Abstract
Abnormal expansion of a polyglutamine tract in huntingtin (Htt) protein results in Huntington's disease (HD), an autosomal dominant neurodegenerative disorder involving progressive loss of motor and cognitive function. Contrasting with the ubiquitous tissue expression of polyglutamine-expanded Htt (polyQ-Htt), HD pathology is characterized by the increased vulnerability of specific neuronal populations within the striatum and the cerebral cortex. Morphological, biochemical, and functional characteristics of neurons affected in HD that might render these cells more vulnerable to the toxic effects of polyQ-Htt are covered in this review. The differential vulnerability of neurons observed in HD is discussed in the context of various major pathogenic mechanisms proposed to date, and in line with evidence showing a “dying-back” pattern of degeneration in affected neuronal populations.
Keywords: Huntington's disease, medium-sized spiny neurons, huntingtin, axonal transport, dying back degeneration
Introduction
Polyglutamine (polyQ) expansion diseases including Huntington's disease (HD), spinal and bulbar muscular atrophy (SBMA) and spinocerebellear ataxias are heritable neurological diseases characterized by abnormal expansion of CAG repeats in the coding region of structurally unrelated genes (Orr & Zoghbi 2007). Among these, HD represents the most commonly inherited neurological disorder, involving progressive loss of motor and cognitive function that mainly results from degeneration of selected neuronal populations within the basal ganglia and the cerebral cortex (reviewed in (Walker 2007). In recent years, much progress has been made regarding the etiology of HD, and a growing body of evidence suggests the involvement of multiple pathogenic pathways in this disease. However, little is known about mechanisms underlying the increased vulnerability of selected neuronal populations in HD. Here, we provide a detailed summary of the unique pathological topography of HD, and discuss the contribution of cell type-specific characteristics to this topography in the context of various pathogenic mechanisms proposed for HD. These mechanisms are discussed keeping in mind the “dying-back” pattern of degeneration of neurons affected.
HD etiology
HD is inherited in an autosomal-dominant manner and typically presents in adulthood although juvenile forms of HD exist. Molecular mechanisms underlying HD remain elusive, and thus no effective therapeutic treatments are currently available beyond clinical symptomatic management of the movement disorder portion of this disease. HD patients typically die within 17 years of diagnosis from various complications such as accidents, aspiration and dysphagia (Walker 2007).
HD results from expansion of a polymorphic polyQ tract located near the N-terminus of the huntingtin (Htt) protein (HDCRG 1993). The polyQ tract in wild-type, non-pathogenic Htt (WT-Htt) ranges from 6 to 35 glutamines, but Htt variants with 36 or more Qs define a HD allele encoding pathogenic Htt (polyQ-Htt) (Brinkman et al. 1997, Snell et al. 1993). As in other polyQ diseases, the number of glutamine repeats in Htt correlates inversely with the age of HD onset (Andrew et al. 1993, Duyao et al. 1993). The temporal pattern of neuropathological features also relates to the number of glutamines in Htt, with the most damage seen in the brains of age-matched HD patients bearing longer polyQ tracts (Penney et al. 1997). However, HD is universally fatal and the ultimate pathological outcome is similar for all patients showing pathogenic expansions of the polyQ tract.
The widespread distribution of Htt (see below) and the lack of sequence homology with other proteins did not reveal significant information on the normal physiological function of this protein. Deletion of Htt in mice results in embryonic lethality, suggesting a critical, yet unidentified role of Htt during normal development (Nasir et al. 1995, MacDonald et al. 1996). Initial transgenic HD mouse models were illuminating in several respects (Beal & Ferrante 2004). These mice express randomly inserted Htt truncation constructs bearing unusually long polyQ stretches (>115 CAG repeats) that often result in severe early-onset neuropathology and behavioral syndromes (reviewed in (Ramaswamy et al. 2007b). In contrast, various knock-in mouse models expressing pathogenic, polyQ-expanded versions of full-length Htt at endogenous levels are viable, displaying a late onset phenotype with pathological features reminiscent of HD (Menalled 2005). Viability of these knock-in mice indicated that aspects of Htt functionality relevant to embryonic development are not compromised by polyQ tract expansion. The precise contribution that decreased Htt function plays in HD pathogenesis remains unclear, but the autosomal dominant pattern of HD inheritance and other genetic evidence strongly indicates that polyQ expansion confers a toxic gain of function upon Htt (Orr & Zoghbi 2007, Morfini et al. 2005). Consistent with this idea, several lines of experimental evidence showed that polyQ-Htt expression alters multiple, critical cellular processes including transcriptional regulation, cell survival, intracellular signaling, mitochondrial function and axonal transport, among others. However, the nature of molecular mechanisms underlying toxic gain of function(s) associated with polyQ-Htt continue to be debated (Morfini et al. 2005).
HD brain pathology
Motor impersistence (a term referring to the inability to maintain voluntary muscle contractions), represents a major clinical feature of HD that correlates well with disease progression (Reilmann et al. 2001). Involuntary, arrhythmic limb movements termed “chorea” represent a common clinical motor phenotype in most, but not all, HD patients (Barbeau et al. 1981), especially early in the disease. These movements were signature features in the original description of the disease as Huntington's chorea (Okun 2003). In addition to the relentless decline in motor function, non-motor disturbances such as cognitive impairments, personality changes, depression, and behavioral disturbances are commonly seen in HD patients and represent the more serious symptoms for their family, friends and caregivers (Walker 2007). These anomalies are believed to represent the phenotypic manifestation of neuronal dysfunction and degeneration in selected areas of the basal ganglia and the cerebral cortex (Reiner et al. 1988, Storey & Beal 1993).
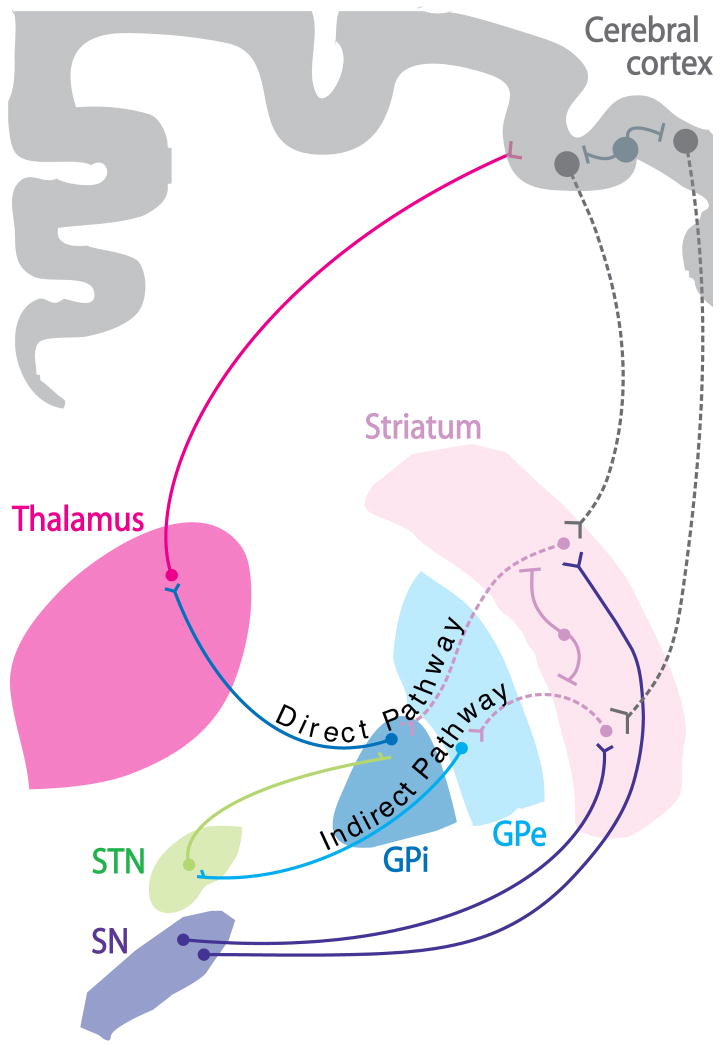
The basal ganglia comprise a set of subcortical brain structures involved in various aspects of motor control and cognition (Graybiel 1990, Mitchell et al. 1999) (Fig. 1). Within the basal ganglia, the neurodegenerative process characteristic of HD typically begins in the striatum (Vonsattel et al. 1985a), which serves the function of “filtering” multiple input pathways originating in different cortical regions (Mitchell et al. 1999). Information processed in the striatum ultimately returns to the cerebral cortex to complete the corticobasal ganglia-thalamocortical loop (Parent & Hazrati 1995) (Fig. 1). Within the striatum, signs of pathology initially appear in the caudate nucleus and putamen, with reactive gliosis and neurons showing neuritic dystrophy. As the disease progresses, these pathologies progressively extend along the caudal-rostral, and dorsal-ventral direction towards the putamen (Vonsattel & DiFiglia 1998). The prominent expansion of the lateral ventricles typical of advanced HD patients results from the dramatic degeneration of neurons within the adjacent caudate nucleus. A significant loss of neurons is also observed in the cerebral cortex of HD patients, including frontal, parietal, and temporal regions (Mann et al. 1993, Heinsen et al. 1994), although these changes are initially less obvious than those seen in the striatum.
Figure 1. Overview of HD pathology.
A subset of projection neurons in the striatum and the cortex (represented by dashed lines) are particularly vulnerable in HD. These include medium spiny neurons (MSNs, pink dashed lines) of the striatum and large pyramidal projection neurons in cortical layers V, VI and III of the cerebral cortex (gray dashed lines). MSNs in the “indirect pathway” of the basal ganglia project to the external segment of the globus pallidus (GPe) and are affected early in the course of the disease. As HD progresses, MSNs projecting to the internal segment of the globus pallidus (GPi) via the “direct pathway” and cortical pyramidal cells projecting to the striatum are also impaired. Remarkably, most interneurons in both the striatum (pink solid lines) and the cerebral cortex (gray solid lines) are largely spared. This morphological and functional difference has been proposed to play a role in the differential vulnerability of neurons observed in HD (Cicchetti et al. 2000), as well as other neurodegenerative diseases (Mattson & Magnus 2006, Morfini et al. 2009a, Morfini et al. 2009b). Abbreviations: STN: subthalamic nucleus; GPe: external globus pallidus; GPi: internal globus pallidus; SN: substantia nigra.
