain reaction for the detection of human papillomaviruses. J Virol Methods. 1990 Jan;27(1):101–112. doi: 10.1016/0166-0934(90)90150-e. [DOI] [PubMed] [Google Scholar]
- Cole S. T., Danos O. Nucleotide sequence and comparative analysis of the human papillomavirus type 18 genome. Phylogeny of papillomaviruses and repeated structure of the E6 and E7 gene products. J Mol Biol. 1987 Feb 20;193(4):599–608. doi: 10.1016/0022-2836(87)90343-3. [DOI] [PubMed] [Google Scholar]
- Dermody T. S., Nibert M. L., Bassel-Duby R., Fields B. N. Sequence diversity in S1 genes and S1 translation products of 11 serotype 3 reovirus strains. J Virol. 1990 Oct;64(10):4842–4850. doi: 10.1128/jvi.64.10.4842-4850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürst M., Gissmann L., Ikenberg H., zur Hausen H. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3812–3815. doi: 10.1073/pnas.80.12.3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L., Langaney A. Origin and differentiation of human mitochondrial DNA. Am J Hum Genet. 1989 Jan;44(1):73–85. [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- Gloss B., Chong T., Bernard H. U. Numerous nuclear proteins bind the long control region of human papillomavirus type 16: a subset of 6 of 23 DNase I-protected segments coincides with the location of the cell-type-specific enhancer. J Virol. 1989 Mar;63(3):1142–1152. doi: 10.1128/jvi.63.3.1142-1152.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein D. J., Finbow M. E., Andresson T., McLean P., Smith K., Bubb V., Schlegel R. Bovine papillomavirus E5 oncoprotein binds to the 16K component of vacuolar H(+)-ATPases. Nature. 1991 Jul 25;352(6333):347–349. doi: 10.1038/352347a0. [DOI] [PubMed] [Google Scholar]
- Ho L., Chan S. Y., Chow V., Chong T., Tay S. K., Villa L. L., Bernard H. U. Sequence variants of human papillomavirus type 16 in clinical samples permit verification and extension of epidemiological studies and construction of a phylogenetic tree. J Clin Microbiol. 1991 Sep;29(9):1765–1772. doi: 10.1128/jcm.29.9.1765-1772.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland J., Spindler K., Horodyski F., Grabau E., Nichol S., VandePol S. Rapid evolution of RNA genomes. Science. 1982 Mar 26;215(4540):1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980 Dec;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kjaer S. K., de Villiers E. M., Haugaard B. J., Christensen R. B., Teisen C., Møller K. A., Poll P., Jensen H., Vestergaard B. F., Lynge E. Human papillomavirus, herpes simplex virus and cervical cancer incidence in Greenland and Denmark. A population-based cross-sectional study. Int J Cancer. 1988 Apr 15;41(4):518–524. doi: 10.1002/ijc.2910410408. [DOI] [PubMed] [Google Scholar]
- Li W. H., Gojobori T., Nei M. Pseudogenes as a paradigm of neutral evolution. Nature. 1981 Jul 16;292(5820):237–239. doi: 10.1038/292237a0. [DOI] [PubMed] [Google Scholar]
- Martin P., Vass W. C., Schiller J. T., Lowy D. R., Velu T. J. The bovine papillomavirus E5 transforming protein can stimulate the transforming activity of EGF and CSF-1 receptors. Cell. 1989 Oct 6;59(1):21–32. doi: 10.1016/0092-8674(89)90866-0. [DOI] [PubMed] [Google Scholar]
- Miyata T., Yasunaga T., Nishida T. Nucleotide sequence divergence and functional constraint in mRNA evolution. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7328–7332. doi: 10.1073/pnas.77.12.7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münger K., Werness B. A., Dyson N., Phelps W. C., Harlow E., Howley P. M. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989 Dec 20;8(13):4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny D. Towards a basis for classification: the incompleteness of distance measures, incompatibility analysis and phenetic classification. J Theor Biol. 1982 May 21;96(2):129–142. doi: 10.1016/0022-5193(82)90216-8. [DOI] [PubMed] [Google Scholar]
- Rico-Hesse R. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology. 1990 Feb;174(2):479–493. doi: 10.1016/0042-6822(90)90102-w. [DOI] [PubMed] [Google Scholar]
- Romanczuk H., Thierry F., Howley P. M. Mutational analysis of cis elements involved in E2 modulation of human papillomavirus type 16 P97 and type 18 P105 promoters. J Virol. 1990 Jun;64(6):2849–2859. doi: 10.1128/jvi.64.6.2849-2859.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sakaoka H., Saito H., Sekine K., Aomori T., Grillner L., Wadell G., Fujinaga K. Genomic comparison of herpes simplex virus type 1 isolates from Japan, Sweden and Kenya. J Gen Virol. 1987 Mar;68(Pt 3):749–764. doi: 10.1099/0022-1317-68-3-749. [DOI] [PubMed] [Google Scholar]
- Seedorf K., Krämmer G., Dürst M., Suhai S., Röwekamp W. G. Human papillomavirus type 16 DNA sequence. Virology. 1985 Aug;145(1):181–185. doi: 10.1016/0042-6822(85)90214-4. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. Understanding the origins of AIDS viruses. Nature. 1988 Nov 24;336(6197):315–315. doi: 10.1038/336315a0. [DOI] [PubMed] [Google Scholar]
- Shibata D. K., Arnheim N., Martin W. J. Detection of human papilloma virus in paraffin-embedded tissue using the polymerase chain reaction. J Exp Med. 1988 Jan 1;167(1):225–230. doi: 10.1084/jem.167.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. F., Srinivasan A., Schochetman G., Marcus M., Myers G. The phylogenetic history of immunodeficiency viruses. Nature. 1988 Jun 9;333(6173):573–575. doi: 10.1038/333573a0. [DOI] [PubMed] [Google Scholar]
- Ustav M., Stenlund A. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 1991 Feb;10(2):449–457. doi: 10.1002/j.1460-2075.1991.tb07967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa L. L., Franco E. L. Epidemiologic correlates of cervical neoplasia and risk of human papillomavirus infection in asymptomatic women in Brazil. J Natl Cancer Inst. 1989 Mar 1;81(5):332–340. doi: 10.1093/jnci/81.5.332. [DOI] [PubMed] [Google Scholar]
- Werness B. A., Levine A. J., Howley P. M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990 Apr 6;248(4951):76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]



