Abstract
Background
Although integrase inhibitors are highly effective in the management of drug-resistant HIV, some patients fail to achieve durable viral suppression. The long-term consequences of integrase inhibitor failure have not been well-defined.
Methods
We identified 29 individuals who exhibited evidence of incomplete viral suppression on a regimen containing an integrase inhibitor (23 raltegravir, 6 elvitegravir). Prior to initiating the integrase inhibitor-based regimen, the median CD4+ T cell count and plasma HIV RNA levels were 62 cells/mm3 and 4.65 log10 copies/mL, respectively.
Results
At the first failure time-point, the most common integrase resistance pattern for subjects taking raltegravir was wild-type, followed in order of frequency by Q148H/K/R+G140S, N155H, and Y143R/H/C. The most common resistance pattern for subjects taking elvitegravir was E92Q. Long-term failure was associated with continued viral evolution, emergence of high-level phenotypic resistance, and a decrease in replicative capacity.
Conclusions
Although wild-type failure during early integrase inhibitor failure is common, most patients eventually develop high level phenotypic drug resistance. This resistance evolution is gradual and associated with declines in replicative capacity.
Keywords: integrase inhibitors, drug resistance, virologic failure
INTRODUCTION
The management of drug-resistant HIV has improved dramatically with the recent approval of a number of highly effective antiretroviral drugs 1–4. Among these is raltegravir 5, the first clinically available inhibitor of HIV integrase. When used in combination with other active drugs, raltegravir has proven to be very potent, well-tolerated, and highly effective 6–9.
Due to the limited number of patients exhibiting virologic failure in clinical trials of raltegravir, the long-term consequences of incomplete viral suppression while taking an integrase inhibitor have not been well defined. Among the subjects who had evidence of virologic failure in the phase III raltegravir studies (BENCHMRK), 68% had at least one raltegravir resistance mutation 8. However, the longitudinal changes in integrase inhibitor resistance were not described in this study. We present here a cohort of treatment-experienced subjects who exhibited evidence of incomplete viral suppression while taking an integrase inhibitor.
METHODS
Study Participants
All subjects were enrolled in an ongoing, observational cohort based in San Francisco (SCOPE cohort). Subjects who received an integrase inhibitor (raltegravir or elvitegravir) were identified. Subjects enrolled in SCOPE are typically seen every four months. At each visit, an extensive interview is performed and plasma and other biologic specimens are obtained and stored. Adherence was measured by asking subjects the number of missed doses of each antiretroviral drug in the previous 30 days. Virologic failure was defined as either: 1) developing detectable viremia after achieving viral suppression, or 2) failure to achieve viral suppression. The SCOPE study was approved by the University of California San Francisco Committee on Human Research. All subjects provided written informed consent.
Integrase inhibitor resistance testing
All resistance testing was performed at Monogram Biosciences. Genotypic and phenotypic resistance testing was performed in subjects with detectable viremia. Genotypic resistance measurements were performed using the GeneSeqR Integrase assay, in which patient-derived integrase coding sequences were determined using conventional dye-deoxy chain terminator chemistry (ABI, Foster City, CA). Mutations were identified based on the most recent International AIDS Society-USA guidelines 10. Primary integrase inhibitor mutations were defined as Y143R/H/C, Q148H/K/R, or N155H. Secondary mutations were defined as L74M, E138A/K, G140S, E92Q, T97A, G163K/R, V151I, or D232N. Integrase sequences have been submitted to GenBank.
Phenotypic susceptibility to raltegravir was determined using PhenoSense® Integrase. Briefly, integrase sequences were amplified from patient plasma by RT-PCR and transferred into a resistance test vector containing a luciferase reporter gene. Pseudotyped virus containing patient-derived integrase was used to infect HEK293 cells in the presence and absence of serial dilutions of raltegravir. Susceptibility was reported as the fold-change in IC50 (ratio of IC50 of the sample virus to the IC50 of a reference strain [NL4-3]). In addition, replication capacity was determined and expressed as the percent of viral infectivity as measured by luciferase production, in the absence of drug compared to a reference strain (NL4-3).
