Abstract
The Vesicle Inducing Protein in Plastids 1 (Vipp1) was suggested to be involved in thylakoid membrane formation in both chloroplasts and cyanobacteria. The protein shows sequence homology to the Phage Shock Protein A (PspA) from bacteria, and both proteins have similar secondary structures. 2D-structures of PspA and of Vipp1 have been determined by electron microscopy in the recent years. Both PspA and Vipp1 form large homooligomeric rings with high molecular masses but their ring dimensions differ significantly. Furthermore, Vipp1 forms rings with different rotational symmetries whereas PspA appears to form rings with singular rotational symmetry. In this article addendum we compare the structures of PspA and Vipp1. Furthermore, we suggest a spatial structural model of the observed Vipp1 rings.
Key words: PspA, Vipp1, ring, structure, function
In 2001 a gene was identified in Arabidopsis thaliana, deletion of which resulted in thylakoid membrane perturbations as well in disturbed vesicle formation.1 Due to this observation, the encoded protein was named Vesicle Inducing Protein in Plastids 1 (Vipp1). Homologous vipp1 genes were found in the genomes of cyanobacteria, and while the gene in the cyanobacterium Synechocystis sp. PCC 6803 could not be completely deleted, depletion of the gene product resulted in a severe reduction of internal thylakoid membranes and in decreased levels of active photosystems.2–4 While the protein appears to operate similar in cyanobacteria and chloroplasts, its exact physiological functions remain mystic. Interestingly, the amino acid sequence of Vipp1 shows similarity to the Phage Shock Protein A (PspA) from bacteria.3 Deletion experiments indicated that PspA is not essential in bacteria whereas Vipp1 is essential for normal development of both cyanobacteria and chloroplasts.1–3,5,6 Although the exact physiological function of PspA is essentially elusive, recent observations indicate that the protein is involved in maintaining the integrity of the E. coli cytoplasmic membrane.7 PspA from E. coli binds to specific lipids of the cytoplasmic membrane,7 and also Vipp1 associates tightly with both the cytoplasmic or inner envelope and the thylakoid membrane in cyanobacteria and chloroplasts.1,8,9
A 2D projection structure of the 1 MDa homooligomeric PspA ring from E. coli has been determined by electron microscopy (EM), and more recently the structure of Vipp1 rings were reported.8,10 Although both structures have similar arrangements, there are significant differences, which could correlate to separate physiological roles.
The amino acid sequences of PspA from E. coli as well as for PspA and Vipp1 from Synechocystis align from their N-terminus up to amino acid residue 218 without insertions or deletions (Fig. 1). The two PspA proteins share a sequence identity of 27% and a sequence similarity of 51%. In line with rather low sequence conservation, the Synechocystis PspA protein and Synechocystis Vipp1 also share just 31% sequence identity (51% similarity). While the amino acid sequence of the proteins is not strictly conserved, the proteins have a highly conserved secondary structure. The predicted secondary structure for all proteins is essentially purely α-helical, apart from some coil interruptions, which is in excellent agreement with experimental determination of the Vipp1 secondary structure.8 Both PspA and Vipp1 contain large regions likely to form coiled-coils of helices when analyzed with the program PCOILS.11 In contrast to PspA, Vipp1 contains a C-terminal extension of 45 amino acids, which forms an additional α-helix. This domain, which is not involved in oligomer formation, appears to be a unique feature of Vipp1 when compared to PspA.
Figure 1.
Amino acid sequence alignment of Vipp1 from Synechocystis and PspA from Synechocystis and E. coli with Clustal X.14 Identical (*), strongly similar (:) and weakly identical (.) amino acid residues are indicated. The grey regions represent α-helical domains predicted with PSIPRED, V2.6.15
Both PspA and Vipp1 form large homooligomeric rings. The structural details of these rings, including dimensions and observed symmetry are compared in Table 1. While PspA forms only a single ring population with 9-fold rotational symmetry, Vipp1 rings have been observed with 12 to 17-fold rotational symmetry and increased ring diameters. The unique PspA ring is formed by 36 PspA molecules, whereas 48 to 68 Vipp1 monomers contribute to the formation of the various Vipp1 rings. The PspA ring has a height of ∼8.5 nm which is considerable smaller than the Vipp1 rings which all have a constant height of ∼22 nm.