Neuronal cell types affected in HD
At the cellular level, HD is characterized by differential vulnerability of specific neuronal subpopulations within the striatum and cerebral cortex (Fig. 1). The striatum, comprised of the caudate nucleus and putamen, represents the major “input” stage of the basal ganglia, being mainly composed of projection neurons (up to 95% of total striatal neurons) and a much smaller number of interneurons (approx. 5%). Golgi staining methods and electron microscopic studies identified and classified various subtypes of projection neurons and interneurons in the striatum with unique morphological and biochemical characteristics (DiFiglia et al. 1976, Difiglia et al. 1980, Parent et al. 1984). Striatal projection neurons (also known as Golgi type I cells) are all GABAergic and morphologically characterized by a long axon, medium-sized cell bodies, and spiny dendrites, hence the commonly used term of medium spiny neurons (MSNs) (Gerfen 1988, DiFiglia et al. 1976, Difiglia et al. 1980). MSNs project their axons over relatively long distances to targets in the globus pallidus (GP) and the substantia nigra pars reticulata (SNr), main “output” structures of the basal ganglia (Fig. 1). Striatal interneurons (also known as Golgi type II cells) represent key elements of the local striatal circuitry, displaying a wide range of morphological and biochemical heterogeneity (Parent et al. 1984, Parent et al. 1980). Striatal interneurons have unusually short axons, medium to very large-sized cell bodies, and display extensive dendritic arborization, making abundant synaptic contacts with multiple MSNs (Kawaguchi 1997, Kawaguchi et al. 1995). Intriguingly, MSNs represent the main and earliest striatal cell type affected in HD, whereas striatal interneurons are typically unaffected or only mildly affected at late stages of the disease.
The differential vulnerability of MSNs within the HD striatum extends to specific MSNs subtypes, as defined by their projection targets and neurochemical content (Mitchell et al. 1999) (Figs. 1-2). Based on their projection targets, MSNs in the striatum can be divided into two main groups: a) MSNs in the “direct” (or striatonigral) pathway, which project axons monosynaptically to the internal segment of the GP (GPi) or to the SNr (Smith et al. 1998), These neurons preferentially express the peptide enkephalin (ENK, a pentapeptide derived from the preproenkephalin gene). b) MSNs in the “indirect” (or striatopallidal) pathway, which project axons that polysynaptically contact the external segment of the GP (GPe). These neurons preferentially express substance P (SP). Intriguingly, MSNs in the “indirect pathway” are affected at earlier stages and to a greater extent than MSNs in the “direct pathway” (Reiner et al. 1988, Albin et al. 1992a, Richfield et al. 1995b). Early functional abnormalities in the indirect pathway have been associated with development of the chorea-like movements in HD (Crossman et al. 1988, Crossman 1987). Degeneration of MSNs of the direct pathway late in the course of HD manifests as rigidity and bradykinesia (Berardelli et al. 1999).
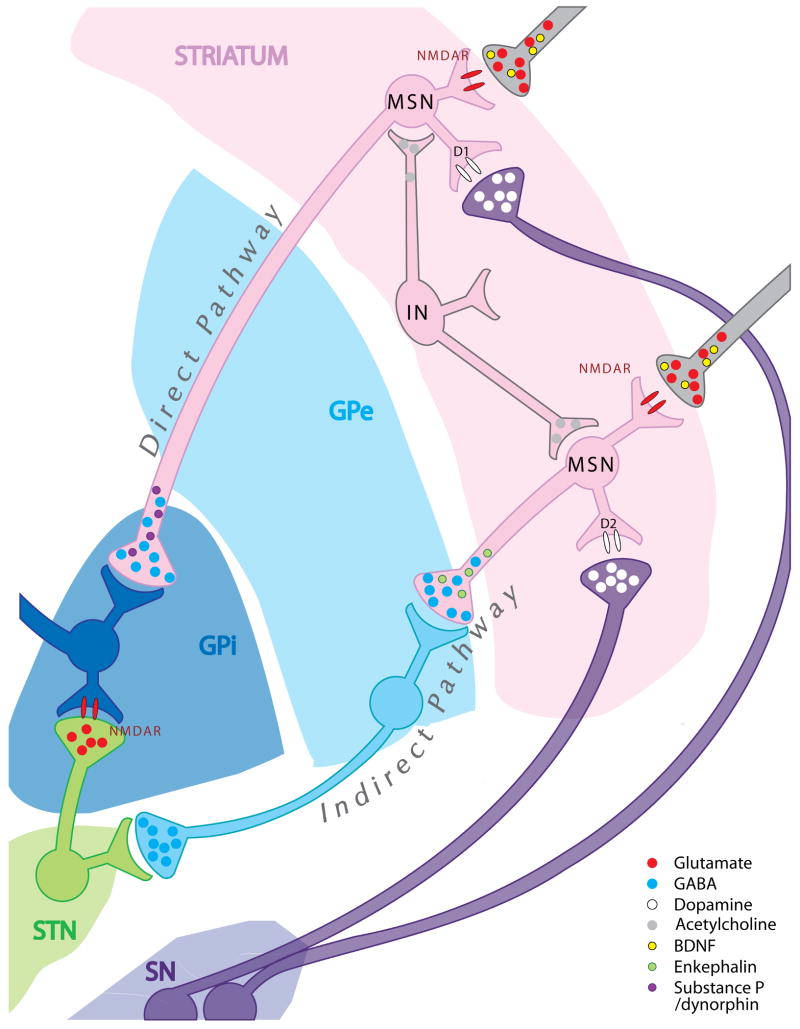
Figure 2. Cell type-specific characteristics of neuronal populations affected in HD.
Cumulative pathological and experimental evidence indicates that cell type-specific characteristics modulate the vulnerability of specific neuronal populations to mutant huntingtin expression. Striatal medium spiny neurons (MSN) and cortical neurons affected in HD project axons to anatomically distant target structures outside the striatum and the cortex, respectively. In contrast, striatal and cortical interneurons (IN) that are largely spared in HD bear short axons that remain within the boundaries of these brain structures. Differences in biochemical content may also contribute to the differential vulnerability of these neuronal populations, but the underlying pathogenic mechanisms remain unknown. MSNs are GABA-ergic projection neurons (GABA, blue circles) and receive brain-derived neurotrophic factor (BDNF, yellow circles) from cortical glutamatergic (Glutamate, red circles), as well as dopaminergic afferents (Dopamine, white circles) from the substantia nigra (SN). Interestingly, more vulnerable MSNs in the “indirect pathway” express enkephalin peptides (Enkephalin, green circles) and D2 dopamine receptors (white ovals, bottom), while less vulnerable MSNs in the “direct pathway” express substance P/dynorphin (purple circles) and D1 receptors (white ovals, top). Abbreviations: MSN: medium spiny neuron; IN: interneuron; BDNF: brain-derived neurotrophic factor; D1: dopamine receptor subtype 1; D2: dopamine receptor subtype 2; NMDAR: N-methyl-D-aspartic acid receptor; GABA: γ-aminobutyric acid; GPe: external globus pallidus; GPi: internal globus pallidus; SN: substantia nigra; STN: subthalamic nucleus.
Albeit less pronounced than in the striatum, differential vulnerability and loss of selected neuronal populations is also readily observed in the cerebral cortex of HD patients. Specifically, large pyramidal projection neurons in cortical layers V, VI and to a lesser extent, layer III, are preferentially lost (Cudkowicz & Kowall 1990, Hedreen et al. 1991). Long axons emanating from cortical projection neurons in layers V and VI innervate the striatum. In primates, these axons are thin and unbranched with a single target (Parent & Parent 2006). As in the striatum, there is remarkable preservation of small cortical interneurons (i.e., layer IV granule cells) in HD (Cudkowicz & Kowall 1990, Sotrel et al. 1991).
Vulnerability of different neuronal cell populations to Htt mutations: Selective or differential?
The earlier and more pronounced degeneration of specific neuronal populations within the striatum and cortex observed in HD prompted the phrase “selective neuronal vulnerability” to be used by a number of investigators (Sieradzan & Mann 2001, Perez-Navarro et al. 2006, Cowan & Raymond 2006). However, the unique cellular topography of HD pathology does not mean that the toxic effects of polyQ-Htt are limited to selected neuronal populations. Supporting this idea, a large body of pathological and experimental evidence demonstrates that polyQ-Htt expression can elicit toxic effects in additional neuronal cell types and even in some nonneuronal cells. Pathological examination in advanced HD patients revealed degeneration of neurons in other brain structures including the hippocampus, the angular gyrus in the parietal lobe, and the lateral tuberal nuclei of the hypothalamus (Lange et al. 1976, Vonsattel et al. 1985b, Oyanagi et al. 1989, Heinsen et al. 1999). Further, pronounced neuronal loss has been observed in the cerebellum of patients with juvenile HD onset (Byers et al. 1973, Rodda 1981). Although some of these neuronal losses may reflect secondary neuronal damage due to deafferentation, experiments involving tissue-specific overexpression of polyQ-Htt in vivo demonstrated that polyQ-Htt expression promotes functional abnormalities in many neurons not typically affected in HD (Senut et al. 2000, de Almeida et al. 2002). Additionally, nonneuronal cells appear affected in various peripheral tissues of HD patients and animal HD models (reviewed in (Sassone et al. 2009)). Finally, a plethora of studies documented functional abnormalities and decreased cell survival associated with the expression of pathogenic polyQ-Htt constructs across a range of cultured cell types including mouse (Ye et al. 2008, Lunkes & Mandel 1998) and human (Carmichael et al. 2002) neuroblastoma cells, PC12 cells (Li et al. 1999b), and kidney cells (COS-7) (Carmichael et al. 2002), among many others. Taken together, these observations indicate that the toxic effects of polyQ-Htt do not selectively affect specific neuronal populations. Instead, cell type-specific features might differentially render specific neuronal populations increasingly vulnerable to polyQ-Htt-induced toxicity.