RESULTS
Baseline characteristics prior to integrase inhibitor therapy
A total of 79 subjects enrolled in the SCOPE cohort initiated an integrase inhibitor based regimen (73 raltegravir and 6 elvitegravir). Integrase genotypic and phenotypic resistance testing was performed prior to integrase inhibitor therapy in 45/79 subjects; the median fold-change in IC50 to raltegravir was 0.89. The median replicative capacity was 114%. Four of 45 subjects had a secondary integrase inhibitor associated mutation at baseline (1 subject with E138K, 3 subjects with V151I) 10; there were no primary integrase mutations.
Integrase inhibitor resistance at first virologic failure
Among the 79 subjects who initiated an integrase inhibitor based regimen, 50 had evidence of durable viral suppression. The remaining 29 subjects (23 raltegravir, 6 elvitegravir) failed to achieve and maintain an undetectable viral load and were included in the failure analyses. Prior to initiating an integrase inhibitor based regimen, the median baseline CD4+ T cell count and plasma HIV RNA level was 62 cells/mm3 and 4.65 log10 copies/mL, respectively (Table 1). The cohort was highly treatment-experienced and was followed for a median 15.8 months. The median CD4+ T cell count and plasma HIV RNA level at the time of first virologic failure was 118 cells/mm3 and 4.44 log10 copies RNA/mL, respectively. Subjects had been on an integrase inhibitor for a median 4.9 months at the time of first virologic failure.
TABLE 1.
Baseline Characteristics, Integrase Inhibitor Failure Cohort (n=29)
| Gender | 93% Male, 3% Female, 3% Male-to-female |
| Age (years) | 47 (45–50) |
| CD4+ T cell count (cells/mm3) | 62 (8–268) |
| Plasma HIV RNA (log10 copies/mL) | 4.65 (4.12–5.36) |
| Number of prior ARV | 12 (9–16) |
| Number of prior ARV drug classes | 4 (3–4) |
| Duration of follow-up (months) | 15.8 (12.4–17.5) |
Data are medians with interquartile ranges (IQR). ARV=antiretroviral drugs.
Nineteen of the 23 subjects who exhibited incomplete viral suppression on a raltegravir-based regimen had at least one genotypic/phenotypic resistance tested performed after a median 4.1 months of therapy. At the first failure time-point, the most common integrase genotype was wild-type (no known primary mutations, n=13), followed in order of frequency by Q148H/K/R (n=3), N155H (n=3), and Y143R/H/C (n=1) (notably, one subject had a mixture of resistant virus subpopulations that contained mutations at either codon 148 or 155). The G140S was present in four subjects, typically in association with Q148H/K/R. The E92Q mutation was observed in one raltegravir failure, which also contained mixtures of resistant virus at codons 148, 155, and 140. The median viral load in subjects failing with wild-type versus resistant integrase was 3.86 and 4.28 log10 copies/mL, respectively (p=0.177).
We also identified six subjects who were participants in a phase II study of elvitegravir. The most common integrase resistance mutation at the first failure time-point was E92Q (3/5 subjects with available genotypes, while the remaining 2 subjects had wild-type virus). The E92Q mutation was associated with low-level phenotypic resistance to raltegravir.
Absence of integrase inhibitor resistance in “blippers”
Six of 50 subjects who had viral suppression on a raltegravir-based regimen had episodes of detectable viremia < 1000 copies/mL during otherwise effective therapy (median 177 copies/mL, IQR 71–300). Virus was successfully amplified from at least one time-point for 5/6 of these subjects and found to have no genotypic or phenotypic evidence of resistance to raltegravir.
Evolution of integrase inhibitor resistance over time
The genotypic and phenotypic level of integrase resistance increased gradually over time among subjects who remained on an integrase inhibitor-based regimen despite persistent viremia. The proportion of viruses lacking primary or secondary integrase resistance mutations declined at each subsequent time-point, while the proportion of viruses with more than one integrase mutation increased (Fig. 1A). Similar changes were observed in the evolution of phenotypic resistance (Fig. 1B). Moreover, the level of phenotypic resistance to raltegravir in subjects failing with either the Q148H/K/R or Y143R/H/C mutations was consistently high (> 100 fold-change in IC50), whereas the level of phenotypic resistance to raltegravir was much lower in those failing with the N155H mutation (< 50 fold-change in IC50).
Figure 1.