Table 1.
A comparison of structure and symmetry of PspA and Vipp1 rings, as determined by electron microscopy and single particle image analysis
| Width (nm) | Height (nm) | Symmetry | Copies/ring | Reference | ||
| PspA | 20 | 8.5 | 9 fold | 36 | 10 | |
| Vipp1 | 25—33 | 22 | 12–17 | fold | 48–68 | 8 |
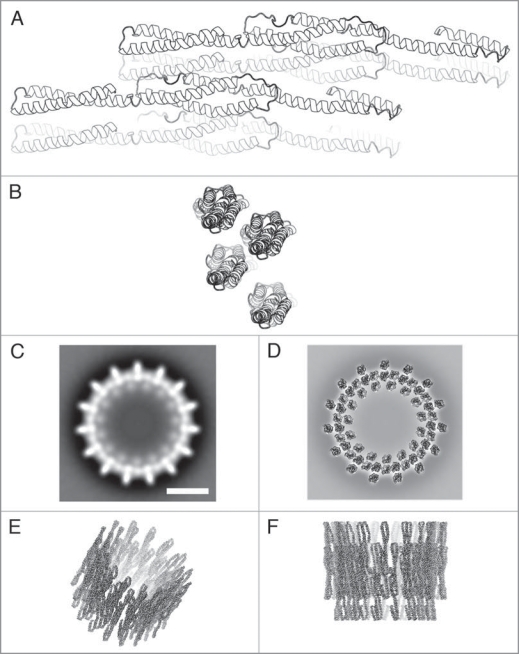
So far, no crystal structure for Vipp1 is available but a potential tertiary structure can be obtained by 3D structure prediction. For this purpose the Synechocystis Vipp1 sequence was submitted to the PHYRE web server. A description about and case study using PHYRE is given by Kelley and Sternberg.12 All generated 3D models had either one long α-helix or coiled-coil structures. The model based on the 3D structure of smooth muscle α-actinin (pdb entry 1sjj) was chosen for further modelling of the oligomeric Vipp1 ring. This starting model (Fig. 2A) contains amino acids 4–253 from Vipp1 (∼93% of the total Vipp1 sequence).
Figure 2.
Model of the Vipp1 structure. (A and B) Side and top view ribbon representations of four predicted Vipp1 monomers arranged in a dimer of dimers configuration. (C) Projection map showing a Vipp1 ring with 15-fold symmetry. (D–F) Top, tilted and side view of the modelled Vipp1 ring, respectively. In (D) the model is placed on top of the projection map shown in (C). The scale bar in (C) equals 10 nm which is also valid for (D–F).
The Vipp1 starting model has a length of 17 nm and a diameter of ∼2.5 nm. Partly, it is a three-start coiled-coil in the central part of the structure as shown in Figure 2A.
Already at this stage it becomes obvious that the tertiary structure of PspA and Vipp1 must significantly differ. The suggested Vipp1 model has a length of 17 nm which later on (compared below) translates to a ring height of >17 nm. Since the PspA ring has only a height of 8.5 nm, the generated model cannot be used to derive the 3D structure of the PspA ring. It is e.g., possible that the long α-helices in the PspA monomer are kinked to form a higher-ordered helix-bundle with a length of ∼8 to 9 nm. This difference in tertiary structure may determine the specificity of PspA vs. Vipp1.