Neurons affected in HD follow a “dying-back” pattern of degeneration
A discussion of cellular features modulating polyQ-Htt toxicity requires a revision of the sequence of pathogenic events in affected neurons. Historically, our understanding of the neurodegenerative process in HD was limited to the post mortem study of brain tissue harvested from HD patients. The marked loss of neurons observed in these tissues logically focused research efforts into cell death-related mechanisms (Vila & Przedborski 2003, Portera-Cailliau et al. 1995), and the development of HD animal models based on acute intoxication and induction of cell death in the striatum. More recently, the development of various rodent HD models revealed important information on earlier pathogenic events (Menalled 2005). These animal models accurately reproduce the autosomal dominant pattern of inheritance as well as the major pathological characteristics of HD, including formation of Htt aggregates (see below), the development of motor and behavioral symptoms, and the differential vulnerability of discrete neuronal populations [reviewed in (Ramaswamy et al. 2007b)]. Variations in disease onset and severity among HD models can be attributed to differences in length of the polyQ tract, promoters driving transgene expression, and size of the exogenous Htt transgene introduced (Ramaswamy et al. 2007b, Vonsattel 2008, Menalled 2005). However, a common theme emerged from detailed pathological, electrophysiological and behavioral analysis of these animals. All models analyzed thus far have shown various degrees of behavioral and motor abnormalities well before apparent neuronal degeneration (Levine et al. 2004, Tobin & Signer 2000, Menalled 2005). For example, behavioral defects reminiscent of the HD human phenotype (i.e., increased motor activity, gait abnormalities, and altered stride length) were observed in HD transgenic mice models expressing truncated polyQ-Htt constructs, including R6/2 mice (Lione et al. 1999, Luesse et al. 2001, Murphy et al. 2000), N171-82Q mice (Klivenyi et al. 2006), and a transgenic rat model expressing a truncated Htt with 51 CAG repeats (von Horsten et al. 2003). Moreover, similar defects have been reported in various knock-in HD animal models expressing polyQ-Htt at endogenous levels in its appropriate genomic context (Menalled 2005). Remarkably, behavioral abnormalities in knock-in HD animal models were detected in the absence of neuronal loss (Menalled 2005) and prior to formation of polyQ-Htt aggregates, suggesting that aggregate formation may not be required for functional changes to occur in affected neurons. The relevance of these observations to HD is highlighted by functional MRI imaging studies revealing early changes in neuronal function in presymptomatic HD patients (Reading et al. 2004), and studies suggesting that the early manifestation of chorea does not result from neuronal cell loss (Rosenblatt et al. 2003).
Providing a structural basis for these early alterations in neuronal function, histopathological studies documented a marked reduction in the number of axonal fibers and synaptic proteins early in the course of HD (DiProspero et al. 2004b, Reiner et al. 1988, Levine et al. 2004, Li et al. 2001). These alterations in axonal connectivity and synaptic function were consistent with the progressive electrophysiological disturbances reported in association with polyQ-Htt expression (Klapstein et al. 2001, Bibb et al. 2000, Cepeda et al. 2001, Laforet et al. 2001). Diffusion tensor imaging studies further demonstrated early signs of axonal degeneration in white matter of living, presymptomatic HD patients (Weaver et al. 2009, Rosas et al. 2009). Together, these observations suggested the existence of critical pathogenic events affecting neuronal functionality prior to cell death in HD. The accumulated evidence indicates that alterations in neuronal connectivity play a major role in HD pathology, and that neurons affected in HD follow a “dying back” pattern of degeneration (Fig. 3) (Li et al. 2001, Ferrante et al. 1991, Morfini et al. 2005, Wade et al. 2008). This emerging concept has far reaching implications regarding experimental therapeutic strategies. For instance, trophic factor therapies, which have been proposed for clinical testing (Ramaswamy et al. 2007a), have primarily focused on preserving striatal perikarya. However, few of these studies (Emerich et al. 1997) have employed tools to demonstrate that basal ganglia circuitry, including the essential aspect of sustained innervation and appropriate connectivity, was maintained following trophic factor delivery.
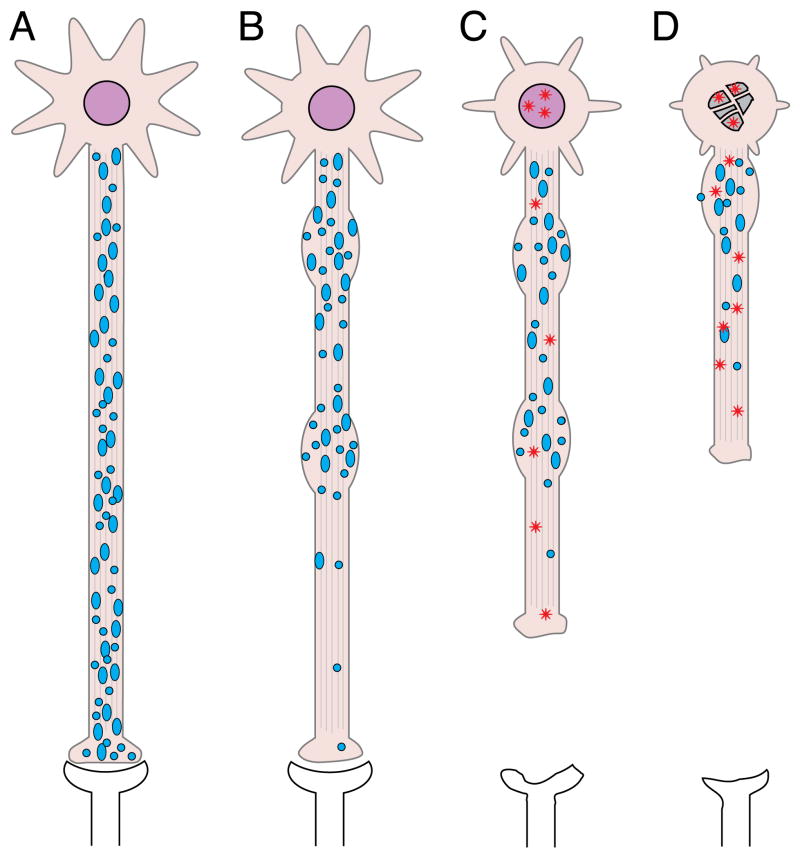
Figure 3. “Dying back” pattern of neuronal degeneration in HD.
A) Neurons undergo normal development, retaining normal connectivity and functionality prior to disease state. B) Affected neurons begin to exhibit signs of synaptic and axonal alterations early in the disease process, including abnormalities in the phosphorylation of axonal proteins, abnormal accumulation of membrane-bounded organelles (blue circles) in axons (Davies & Scherzinger 1997, DiFiglia et al. 1997a, Sapp et al. 1999, Davies et al. 1997), and loss of synaptic proteins (DiProspero et al. 2004b). These changes correspond to functional impairments in synaptic function that appear very early on, even in presymptomatic stages (Cepeda et al. 2003, Levine et al. 2004). C) Axonal degeneration steadily advances in a retrograde fashion, and nuclear/neuritic Htt aggregates (red stars) become evident. As HD progresses, dysfunction of striatal and corticostriatal projection neurons manifest in clinical symptoms such as motor deficits and cognitive decline long before evidence of cell death (Mizuno et al. 2000). D) Disruption of functional synaptic connectivity and eventual loss of appropriate trophic support (Zuccato & Cattaneo 2007) ultimately result in cell death, likely by apoptosis-related mechanisms (Vis et al. 2005).
Cellular factors influencing mutant huntingtin-induced toxicity
Projection neurons in the striatum and the cerebral cortex are major regions of the HD pathology, whereas interneurons within these structures largely survive the neurodegenerative process (Albin et al. 1992b, Reiner et al. 1988, Richfield et al. 1995a). In addition, MSNs in the direct pathway are affected earlier and to a larger extent than MSNs in the indirect pathway. Differences in biochemical content, morphology and connectivity among these neuronal cell types could provide clues towards an explanation of their differential vulnerabilities in HD. In the case of striatal neurons, one or more unique characteristics of MSNs could exacerbate the toxic effects of polyQ-Htt in these cells, rendering an otherwise “manageable” insult into a deadly one. Alternately, a unique set of protective characteristics might confer upon striatal interneurons an advantage to deal with the toxic effects of polyQ-Htt. Similar speculations can apply to projection neurons and interneurons in the cortex, but mechanisms underlying the differential vulnerability of cortical neurons are rarely addressed (Cepeda et al. 2007, Levine et al. 2004).
Because a molecular basis for the toxic gain of function associated with polyQ-Htt has been unclear, the contribution of cell type-specific traits in the modulation of polyQ-Htt-induced toxicity can only be speculative. Below, we discuss the potential contributions of various cell type-specific factors to the differential vulnerability of neurons characteristic of HD (Table I). These factors are discussed within the context of several major pathogenic mechanisms proposed for HD.
Table I. Differential vulnerability of specific cell populations in HD and its relationship to morphological and biochemical characteristics.