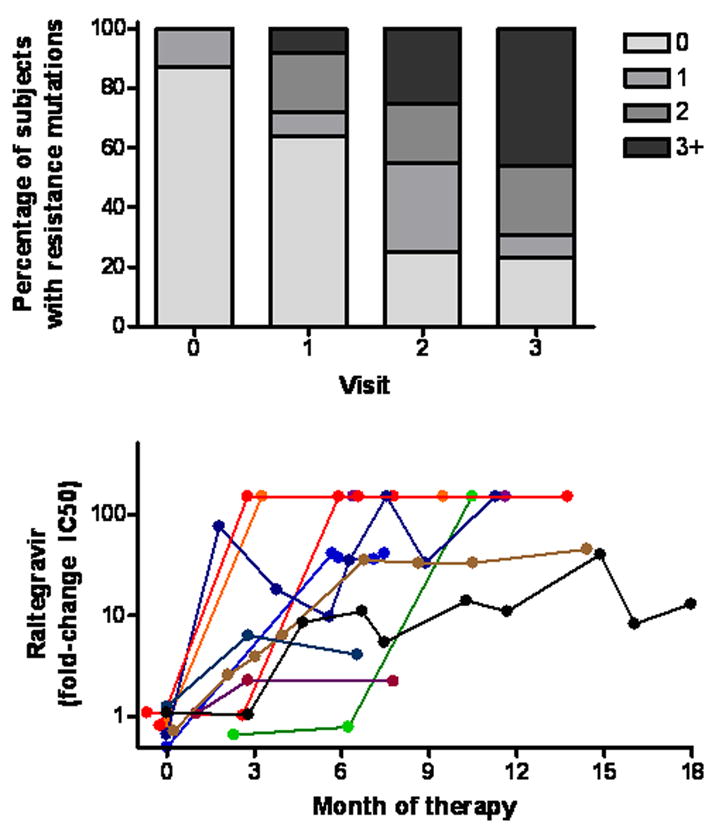
Incomplete viral suppression on an integrase inhibitor based regimen is associated with continued viral evolution, as defined by the number of primary and secondary integrase mutations present at each consecutive visit (Panel A) and by the level of phenotypic resistance (Panel B). All subjects who had incomplete viral suppression and who remained on an integrase inhibitor-based regimen and who had interpretable genotypic resistance data are shown in Panel A (n=23, 25, 20 and 13 at baseline and visits 1–3, respectfully). All subjects who eventually developed evidence of integrase resistance are shown in Panel B (n=11).
Role of adherence in predicting resistance during failure
We next assessed the relationship between adherence and the risk of virologic failure and integrase inhibitor resistance. The cohort was divided into three groups: subjects who had durable virologic success (including those with episodes of detectable viremia < 1000 copies/mL during otherwise effective therapy), subjects who exhibited virologic failure with at least one primary integrase inhibitor associated mutation, and subjects who failed without any integrase inhibitor mutations. The average self-reported level of adherence in the previous 30 days to all antiretroviral drug doses was 97.8% in subjects who had durable virologic success, 98.7% in subjects who failed with integrase inhibitor resistance, and 90.7% in subjects who failed with wild-type virus (Wilcoxon rank-sum test: p=0.004 for wild-type failure vs. success, p=0.007 for wild-type failure vs. resistance failure). The findings were similar when the analyses were limited to adherence to integrase inhibitor doses (data not shown).
Replicative capacity
There were no obvious differences in the replicative capacity of viruses containing the Y143R/H/C, Q148H/K/R, or N155H mutations (data not shown), although the number of subjects in each group was limited. Among the subjects who developed clear evidence of integrase inhibitor resistance (n=11, Fig. 1B), there appeared to be a decline in integrase replicative capacity over time (Fig. 2B). In contrast, there was no consistent change in replicative capacity among those who failed without evidence of integrase inhibitor resistance (data not shown). Moreover, for the 10 subjects who developed phenotypic/genotypic evidence of integrase inhibitor resistance and had pre-/post-failure samples available, the median decrease in replicative capacity between the pre-integrase inhibitor baseline and the last failure time-point was 45% (p= 0.0002; Fig. 2A).
Figure 2.