Ring structures with 15 outward and 15 inward pointing spikes (15-fold rotational symmetry) are in the middle of the observed range of Vipp1 symmetries. To each spike two densities can be attributed, both with a diameter of ∼2.5 nm. Since the diameters of the observed 2.5 nm densities and of the predicted Vipp1 monomer are roughly identical, we speculate that each density could originate from a Vipp1 monomer. Thus, the spikes, as seen in the EM projection maps, are each occupied by two Vipp1 proteins. Two Vipp1 monomers were arranged manually such that the total dimer length was ∼22 nm, which corresponds to the ring height determined by single particle analysis.8 Furthermore, two dimers were placed next to each other based on the spikes in the EM projection map with 15-fold symmetry (Fig. 2B–D). Biochemical studies have also indicated that the smallest repeating unit within the Vipp1 rings is a dimer of dimers.8 The 15-fold ring model (Fig. 2D–F below) was generated from this tetramer using routines from the CCP4 package.13 The α-helical configurations of the model are in good agreement with the overall ring dimensions, and the presented model can give an impression of how the Vipp1 ring structure could look like.
Acknowledgements
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (SCHN 690/3-1) and the Council of Chemical Sciences (CW) of the Netherlands Science Foundation NWO. We thank Marcel Bokhove (Department of Biophysical Chemistry, University of Groningen) for help with modelling.
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/10529
References
- 1.Kroll D, Meierhoff K, Bechtold N, Kinoshita M, Westphal S, Vothknecht UC, et al. VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc Natl Acad Sci USA. 2001;98:4238–4242. doi: 10.1073/pnas.061500998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuhrmann E, Gathmann S, Rupprecht E, Golecki J, Schneider D. Thylakoid membrane reduction affects the photosystem stoichiometry in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 2009;149:735–744. doi: 10.1104/pp.108.132373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Westphal S, Heins L, Soll J, Vothknecht UC. Vipp 1 deletion mutant of Synechocystis: a connection between bacterial phage shock and thylakoid biogenesis? Proc Natl Acad Sci USA 2001. 98:4243–4248. doi: 10.1073/pnas.061501198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao H, Xu X. Depletion of Vipp1 in Synechocystis sp. PCC 6803 affects photosynthetic activity before the loss of thylakoid membranes. FEMS Microbiol Lett. 2009;292:63–70. doi: 10.1111/j.1574-6968.2008.01470.x. [DOI] [PubMed] [Google Scholar]
- 5.Weiner L, Model P. Role of an Escherichia coli stress-response operon in stationary-phase survival. Proc Natl Acad Sci USA. 1994;91:2191–2195. doi: 10.1073/pnas.91.6.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aseeva E, Ossenbuhl F, Sippel C, Cho WK, Stein B, Eichacker LA, et al. Vipp1 is required for basic thylakoid membrane formation but not for the assembly of thylakoid protein complexes. Plant Physiol Biochem. 2007;45:119–128. doi: 10.1016/j.plaphy.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi R, Suzuki T, Yoshida M. Escherichia coli phage-shock protein A (PspA) binds to membrane phospholipids and repairs proton leakage of the damaged membranes. Mol Microbiol. 2007;66:100–109. doi: 10.1111/j.1365-2958.2007.05893.x. [DOI] [PubMed] [Google Scholar]
- 8.Fuhrmann E, Bultema JB, Kahmann U, Rupprecht E, Boekema EJ, Schneider D. The Vesicle Inducing Protein 1 from Synechocystis sp. PCC 6803 Organizes into Diverse Higher Ordered Ring Structures. Mol Biol Cell. 2009. [DOI] [PMC free article] [PubMed]
- 9.Li HM, Kaneko Y, Keegstra K. Molecular cloning of a chloroplastic protein associated with both the envelope and thylakoid membranes. Plant Mol Biol. 1994;25:619–632. doi: 10.1007/BF00029601. [DOI] [PubMed] [Google Scholar]
- 10.Hankamer BD, Elderkin SL, Buck M, Nield J. Organization of the AAA(+) adaptor protein PspA is an oligomeric ring. J Biol Chem. 2004;279:8862–8866. doi: 10.1074/jbc.M307889200. [DOI] [PubMed] [Google Scholar]
- 11.Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 12.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 13.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 14.Thompson J, Gibson T, Plewniak F, Jeanmougin F, Higgins D. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]