Different subpopulations of neurons in the striatum and cerebral cortex display varying degrees of vulnerability (indicated by a “+” sign) in HD. No common biochemical characteristic have been found among the main neuronal populations affected in HD. However, an analysis of morphological characteristics suggests increased vulnerability of neurons bearing longer and more prominent axons. Accordingly, MSNs and cortical projection neurons represent the main neuronal cell types affected in this disease, while interneurons are largely spared. Other cell type-specific properties including afferents, targets, and biochemical content (i.e., neurotransmitter receptors, peptides, etc.) could further modulate mutant huntington toxicity. Accordingly, several biochemical features characterize more vulnerable MSNs in the “indirect” pathways from those in the “direct” pathway, which are affecter later in the disease.
| Anatomical location |
Cell type | Relative vulnerability |
Morphology | Afferents | Target | NT receptors | NT | Peptides | Other molecular markers |
|---|---|---|---|---|---|---|---|---|---|
| Striatum | MSN (direct pathway) | + + + | projection neuron, long axon | Cortex (Glu), SNc (DA), Thalamus (Glu) | GPi, SNr | D1, NMDA, AMPA | GABA | Substance P/Dynorphin | DARPP-32, GAD |
| MSN (indirect pathway) | + + + + + | projection neuron, long axon | Cortex (Glu), SNc (DA), Thalamus (Glu) | GPe | D2, A2a, NMDA, AMPA | GABA | Enkephalin | DARPP-32, GAD | |
| Interneurons | + | extensive dendritic network, axon projects locally | MSNs, other interneurons | MSNs, other interneurons | D2, NMDA, AMPA | Ach | neuropeptide Y, parvalbumin | iNOS, somatostatin | |
| Cerebral Cortex | Pyramidal neurons (layers V/VI) | + + + | projection neuron, long axon | Thalamus, brainstem nuclei | Striatum, brainstem, thalamus | Glu, ACh, DA, NE, 5HT | Glu | - | MAP2, CaMK |
| Interneurons | + | extensive dendritic network, axon projects locally | Thalamus | Pyramidal neurons | Glu, GABA | GABA | Somatostatin, neuropeptide Y | GAD |
Htt distribution, expression levels, and somatic instability
In some familial forms of human diseases, the increased vulnerability of cell types affected can be explained by differential expression of the pathogenic gene product. Following the discovery of the Htt gene, multiple studies examined whether heterogeneities in Htt mRNA and/or protein expression could underlie the increased vulnerability of MSNs and cortical neurons in HD. However, extensive mRNA and protein expression analyses indicated that Htt is expressed in nearly all tissues (Trottier et al. 1995, Li et al. 1993, Fusco et al. 1999). Moreover, Htt expression levels were comparable in normal and HD patients (Bhide et al. 1996, Landwehrmeyer et al. 1995a). Immunochemical studies showed widespread distribution of Htt protein throughout the brain (DiFiglia et al. 1995), with no evidence of increased Htt expression in brain regions most affected in HD (Gutekunst et al. 1995, Ide et al. 1995). In fact, detailed studies showed that striatal interneurons, largely unaffected in HD, express higher levels of Htt than MSNs (Bhide et al. 1996), suggesting that differences in mutant Htt expression levels do not account for the increased vulnerability of MSNs.
Recent studies in HD patients and animal HD models showed that the mutant CAG repeat tract in the IT15 gene coding for Htt can undergo both inter- and intra-generational variability in expansion size (Shelbourne et al. 2007, Ishiguro et al. 2001). Studies on end-stage human HD autopsy material indicated that approximately 10% of sampled striatal cells contained hyperexpansions of over 200 repeats (Kennedy et al. 2003). Further, age-dependent instability of Htt was observed in the striatum and the cerebral cortex of knock-in HD mice (Ishiguro et al. 2001, Kennedy & Shelbourne 2000). These findings led to the proposal that increased instability of Htt's CAG expansion might contribute to the marked vulnerability of striatal neurons in HD (Shelbourne et al. 2007). However, greater instability of CAG repeats in the striatum was also observed in other polyQ-expansion diseases displaying little or no striatal pathology (Lopes-Cendes et al. 1996, Watase et al. 2003). Moreover, observations from a bacterial artificial chromosome-based HD mouse model expressing polyQ-Htt with a stable CAA-CAG tract appeared inconsistent with a role of somatic repeat instability in HD pathogenesis (Gray et al. 2008). These observations argued against a direct causal relationship between somatic CAG expansion mosaicism and the increased vulnerability of striatal MSNs. Collectively, the evidence indicates that neither heterogeneities in Htt expression levels nor somatic instability of the Htt gene play a role in the differential vulnerability of striatal neurons affected in HD.
Mutant Htt aggregation
Following the discovery of the Htt gene, various antibodies mapping to different Htt epitopes were generated (Li & Li 1998). These antibodies revealed N-terminal fragments of polyQ-Htt that accumulate to form microscopically visible aggregates (or inclusions) in both the nucleus (DiFiglia et al. 1997b) and neurites (Li et al. 1999a) of some, but not all, neurons affected in HD patients (Kuemmerle et al. 1999, DiFiglia et al. 1997a). These aggregates are also found in most HD rodent models (Li & Li 2004, Perez-Navarro et al. 2006), and thus represent a major histopathological feature of HD.
Initial focus on polyQ-Htt aggregates as a pathogenic agent of HD was fueled by biochemical findings suggesting that elongated polyQ stretches form insoluble structures toxic to cells (Perutz 1996, Rubinsztein & Carmichael 2003). Cellular HD models based on overexpression of truncated polyQ-Htt constructs first proposed a positive correlation between the abundance of polyQ-Htt aggregates and cellular toxicity (Cooper et al. 1998, Hackam et al. 1998). However, Htt toxicity and aggregation were experimentally dissociated in both cellular (Arrasate et al. 2004) and animal (Perrin et al. 2009) HD models. At present, it remains unclear whether polyQ-Htt aggregates promote neuronal dysfunction and death, or whether polyQ-Htt aggregation represent the outcome of endogenous cellular mechanisms conferring protection against soluble, toxic polyQ-Htt species (Truant et al. 2008, Wang et al. 2008).
Despite the large body of research on polyQ-Htt aggregates, little evidence exists linking these structures to the preferential degeneration of MSNs and cortical neurons in HD. As observed in SBMA (La Spada & Taylor 2003), and other polyQ diseases (Michalik & Van 2003), a poor correlation was found between polyQ-Htt aggregation and neuronal vulnerability (Saudou et al. 1998). Moreover, the presence of polyQ-Htt aggregates in surviving neurons of advanced HD patients suggested these structures might even promote neuronal survival (Truant et al. 2008). Experiments in cellular (Arrasate et al. 2004) and animal (Perrin et al. 2009) HD models further supported this view. Finally, prominent polyQ-Htt aggregates have been reported in striatal interneurons (Kuemmerle et al. 1999), and in brain regions largely spared in HD such as the hippocampus (Becher et al. 1998, Gutekunst et al. 1999). The precise contribution of polyQ-Htt aggregation to HD pathogenesis remains unknown, but no definitive evidence exists linking the formation and abundance of polyQ-Htt aggregates to increased neuronal cell vulnerability or survival in HD.
Abnormalities in BDNF signaling
Mechanisms underlying neural degeneration in HD are unknown, but pathology has been linked to alterations in trophic factor function, specifically the function of brain-derived neurotrophic factor (BDNF) (reviewed in (Zuccato & Cattaneo 2007)). BDNF is normally synthesized by neurons in the cortex and the SN, and subsequently transported along axons to the striatum (Baquet et al. 2004). Results from experiments in various knockout mouse models suggested that BDNF supports the survival of MSNs, leading to the proposal that abnormalities in BDNF signaling could contribute to the reduced survival of striatal neurons in HD [reviewed in (Zuccato & Cattaneo 2007)]. Supporting this idea, reductions in BDNF levels were demonstrated in a number of cellular and animal HD models, as well as in HD patients (Zuccato & Cattaneo 2007). Further, genetic studies indicate that reductions in BDNF levels selectively exacerbate polyQ-Htt-induced degeneration of enkephalinergic striatal projection neurons in the indirect pathway (Canals et al. 2004).
Functional studies indicated WT-Htt promotes BDNF expression in both cortex and striatum (Zuccato et al. 2001, Canals et al. 2001). Interestingly, this function of WT-Htt appears compromised by polyQ expansion, raising the possibility that reductions in striatal BDNF levels might result from polyQ-Htt-induced transcriptional changes (Zuccato et al. 2001, Canals et al. 2001). Alternatively, reductions in striatal BDNF could result from polyQ-Htt-induced reductions in anterograde axonal transport in cortical neurons (Gauthier et al. 2004b, Szebenyi et al. 2003a, Trushina et al. 2004), decreased BDNF endocytosis by MSNs (Her & Goldstein 2008), or both. Mechanisms underlying BDNF deficits remain a matter of debate.
Recent studies in a conditional knock out model indicated that total number of striatal neurons remained unaffected by BDNF depletion, challenging a long-standing assumption that BDNF serves as a critical survival factor for these neurons (Rauskolb et al. 2010). Instead, results from studies in these mice suggest BDNF is required for dendritic growth of striatal neurons (Rauskolb et al. 2010). The degree to which defects in BDNF signaling contribute to the degeneration of striatal and cortical projection neurons in HD remains unknown, but deserves further study.
Mitochondrial Alterations
Mitochondria are the chief cellular producers of chemical energy and major regulators of metabolism, as well as crucial modulators of intracellular signaling and survival (reviewed in (McBride et al. 2006)). Observations of cellular metabolism disturbances in HD patients suggested that mitochondrial dysfunction might represent a critical pathogenic component (Sanchez-Pernaute et al. 1999, Jenkins et al. 1993). Post-mortem studies on HD brain tissue further supported this idea, showing decreased activity in complex II, III, and IV of the mitochondrial respiratory chain (Gu et al. 1996)s. Experimental evidence using cultured neurons (Seong et al. 2005) and HD animal models (Browne et al. 1997) showed deficits in mitochondrial respiration in association with polyQ-Htt expression. Consistent with these observations, various mitochondrial inhibitors generate striatal pathology and phenotypes reminiscent of HD when administered systemically (Browne 2008). For example, accidental ingestion of the irreversible mitochondrial inhibitor 3-nitropropionic acid (3-NP) caused chorea, dystonia, and basal ganglia degeneration in humans (Ludolph et al. 1991). Also, systemic administration of 3-NP in rats (Beal et al. 1993) and primates (Palfi et al. 1996) caused striatal cell loss, inducing the accompanying movement deficits. However, unbiased gene expression analysis comparing HD and 3-NP changes in energy metabolism found significant differences and indicated that HD effects on metabolism were extramitochondrial (Lee et al. 2007). Further, any treatment compromising the connectivity and survival of neurons in the striatal pathway would produce similar clinical symptoms. Significantly, functional experiments indicated that activation of the cJun Amino Terminal kinase (JNK) pathway represented an essential step in 3-NP-induced striatal degeneration (Garcia et al. 2002), but mechanisms linking reductions in mitochondrial function and JNK activation in HD remain elusive.