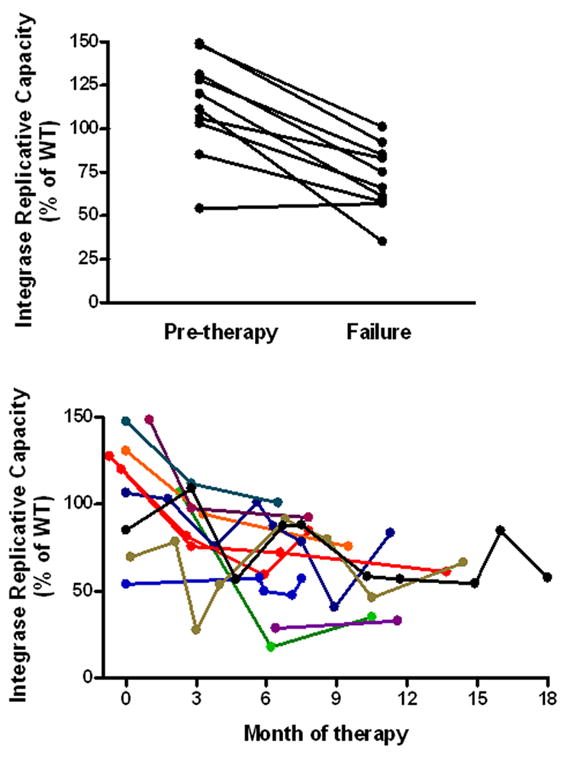
Evolution of integrase replicative capacity. Among subjects failing with integrase inhibitor resistance, there was a decrease in replicative capacity between pre-therapy baseline and the last failure time-point on drug (n=10, Panel A). Long-term virologic failure of an integrase inhibitor based regimen is associated with a decrease in replicative capacity (n=11, Panel B).
Raltegravir treatment interruption and possible continued partial benefit
One individual harboring a virus with the N155H + V151I + D232N mutations (41-fold change in IC50) discontinued raltegravir while remaining on a stable background regimen. In contrast to most of the other subjects in the cohort, this individual never had evidence of advanced HIV disease (pre-integrase inhibitor CD4+ T cell count and plasma viral load of 331 cells/mm3 and 4.07 log10 copies/mL, respectively). After an initial virologic response to raltegravir, the subject’s viral load rebounded to a steady-state level of approximately 4 log10 copies/mL. Because of concerns of ongoing viral evolution and relatively preserved CD4+ T cell counts, raltegravir was interrupted in an attempt to preserve this class for future antiretroviral regimens. Interestingly, plasma HIV RNA levels remained relatively stable for one week in the absence of raltegravir, suggesting limited residual antiviral activity (Fig. 3A). However, viral load subsequently increased by 10-fold as genotypic/phenotypic evidence for raltegravir resistance waned over the next month. In addition, replicative capacity increased as the raltegravir inhibitor mutations waned (Fig. 3B).
Figure 3.
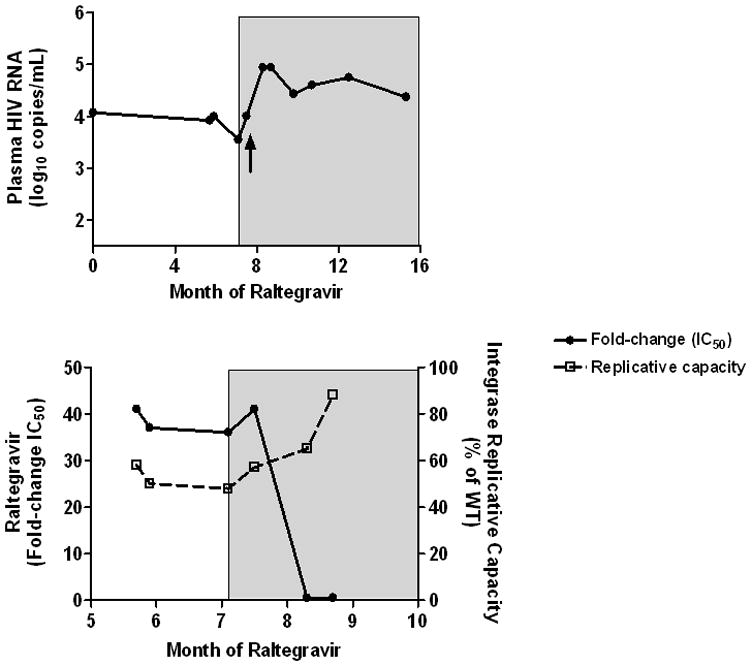
Interruption of raltegravir. One individual harboring a virus with the N155H + V151I + D232N mutations (41-fold change in IC50) discontinued raltegravir while remaining on a stable background regimen. Shaded area indicates the period when raltegravir was stopped. Change in plasma HIV RNA levels are shown in Panel A; the time-point when raltegravir resistance mutations waned is marked by an arrow. Change in raltegravir phenotypic susceptibility and replicative capacity are shown in Panel B.