Several mechanisms have been proposed linking polyQ-Htt to mitochondrial dysfunction in HD (Grunewald & Beal 1999), including polyQ-Htt-induced reductions in ATP generation (Seong et al. 2005), Ca2+ buffering (Reddy et al. 2009, Panov et al. 2002), and mitochondrial trafficking (Li et al. 2010, Reddy et al. 2009, Orr et al. 2008). While tissue-specific patterns of mitochondrial protein composition have been described (Mootha et al. 2003), no evidence exists showing that polyQ-Htt differentially affects mitochondria from specific cell types, making unclear how polyQ-Htt-induced mitochondrial dysfunction might differentially affect selected neuronal populations. Alternatively, it has been proposed that MSNs are uniquely susceptible to defects in mitochondria-dependent energy production (Beal 1992). Unlike other neurons in the basal ganglia, MSNs display unusually low levels of spontaneous discharge (Mitchell et al. 1999). This distinctive electrophysiological characteristic of MSNs could place a higher energy demand on these neurons, because a large portion of their ATP would presumably be used for the maintenance of a hyperpolarized state (Calabresi et al. 1995). The susceptibility of striatal neurons to mitochondrial inhibitors would therefore reflect unique energy requirements by these cells (Lee et al. 2007). However, disruption of mitochondrial function does not typically produce HD-like pathology. For example, systemic application of 3-NP induced neuronal cell pathology in the hippocampus and the thalamus, structures which are normally affected only at late HD stages (Bossi et al. 1993). Further, different mouse strains display differential vulnerability to 3-NP administration (Alexi et al. 1998), and different patterns of neural degeneration have been described for different mitochondrial inhibitors (McLin et al. 2006). Most studies of mitochondrial pathology in HD look at advanced stages of the disease, when mitochondrial-based cell death pathways may be activated as a secondary response to primary pathogenic mechanisms (Bredesen et al. 2006, Jellinger 2006). Such observations indicate that much remains to be learned regarding molecular factors that underlie the selective vulnerability of striatal neurons secondary to mitochondrial changes.
Neurochemical content
Based on their neurochemical content, MSNs can be classified either as part of the “striosomes” (also referred to as “patches”) or as part of the “extrastriasome” (also known as “matrix”) compartments (Graybiel 1990, Gerfen 1992). Projection MSNs represent the main cell type present in striosomes, while striatal interneurons are the dominant cell type at the matrix compartment. According to this classification, MSNs in the striosomal compartment are among the first to degenerate in HD (Hedreen & Folstein 1995). Various biochemical markers have been identified showing differential expression between the striosomal and matrix compartments, including neuropeptides and neurotransmitter receptors (Table I).
Neuropeptide and calcium-binding proteins
Besides expressing the neurotransmitter GABA, striosomal MSNs express higher levels of various neuropeptides including ENK, SP, dynorphin (DYN, a class of opioid peptides derived from the precursor protein prodynorphin) and neurotensin (NT, a 13 amino acid neuropeptide), among others (Holt et al. 1997, Angulo & McEwen 1994). Interestingly, the expression of specific neuropeptides is associated with increased vulnerability of MSN subtypes. As mentioned above, MSN neurons in the “direct pathway” express DYN and/or SP, whereas MSNs in the “indirect pathway” express ENK (Gerfen & Young 1988). However, it remains unclear whether expression of DYN and/or SP exacerbates polyQ-Htt toxicity, or whether ENK expression might provide a protective effect. Similarly, interneurons expressing nitric oxide synthase (NOS), somatostatin and neuropeptide Y (Ferrante et al. 1985) are particularly resistant in HD, but mechanisms making these proteins protective remain elusive and thus such associations are correlative.
A positive association has been observed between the level of expression of some calcium-binding proteins (i.e., calbindin, parvalbumin and caretinin) and the survival of interneurons in HD (Gerfen et al. 1985). Such associations led to the proposal that higher levels of calcium-binding proteins in striatal interneurons may protect these cells from glutamate-mediated excitotoxic mechanisms (Mitchell & Griffiths 2003). However, these proteins are not exclusively distributed in the striasomal compartment (Gerfen et al. 1985), and a small fraction of calbindin or parvalbumin positive interneurons degenerate in HD (Mitchell et al. 1999). Conversely, decreased expression of hippocalcin and other calcium sensor proteins in MSNs was proposed to contribute to the increased vulnerability of these cells in HD (Luthi-Carter et al. 2000). However, functional experiments based on lentivirus-mediated hippocalcin expression yielded results in direct contradiction with this latter hypothesis (Rudinskiy et al. 2009).
Glutamate-related factors
Glutamate is the most abundant excitatory neurotransmitter in mammalian brain, activating both N-methyl-D-aspartate (NMDA) and non-NMDA ionotropic glutamate receptors (i.e., AMPA and kainate receptors) (Gasic & Hollmann 1992). Abnormally sustained stimulation of NMDA receptors by glutamate can lead to prolonged increases in intracellular calcium, triggering various intracellular events including activation of kinases and phosphatases, calcium-dependent proteases, synthesis of nitric oxide synthase (NOS), generation of reactive oxygen species, mitochondrial dysfunction, and activation of apoptotic pathways (reviewed in (Aarts & Tymianski 2004)). Relevant to the subject of this review, both striatal MSNs and cortical neurons receive a rich supply of excitatory glutamatergic inputs (Bouyer et al. 1984). Striatal MSNs in particular are constantly stimulated by these inputs, remaining hyperpolarized much of the time (Wilson & Kawaguchi 1996). These observations led to suggestions that increased exposure of MSNs to cortical glutamate stimulation could render these cells more vulnerable to excitotoxic damage. This idea provided the rationale for the use of NMDAR antagonists in HD (Kieburtz et al. 1996, Bonelli & Hofmann 2004, Handley et al. 2006), but these treatments had no beneficial effects in HD patients (Handley et al. 2006).
Experimental evidence using intrastriatal injection of agonists for NMDA (e.g., quinolinic acid) and non-NMDA (e.g., kainic acid) receptors in rodent and non-primate animals demonstrated greater vulnerability of MSNs to glutamate-induced excitotoxicity, compared to striatal interneurons (McGeer & McGeer 1976, Coyle & Schwarcz 1976, Emerich et al. 1996). However, mechanisms linking polyQ-Htt expression to the increased vulnerability of these neuronal subtypes to glutamate toxicity have not yet been established. Despite this, various HD models have been proposed based on the administration of glutamate receptor agonists (McGeer & McGeer 1976, Coyle & Schwarcz 1976). While these models generally resemble the HD phenotype of striatal dysfunction, their time window for inducing cell death contrasts sharply with the dying-back pattern of degeneration observed in HD (Fig. 3). Further, some studies showed that the pattern of striatal neuron degeneration resulting from systemic quinolonic acid injection differs significantly from that of HD, suggesting different pathogenic mechanisms (Figueredo-Cardenas et al. 1994). Relevant HD animal models need to replicate the sequence of changes in neuronal connections and neuronal populations seen in the disease, including the “dying back” pattern of neuronal degeneration. Since the neurological symptoms in HD reflect loss of functional connections made by affected neurons, any treatment that disrupted these synaptic relationships would be expected to exhibit similar clinical symptoms, without necessarily involving the pathogenic mechanisms initiated by polyQ-Htt.
A molecular basis underlying the differential vulnerability of striatal neurons to glutamate excitotoxicity is currently unknown, but reductions in levels of calcium-binding proteins, heterogeneous expression of NMDA receptor subtypes, and abnormalities in glial function have all been proposed as contributing factors (reviewed in (Mitchell et al. 1999)).
NMDA glutamate receptors exist as heteromeric dimers of NR1 and NR2 (A, B, C and D) subunits (Rigby et al. 1996). In the brain, levels of NR1 expression greatly exceeds that of the other subunits combined, whereas NR2A, B, C and D subunits varied widely (Goebel & Poosch 1999). NR1 subunits are essential for NMDA receptor function, but heteromerization with NR2 subunits increases both the permeability of the channel (over 100 fold), and its deactivation time (Schoepfer et al. 1994). Significantly, differential expression of NMDA subunits have been observed among striatal neurons (Cicchetti et al. 2000), with MSN projection neurons reportedly expressing higher levels of the NR2B subunit, and striatal interneurons predominantly expressing NR2D (Landwehrmeyer et al. 1995b, Kuppenbender et al. 2000). While differences in NMDA receptor subtype expression could help explain the increased vulnerability of MSNs over striatal interneurons to NMDA agonists, these do not explain the minimal HD pathology observed in the hippocampus and olfactory bulb, two other CNS regions showing higher levels of NR2B expression than the striatum (Goebel & Poosch 1999, Laurie et al. 1997). Recently, experiments involving selective expression of polyQ-Htt in astrocytes suggested that reduced expression of glutamate transporter in these cells might contribute to increases in extracellular glutamate levels (Bradford et al. 2010). Currently, no consensus exists on mechanisms by which polyQ-Htt expression might promote imbalances in glutamate signaling (Mitchell et al. 1999, Sieradzan & Mann 2001, Estrada Sanchez et al. 2008), nor is clear how the contribution of glutamate signaling relates to the early axonal and synaptic degeneration phenotype of HD.