DISCUSSION
Although experience from clinical trials indicates that the vast majority of patients receiving a raltegravir-based regimen do well, there remains a poorly defined subset of patients for whom these drugs fail to fully suppress viral replication. Here, we report on our experiences with patients who exhibited incomplete viral suppression on an integrase inhibitor based regimen. As has been reported before, certain key “signature” mutations were commonly observed (Q148H/K/R > N155H > Y143R/H/C) 10, and when present were associated with marked decreases in phenotypic susceptibility. However, in contrast to expectations for a drug which is considered to have a low “genetic barrier” to resistance, resistance to this drug emerged gradually, with most subjects having no genotypic or phenotypic evidence of integrase inhibitor resistance during early virologic failure. Among those patients who remained on an integrase inhibitor despite detectable viremia, high-level resistance invariably emerged. This viral evolution is consistent with what has been observed with other antiretroviral drugs (particularly some of the nucleoside analogues and protease inhibitors), and suggests that early efforts to modify therapy (or at least interrupt the integrase inhibitor) may preserve future options within this class.
The complex interaction between resistance, viral fitness, and persistent antiviral activity differs between each of the therapeutic drug classes. Some antiretroviral drugs exhibit clear benefit in the face of drug resistance (e.g, nucleoside analogues), while other drugs have limited or no residual activity (e.g., first generation non-nucleoside reverse transcriptase inhibitors). Knowledge regarding how to use integrase inhibitors in the context of resistance is necessary, as there will always likely be a subset of patients who have failed all drug classes and have limited options for complete viral suppression. In our study, we observed that long-term failure of integrase inhibitors was associated with sustained decreases in integrase replicative capacity. This decrease may in theory confer some benefit in a partially suppressive regimen. We also observed a rapid increase in viremia after removal of raltegravir in a patient who had developed resistance (N155H) to this drug. This increase in viremia appeared to occur as genotypic/phenotypic evidence for raltegravir resistance waned, suggesting a possible “fitness benefit.” Notably, this is consistent with observations from the SIV-infected macaque model, where removal of an integrase inhibitor in animals harboring the N155H mutation was associated with initial stable viremia, followed by increases in viremia as the mutation waned 11. Our data, however, are inconsistent with a recent report of 5 subjects with raltegravir failure whose plasma HIV RNA levels remained stable after discontinuation of raltegravir 12. Clearly, more data are necessary to define to what degree integrase inhibitors have residual activity in the context of resistance.
As has been reported for protease inhibitors in the past, integrase inhibitor resistance appeared to occur only in the presence of high levels of adherence 13. Since self-report adherence measurements are often imprecise, it is possible that the subjects failing with wild-type were taking less of their therapy than stated, but it is unlikely that most of these subjects had completely interrupted their medication. Given the fitness costs associated with integrase inhibitor-associated mutations, it is possible that the negative impact of genotypic mutations on replicative capacity may outweigh the positive impact of these mutations on drug-susceptibility in the context of low-to-moderate levels of drug exposure 13.
In summary, in this cohort of highly treatment-experienced subjects failing an integrase inhibitor containing regimen, wild-type failure was relatively common (particularly during early failure), but high-level genotypic and phenotypic resistance eventually emerged in most subjects who remained on therapy. Whether early modification of a failing regimen can preserve raltegravir (and other integrase inhibitors) remains to be defined. The challenging decision of whether to continue raltegravir in patients who have limited options for complete viral suppression will for now depend upon a patient’s specific clinical scenario.
Acknowledgments
Financial support. This work was supported by grants from the NIAID (K23AI075985, K24 AI069994), the UCSF/Gladstone Center for AIDS Research (P30 AI27763, P30 MH59037), the Center for AIDS Prevention Studies (P30 MH62246), and the UCSF Clinical and Translational Science Institute(UL1 RR024131-01). The development of the PhenoSense® Integrase assay was partially supported by NIH NIAID SBIR- R43 AI057074-01.
Footnotes
This study was presented in part at the Drug Resistance Workshop, June 2008, Sitges, Spain (abstract 10); and at the 16th Conference on Retroviruses and Opportunistic Infections, February 2009, Montreal, Canada (abstract 621).