Dopamine signaling
Like glutamate, dopamine (DA) is a key neurotransmitter in several CNS circuits, and the striatum is the brain region most heavily innervated by dopaminergic afferents. These fibers originate from the SN (Albin et al. 1989) (Figs. 1-2). Interestingly, the striatal DA content exhibits a dorsal to ventral gradient (Cass 1997) that is consistent with the progression of pathology in HD (Vonsattel & DiFiglia 1998). Based on these observations, and findings of DA signaling deregulation in HD models and patients (Tang et al. 2007), a role of DA in the increased vulnerability of MSNs has been proposed (Jakel & Maragos 2000). DA in the striatonigral circuit exerts excitatory signals by activation of D1 receptors, and inhibitory signals by activation of D2 receptors (Albin et al. 1989) (Fig. 2). MSNs of the “indirect pathway” express high levels of D2, in contrast with the MSNs in the “direct pathway”, which express high levels of D1 (Gerfen et al. 1990), suggesting that differences in DA-induced signaling could modulate the toxic action of polyQ-Htt. Consistent with this idea, activation of the JNK pathway induced by polyQ-Htt (Phelan et al. 2001, Apostol et al. 2006, Morfini et al. 2009b) was reportedly exacerbated by D2 receptor activation (Luo et al. 1998, Charvin et al. 2005). However, both direct and indirect pathways are eventually affected in HD. Again, the loss of synaptic connections early in the disease make it difficult to determine whether altered dopamine signaling represents a primary or secondary element in HD pathogenesis.
The cumulative evidence indicates that dopamine and perhaps other neurotransmitters might modulate the toxic effects of polyQ-Htt and thus may represent factors contributing to the differential vulnerabilities of striatal neurons in HD. While most studies have focused in altered calcium homeostasis and induction of apoptosis as a consequence of abnormal neurotransmitter signaling (Tang et al. 2007, Sieradzan & Mann 2001), little emphasis has been placed on well-established events downstream of neurotransmitter receptor activation, including the modulation of kinase-dependent signaling pathways (Luo et al. 1998, Zhen et al. 1998). Indeed, specific neurotransmitters and neuropeptides might act synergistically with polyQ-Htt-induced pathogenic kinase-mediated pathways (i.e., JNK activation) to increase the vulnerability of specific neuronal populations (Morfini et al. 2005). Alternatively, some neurotransmitters could promote activation of kinase pathways (e.g., the Akt pathway) that counteracting toxic ones elicited by polyQ-Htt (Humbert et al. 2002, Kim et al. 2002). Accordingly, cell type-specific changes in kinase activity and patterns of protein phosphorylation have been found in association with drugs that target the striatum (Bateup et al. 2008). Additional studies are needed to illuminate molecular mechanisms by which specific neurotransmitters might modulate polyQ-Htt-induced toxicity. Such studies will help determine the precise contribution of each neurotransmitter system to the differential vulnerability of neuronal subtypes in HD.
Alterations in Gene Expression
A plethora of gene studies in transgenic HD models and HD human tissue initially described polyQ-Htt-induced transcriptional alterations (reviewed in (Thomas 2006, Cha 2007)). Major advances in molecular biological and computer technology allowed for the screening of large numbers of candidate genes in an unbiased fashion. Specifically, microarray studies screened HD-related changes in the expression levels of thousands of RNAs encoding molecular components involved in various cellular processes including calcium homeostasis, intracellular signaling, energy metabolism and the transcriptional machinery (Thomas 2006, Cha 2007). Changes in the expression levels of mRNAs encoding neurotransmitters and their receptors, synaptic proteins, energy metabolism and mediators of intracellular signaling have all been described (reviewed in (Cha 2007)). However, data obtained thus far have not provided an explanation for the differential cellular vulnerability of HD. Further, it is unclear whether changes in gene expression identified thus far play a direct role on HD pathogenesis or represent a secondary response to primary pathogenic events. Changes in the levels of specific genes could result from compensatory mechanisms, making difficult to establish the relevance of such changes to HD pathogenesis. Additionally, most gene expression studies screened for transcriptional changes in the striatum or cortex, but few established quantitative comparisons between the most affected MSNs or pyramidal neurons and mildly affected or unaffected cells such as striatal and cortical interneurons. No signature pattern of gene expression changes has been defined for neurons affected in HD. Currently, there is no direct evidence linking changes in the transcription of specific genes to the differential vulnerability of neuronal populations in HD.
Deficits in axonal transport
Although no common biochemical features have been found among the most vulnerable neuronal cell types in HD, an analysis of their morphological characteristics does reveal a common theme. Specifically, neurons affected in HD within the striatum and the cortex are principally projection neurons (Cicchetti et al. 2000) (Table I). Indeed, MSNs and cortical neurons affected in HD project axons to anatomically distant target structures outside the striatum and the cortex, respectively (Fig. 2). In contrast, striatal and cortical interneurons that are mainly spared in HD bear short axons that remain within the boundaries of these brain structures. This morphological and functional difference has been proposed to play a role in the differential vulnerability of neurons observed in HD (Cicchetti et al. 2000), as well as other neurodegenerative diseases (Mattson & Magnus 2006, Morfini et al. 2009a, Morfini et al. 2009b). The lack of protein synthesis machinery in axons and the enormous distances separating the cell body from the axonal and synaptic domains impose a unique set of challenges to neuronal cells. One such challenge includes the transport and delivery of proteins and lipid components along axons, a process collectively referred to as axonal transport (AT) [reviewed in (Morfini et al. 2006b, Morfini et al. 2005)]. In mature neurons, AT of membrane-bounded organelles (MBOs, including mitochondria, synaptic vesicles, and plasma membrane components) from their sites of synthesis in the neuronal cell body to their final destination in axons is mainly executed by the microtubule-based motor protein conventional kinesin. The multi-subunit motor cytoplasmic dynein on the other hand, translocates signaling endosomes, multivesicular bodies, and lysosomes from axons back to the neuronal cell body [reviewed in (Morfini et al. 2006b)]. The crucial dependence of neuronal function and axonal maintenance on AT is highlighted by observations linking loss of function mutations in these molecular motors above to various neurological diseases (Reid et al. 2002, Hafezparast et al. 2003, Roy et al. 2005). Significantly, pathological studies revealed a “dying-back” pattern of neuronal degeneration in these diseases, suggesting that alterations in AT might also represent a critical pathogenic event in HD (Roy et al. 2005, Morfini et al. 2005, Morfini et al. 2009a).
Multiple independent studies provided evidence of AT deficits in HD. Ultrastructural observations first showed reduced number of synaptic vesicles, and abnormal MBO profiles within axons of affected neurons (Li 1999, Li et al. 2001, Li & Li 2004). Subsequently, reports from various experimental models demonstrated that inhibition of AT represents an important pathogenic event in HD (Gunawardena et al. 2003, Szebenyi et al. 2003b, Lee et al. 2004, Sinadinos et al. 2009, Morfini et al. 2009b) and other polyQ-expansion diseases (Morfini et al. 2006a). Studies in isolated squid axoplasm first showed that polyQ-Htt inhibits AT in a manner independent of alterations in the neuronal cell body (Szebenyi et al. 2003b, Morfini et al. 2009b). Subsequent experiments in cultured cells (Gauthier et al. 2004a, Her & Goldstein 2008) and Drosophila neurons (Sinadinos et al. 2009, Lee et al. 2004) further documented reductions in AT and accumulation of axonal vesicle cargos in association with polyQ-Htt expression. These observations, the absolute reliance of neurons on AT, and the “dying-back” pattern of neuronal degeneration seen in HD all support the concept that AT deficits might represent a major pathogenic event underlying the increased vulnerability of projection neurons in this disease (Morfini et al. 2009b).
Reductions in AT observed in various HD experimental models raised the question of how polyQ-Htt inhibits AT. An explanation to this question appears to be that polyQ-Htt inhibits AT by promoting alterations in the activity of kinases involved in the phosphorylation of molecular motor proteins (Morfini et al. 2005, Morfini et al. 2009b). Consistent with this notion, aberrant patterns of proteins phosphorylation including neurofilaments (Nihei & Kowall 1992, DiProspero et al. 2004a) and synapsin (Lievens et al. 2002), as well as increased activation of kinases all represent well-established HD features (Apostol et al. 2006, Garcia et al. 2004, Liu 1998). Expression of polyglutamine-expanded huntingtin activates the SEK1-JNK pathway and induces apoptosis in a hippocampal neuronal cell line (Liu 1998). Accordingly, studies in isolated squid axoplasm and a knock-in mouse model further showed polyQ-Htt inhibits AT through a mechanism involving activation of the JNK pathway and phosphorylation of the molecular motor protein conventional (Morfini et al. 2009b). Three JNK isoforms exist in mammals (JNK1, JNK2 and JNK3)(Brecht et al. 2005). Neuron-specific JNK3 (Yang et al. 1997), but not ubiquitously expressed JNK1, mediated the inhibitory effects of polyQ-Htt on AT, thus providing a partial explanation for the increased vulnerability of neurons to polyQ-Htt (Morfini et al. 2009b). Increased activation of JNK3 in HD would promote reductions in AT of various MBO cargoes, leading to deficits in synaptic and axonal function and maintenance. Increased activation of the JNK pathway in HD is also consistent with the changes in gene transcription and activation of apoptotic pathways widely reported in association with polyQ-Htt expression (Phelan et al. 2001, Merienne et al. 2003).
Deficits in AT appear to play a role in the degeneration of neurons observed in HD. Obviously, longer axons characteristic of projection neurons mean that larger cellular volumes and surface area must be maintained, placing greater demands on AT-related mechanism. However, why deficits in AT in HD would affect projection neurons in the striatum and the cortex to a greater extent than projection neurons in other brain structures remains to be established. Several possibilities exist. Among these, MSNs and cortical projection neurons affected in HD may have unique AT requirements or specializations that makes them more vulnerable to polyQ-Htt-induced toxicity. Consistent with this idea, cytotypic differences in AT have been documented in vivo for different neuronal populations (Oblinger et al. 1987). Genetic evidence also demonstrates cell type-specific AT specializations. For example, whereas some mutations in the molecular motor protein subunit dynactin primarily cause motor neuron disease (Puls et al. 2005, Puls et al. 2003), a different set of mutations in this protein result in Perry's syndrome, which lack motor neuron pathology and instead is characterized by degeneration of cortical and extrapyramidal neurons (Farrer et al. 2009). Alternatively, one or more components mediating the effects of polyQ-Htt on AT (i.e., JNK3) might be expressed at higher levels in neurons affected in HD. An exhaustive examination of JNK isoforms in the brain has not been done, but some reports suggest differential expression of each isoform in different areas (Brecht et al. 2005). A molecular basis underlying the increased vulnerability of MSNs and cortical projection neurons to polyQ-Htt-induced alterations in AT is currently unknown, but likely reflects unique AT specializations of these neuronal cell types (Oblinger et al. 1987, Morfini et al. 2001, Her & Goldstein 2008).
Conclusions
In HD, the ubiquitous pattern of polyQ-Htt expression contrasts sharply with the increased vulnerability of specific populations of neurons in the striatum and cerebral cortex. The consistent loss of these neuronal populations has been proposed to result from “selective” vulnerability of these cells to the toxic gain of function associated with polyQ expansion in Htt. However, multiple lines of evidence clearly indicate that the toxic effects of polyQ-Htt are not restricted to specific neuronal populations. Instead, the evidence suggests that cell type-specific features might render such populations increasingly vulnerable to polyQ-Htt-induced toxicity.
Several scenarios, not mutually exclusive, appear consistent with the unique cellular topography of neuronal degeneration in HD. One or more molecular components involved in polyQ-Htt toxicity could be enriched in affected cells, but the identity of such component(s) remains elusive. Alternatively, cell type-specific features could render some neurons more vulnerable to the toxic effects of polyQ-Htt. Such features would represent the “weakest link in the chain” and likely be associated with critical functional needs of affected cells. In this regard, it is noteworthy that thus far, the presence of long axons constitutes the only common characteristic observed among neuronal cell types affected in HD. Indeed, it is striatal and cortical projections neurons bearing axons that project to anatomically remote structures that mainly degenerate in HD, whereas interneurons bearing short axons are largely spared. Significantly, polyQ-Htt has been shown to induce decrements in axonal transport, a cellular process critical for the function and maintenance of axons. Moreover, degeneration of neurons with defective axonal transport follows a “dying back” pattern that is consistent with that of neurons affected in HD. At present, it is unclear why deficits in axonal transport induced by polyQ-Htt would affect projection neurons in the striatum and the cortex to a greater extent than projection neurons in other brain structures, but differences in the composition, amounts, and regulation of axonal transport have all been documented.
An understanding of the interactions between polyQ-Htt and cell type-specific characteristics should provide a novel conceptual framework for the development of effective therapeutic strategies in HD. Such knowledge could yield treatments that specifically address neuronal populations affected in this disease. Regardless, preventing loss of neuronal connectivity should represent a primary goal of any effective therapeutic strategy.
Acknowledgments
This work was supported by NIH (RNS066942A) and Huntington's Disease Society of America grants to G.M.; NIH (NS23868, NS23320, NS41170) grants to S.T.B.; and CHDI grants to J.K.
Bibliography
- Aarts MM, Tymianski M. Molecular mechanisms underlying specificity of excitotoxic signaling in neurons. Curr Mol Med. 2004;4:137–147. doi: 10.2174/1566524043479202. [DOI] [PubMed] [Google Scholar]
- Albin RL, Reiner A, Anderson KD, Dure LS, Handelin B, Balfour R, Whetsell WO, Jr, Penney JB, Young AB. Preferential loss of striato-external pallidal projection neurons in presymptomatic Huntington's disease. Ann Neurol. 1992a;31:425–430. doi: 10.1002/ana.410310412. [DOI] [PubMed] [Google Scholar]
- Albin RL, Reiner A, Anderson KD, Dure LSt, Handelin B, Balfour R, Whetsell WO, Jr, Penney JB, Young AB. Preferential loss of striato-external pallidal projection neurons in presymptomatic Huntington's disease. Ann Neurol. 1992b;31:425–430. doi: 10.1002/ana.410310412. [DOI] [PubMed] [Google Scholar]
- Albin RL, Young AB, Penney JB. The functional anatomy of basal ganglia disorders. Trends Neurosci. 1989;12:366–375. doi: 10.1016/0166-2236(89)90074-x. [DOI] [PubMed] [Google Scholar]
- Alexi T, Hughes PE, Knusel B, Tobin AJ. Metabolic compromise with systemic 3-nitropropionic acid produces striatal apoptosis in Sprague-Dawley rats but not in BALB/c ByJ mice. Exp Neurol. 1998;153:74–93. doi: 10.1006/exnr.1998.6842. [DOI] [PubMed] [Google Scholar]
- Andrew SE, Goldberg YP, Kremer B, et al. The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington's disease. Nat Genet. 1993;4:398–403. doi: 10.1038/ng0893-398. [DOI] [PubMed] [Google Scholar]
- Angulo JA, McEwen BS. Molecular aspects of neuropeptide regulation and function in the corpus striatum and nucleus accumbens. Brain Res Brain Res Rev. 1994;19:1–28. doi: 10.1016/0165-0173(94)90002-7. [DOI] [PubMed] [Google Scholar]
- Apostol BL, Illes K, Pallos J, et al. Mutant huntingtin alters MAPK signaling pathways in PC12 and striatal cells: ERK1/2 protects against mutant huntingtin-associated toxicity. Hum Mol Genet. 2006;15:273–285. doi: 10.1093/hmg/ddi443. [DOI] [PubMed] [Google Scholar]
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Baquet ZC, Gorski JA, Jones KR. Early striatal dendrite deficits followed by neuron loss with advanced age in the absence of anterograde cortical brain-derived neurotrophic factor. J Neurosci. 2004;24:4250–4258. doi: 10.1523/JNEUROSCI.3920-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbeau A, Duvoisin RC, Gerstenbrand F, Lakke JP, Marsden CD, Stern G. Classification of extrapyramidal disorders. Proposal for an international classification and glossary of terms. J Neurol Sci. 1981;51:311–327. doi: 10.1016/0022-510x(81)90109-x. [DOI] [PubMed] [Google Scholar]
- Bateup HS, Svenningsson P, Kuroiwa M, Gong S, Nishi A, Heintz N, Greengard P. Cell type-specific regulation of DARPP-32 phosphorylation by psychostimulant and antipsychotic drugs. Nat Neurosci. 2008;11:932–939. doi: 10.1038/nn.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal MF. Does impairment of energy metabolism result in excitotoxic neuronal death in neurodegenerative illnesses? Ann Neurol. 1992;31:119–130. doi: 10.1002/ana.410310202. [DOI] [PubMed] [Google Scholar]
- Beal MF, Brouillet E, Jenkins BG, et al. Neurochemical and histologic characterization of striatal excitotoxic lesions produced by the mitochondrial toxin 3-nitropropionic acid. J Neurosci. 1993;13:4181–4192. doi: 10.1523/JNEUROSCI.13-10-04181.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal MF, Ferrante RJ. Experimental therapeutics in transgenic mouse models of Huntington's disease. Nat Rev Neurosci. 2004;5:373–384. doi: 10.1038/nrn1386. [DOI] [PubMed] [Google Scholar]
- Becher MW, Kotzuk JA, Sharp AH, Davies SW, Bates GP, Price DL, Ross CA. Intranuclear neuronal inclusions in Huntington's disease and dentatorubral and pallidoluysian atrophy: correlation between the density of inclusions and IT15 CAG triplet repeat length. Neurobiol Dis. 1998;4:387–397. doi: 10.1006/nbdi.1998.0168. [DOI] [PubMed] [Google Scholar]
- Berardelli A, Noth J, Thompson PD, et al. Pathophysiology of chorea and bradykinesia in Huntington's disease. Mov Disord. 1999;14:398–403. doi: 10.1002/1531-8257(199905)14:3<398::aid-mds1003>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Bhide PG, Day M, Sapp E, et al. Expression of normal and mutant huntingtin in the developing brain. J Neurosci. 1996;16:5523–5535. doi: 10.1523/JNEUROSCI.16-17-05523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb JA, Yan Z, Svenningsson P, Snyder GL, Pieribone VA, Horiuchi A, Nairn AC, Messer A, Greengard P. Severe deficiencies in dopamine signaling in presymptomatic Huntington's disease mice. Proc Natl Acad Sci U S A. 2000;97:6809–6814. doi: 10.1073/pnas.120166397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonelli RM, Hofmann P. A review of the treatment options for Huntington's disease. Expert Opin Pharmacother. 2004;5:767–776. doi: 10.1517/14656566.5.4.767. [DOI] [PubMed] [Google Scholar]
- Bossi SR, Simpson JR, Isacson O. Age dependence of striatal neuronal death caused by mitochondrial dysfunction. Neuroreport. 1993;4:73–76. doi: 10.1097/00001756-199301000-00019. [DOI] [PubMed] [Google Scholar]
- Bouyer JJ, Park DH, Joh TH, Pickel VM. Chemical and structural analysis of the relation between cortical inputs and tyrosine hydroxylase-containing terminals in rat neostriatum. Brain Res. 1984;302:267–275. doi: 10.1016/0006-8993(84)90239-7. [DOI] [PubMed] [Google Scholar]
- Bradford J, Shin JY, Roberts M, Wang CE, Sheng G, Li S, Li XJ. Mutant huntingtin in glial cells exacerbates neurological symptoms of Huntington disease mice. J Biol Chem. 2010 doi: 10.1074/jbc.M109.083287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecht S, Kirchhof R, Chromik A, et al. Specific pathophysiological functions of JNK isoforms in the brain. Eur J Neurosci. 2005;21:363–377. doi: 10.1111/j.1460-9568.2005.03857.x. [DOI] [PubMed] [Google Scholar]
- Bredesen DE, Rao RV, Mehlen P. Cell death in the nervous system. Nature. 2006;443:796–802. doi: 10.1038/nature05293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman RR, Mezei MM, Theilmann J, Almqvist E, Hayden MR. The likelihood of being affected with Huntington disease by a particular age, for a specific CAG size. Am J Hum Genet. 1997;60:1202–1210. [PMC free article] [PubMed] [Google Scholar]