Contributions of authors. HH: Study design, statistical analysis, authorship of manuscript. HL, RH, JL: subject recruitment. SF, SG, WH, CP: genotype, phenotype data. JNM, DB, SGD: study design, statistical analysis.
Potential conflicts of interest. SG, SF, WH, and CP are employees of Monogram Biosciences. HH has received research support from Merck and Monogram Biosciences. SD has received research support from Merck, Gilead, and Monogram Biosciences.
References
- 1.Hammer SM, Eron JJ, Jr, Reiss P, et al. Antiretroviral treatment of adult HIV infection: 2008 recommendations of the International AIDS Society-USA panel. JAMA. 2008;300:555–570. doi: 10.1001/jama.300.5.555. [DOI] [PubMed] [Google Scholar]
- 2.Gulick RM, Lalezari J, Goodrich J, et al. Maraviroc for previously treated patients with R5 HIV-1 infection. N Engl J Med. 2008;359:1429–1441. doi: 10.1056/NEJMoa0803152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lazzarin A, Campbell T, Clotet B, et al. Efficacy and safety of TMC125 (etravirine) in treatment-experienced HIV-1-infected patients in DUET-2: 24-week results from a randomised, double-blind, placebo-controlled trial. Lancet. 2007;370:39–48. doi: 10.1016/S0140-6736(07)61048-4. [DOI] [PubMed] [Google Scholar]
- 4.Clotet B, Bellos N, Molina JM, et al. Efficacy and safety of darunavir-ritonavir at week 48 in treatment-experienced patients with HIV-1 infection in POWER 1 and 2: a pooled subgroup analysis of data from two randomised trials. Lancet. 2007;369:1169–1178. doi: 10.1016/S0140-6736(07)60497-8. [DOI] [PubMed] [Google Scholar]
- 5.Hazuda DJ, Felock P, Witmer M, et al. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 6.Markowitz M, Nguyen BY, Gotuzzo E, et al. Rapid and durable antiretroviral effect of the HIV-1 Integrase inhibitor raltegravir as part of combination therapy in treatment-naive patients with HIV-1 infection: results of a 48-week controlled study. J Acquir Immune Defic Syndr. 2007;46:125–133. doi: 10.1097/QAI.0b013e318157131c. [DOI] [PubMed] [Google Scholar]
- 7.Grinsztejn B, Nguyen BY, Katlama C, et al. Safety and efficacy of the HIV-1 integrase inhibitor raltegravir (MK-0518) in treatment-experienced patients with multidrug-resistant virus: a phase II randomised controlled trial. Lancet. 2007;369:1261–1269. doi: 10.1016/S0140-6736(07)60597-2. [DOI] [PubMed] [Google Scholar]
- 8.Cooper DA, Steigbigel RT, Gatell JM, et al. Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N Engl J Med. 2008;359:355–365. doi: 10.1056/NEJMoa0708978. [DOI] [PubMed] [Google Scholar]
- 9.Steigbigel RT, Cooper DA, Kumar PN, et al. Raltegravir with optimized background therapy for resistant HIV-1 infection. N Engl J Med. 2008;359:339–354. doi: 10.1056/NEJMoa0708975. [DOI] [PubMed] [Google Scholar]
- 10.Johnson VA, Brun-Vezinet F, Clotet B, et al. Update of the Drug Resistance Mutations in HIV-1: Spring 2008. Top HIV Med. 2008;16:62–68. doi: 10.1007/s11750-007-0034-z. [DOI] [PubMed] [Google Scholar]
- 11.Hazuda DJ, Young SD, Guare JP, et al. Integrase inhibitors and cellular immunity suppress retroviral replication in rhesus macaques. Science. 2004;305:528–532. doi: 10.1126/science.1098632. [DOI] [PubMed] [Google Scholar]
- 12.Wirden M, Simon A, Schneider L, et al. Raltegravir has no residual antiviral activity in vivo against HIV-1 with resistance-associated mutations to this drug. J Antimicrob Chemother. 2009 doi: 10.1093/jac/dkp310. [DOI] [PubMed] [Google Scholar]
- 13.Bangsberg DR, Acosta EP, Gupta R, et al. Adherence-resistance relationships for protease and non-nucleoside reverse transcriptase inhibitors explained by virological fitness. AIDS. 2006;20:223–231. doi: 10.1097/01.aids.0000199825.34241.49. [DOI] [PubMed] [Google Scholar]