- Browne SE. Mitochondria and Huntington's disease pathogenesis: insight from genetic and chemical models. Ann N Y Acad Sci. 2008;1147:358–382. doi: 10.1196/annals.1427.018. [DOI] [PubMed] [Google Scholar]
- Browne SE, Bowling AC, MacGarvey U, Baik MJ, Berger SC, Muqit MM, Bird ED, Beal MF. Oxidative damage and metabolic dysfunction in Huntington's disease: selective vulnerability of the basal ganglia. Ann Neurol. 1997;41:646–653. doi: 10.1002/ana.410410514. [DOI] [PubMed] [Google Scholar]
- Byers RK, Gilles FH, Fung C. Huntington's disease in children. Neuropathologic study of four cases. Neurology. 1973;23:561–569. doi: 10.1212/wnl.23.6.561. [DOI] [PubMed] [Google Scholar]
- Calabresi P, De Murtas M, Pisani A, Stefani A, Sancesario G, Mercuri NB, Bernardi G. Vulnerability of medium spiny striatal neurons to glutamate: role of Na+/K+ ATPase. Eur J Neurosci. 1995;7:1674–1683. doi: 10.1111/j.1460-9568.1995.tb00689.x. [DOI] [PubMed] [Google Scholar]
- Canals JM, Checa N, Marco S, Akerud P, Michels A, Perez-Navarro E, Tolosa E, Arenas E, Alberch J. Expression of brain-derived neurotrophic factor in cortical neurons is regulated by striatal target area. J Neurosci. 2001;21:117–124. doi: 10.1523/JNEUROSCI.21-01-00117.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canals JM, Pineda JR, Torres-Peraza JF, Bosch M, Martin-Ibanez R, Munoz MT, Mengod G, Ernfors P, Alberch J. Brain-derived neurotrophic factor regulates the onset and severity of motor dysfunction associated with enkephalinergic neuronal degeneration in Huntington's disease. J Neurosci. 2004;24:7727–7739. doi: 10.1523/JNEUROSCI.1197-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael J, Sugars KL, Bao YP, Rubinsztein DC. Glycogen synthase kinase-3beta inhibitors prevent cellular polyglutamine toxicity caused by the Huntington's disease mutation. J Biol Chem. 2002;277:33791–33798. doi: 10.1074/jbc.M204861200. [DOI] [PubMed] [Google Scholar]
- Cass WA. Decreases in evoked overflow of dopamine in rat striatum after neurotoxic doses of methamphetamine. J Pharmacol Exp Ther. 1997;280:105–113. [PubMed] [Google Scholar]
- Cepeda C, Ariano MA, Calvert CR, Flores-Hernandez J, Chandler SH, Leavitt BR, Hayden MR, Levine MS. NMDA receptor function in mouse models of Huntington disease. J Neurosci Res. 2001;66:525–539. doi: 10.1002/jnr.1244. [DOI] [PubMed] [Google Scholar]
- Cepeda C, Hurst RS, Calvert CR, Hernandez-Echeagaray E, Nguyen OK, Jocoy E, Christian LJ, Ariano MA, Levine MS. Transient and progressive electrophysiological alterations in the corticostriatal pathway in a mouse model of Huntington's disease. J Neurosci. 2003;23:961–969. doi: 10.1523/JNEUROSCI.23-03-00961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepeda C, Wu N, Andre VM, Cummings DM, Levine MS. The corticostriatal pathway in Huntington's disease. Prog Neurobiol. 2007;81:253–271. doi: 10.1016/j.pneurobio.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha JH. Transcriptional signatures in Huntington's disease. Prog Neurobiol. 2007;83:228–248. doi: 10.1016/j.pneurobio.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvin D, Vanhoutte P, Pages C, Borrelli E, Caboche J. Unraveling a role for dopamine in Huntington's disease: the dual role of reactive oxygen species and D2 receptor stimulation. Proc Natl Acad Sci U S A. 2005;102:12218–12223. doi: 10.1073/pnas.0502698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchetti F, Prensa L, Wu Y, Parent A. Chemical anatomy of striatal interneurons in normal individuals and in patients with Huntington's disease. Brain Res Brain Res Rev. 2000;34:80–101. doi: 10.1016/s0165-0173(00)00039-4. [DOI] [PubMed] [Google Scholar]
- Cooper JK, Schilling G, Peters MF, et al. Truncated N-terminal fragments of huntingtin with expanded glutamine repeats form nuclear and cytoplasmic aggregates in cell culture. Hum Mol Genet. 1998;7:783–790. doi: 10.1093/hmg/7.5.783. [DOI] [PubMed] [Google Scholar]
- Cowan CM, Raymond LA. Selective neuronal degeneration in Huntington's disease. Curr Top Dev Biol. 2006;75:25–71. doi: 10.1016/S0070-2153(06)75002-5. [DOI] [PubMed] [Google Scholar]
- Coyle JT, Schwarcz R. Lesion of striatal neurones with kainic acid provides a model for Huntington's chorea. Nature. 1976;263:244–246. doi: 10.1038/263244a0. [DOI] [PubMed] [Google Scholar]
- Crossman AR. Primate models of dyskinesia: the experimental approach to the study of basal ganglia-related involuntary movement disorders. Neuroscience. 1987;21:1–40. doi: 10.1016/0306-4522(87)90322-8. [DOI] [PubMed] [Google Scholar]
- Crossman AR, Mitchell IJ, Sambrook MA, Jackson A. Chorea and myoclonus in the monkey induced by gamma-aminobutyric acid antagonism in the lentiform complex. The site of drug action and a hypothesis for the neural mechanisms of chorea. Brain. 1988;111(Pt 5):1211–1233. doi: 10.1093/brain/111.5.1211. [DOI] [PubMed] [Google Scholar]
- Cudkowicz M, Kowall NW. Degeneration of pyramidal projection neurons in Huntington's disease cortex. Ann Neurol. 1990;27:200–204. doi: 10.1002/ana.410270217. [DOI] [PubMed] [Google Scholar]
- Davies SW, Scherzinger E. Nuclear inclusions in Huntington's disease. Trends Cell Biol. 1997;7:422. doi: 10.1016/S0962-8924(97)88136-6. [DOI] [PubMed] [Google Scholar]
- Davies SW, Turmaine M, Cozens BA, et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- de Almeida LP, Ross CA, Zala D, Aebischer P, Deglon N. Lentiviral-mediated delivery of mutant huntingtin in the striatum of rats induces a selective neuropathology modulated by polyglutamine repeat size, huntingtin expression levels, and protein length. J Neurosci. 2002;22:3473–3483. doi: 10.1523/JNEUROSCI.22-09-03473.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiglia M, Pasik P, Pasik T. A Golgi study of neuronal types in the neostriatum of monkeys. Brain Res. 1976;114:245–256. doi: 10.1016/0006-8993(76)90669-7. [DOI] [PubMed] [Google Scholar]
- Difiglia M, Pasik T, Pasik P. Ultrastructure of Golgi-impregnated and gold-toned spiny and aspiny neurons in the monkey neostriatum. J Neurocytol. 1980;9:471–492. doi: 10.1007/BF01204837. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase K, et al. Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron. 1995;14:1075–1081. doi: 10.1016/0896-6273(95)90346-1. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997a;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997b;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- DiProspero NA, Chen EY, Charles V, Plomann M, Kordower JH, Tagle DA. Early changes in Huntington's disease patient brains involve alterations in cytoskeletal and synaptic elements. J Neurocytol. 2004a;33:517–533. doi: 10.1007/s11068-004-0514-8. [DOI] [PubMed] [Google Scholar]
- DiProspero NA, Chen EY, Charles V, Plomann M, Kordower JH, Tagle DA. Early changes in Huntington's disease patient brains involve alterations in cytoskeletal and synaptic elements. J Neurocytol. 2004b;33:517–533. doi: 10.1007/s11068-004-0514-8. [DOI] [PubMed] [Google Scholar]
- Duyao M, Ambrose C, Myers R, et al. Trinucleotide repeat length instability and age of onset in Huntington's disease. Nat Genet. 1993;4:387–392. doi: 10.1038/ng0893-387. [DOI] [PubMed] [Google Scholar]
- Emerich DF, Cain CK, Greco C, Saydoff JA, Hu ZY, Liu H, Lindner MD. Cellular delivery of human CNTF prevents motor and cognitive dysfunction in a rodent model of Huntington's disease. Cell Transplant. 1997;6:249–266. doi: 10.1177/096368979700600308. [DOI] [PubMed] [Google Scholar]
- Emerich DF, Lindner MD, Winn SR, Chen EY, Frydel BR, Kordower JH. Implants of encapsulated human CNTF-producing fibroblasts prevent behavioral deficits and striatal degeneration in a rodent model of Huntington's disease. J Neurosci. 1996;16:5168–5181. doi: 10.1523/JNEUROSCI.16-16-05168.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada Sanchez AM, Mejia-Toiber J, Massieu L. Excitotoxic neuronal death and the pathogenesis of Huntington's disease. Arch Med Res. 2008;39:265–276. doi: 10.1016/j.arcmed.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Farrer MJ, Hulihan MM, Kachergus JM, et al. DCTN1 mutations in Perry syndrome. Nat Genet. 2009;41:163–165. doi: 10.1038/ng.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante RJ, Kowall NW, Beal MF, Richardson EP, Jr, Bird ED, Martin JB. Selective sparing of a class of striatal neurons in Huntington's disease. Science. 1985;230:561–563. doi: 10.1126/science.2931802. [DOI] [PubMed] [Google Scholar]
- Ferrante RJ, Kowall NW, Richardson EP., Jr Proliferative and degenerative changes in striatal spiny neurons in Huntington's disease: a combined study using the section-Golgi method and calbindin D28k immunocytochemistry. J Neurosci. 1991;11:3877–3887. doi: 10.1523/JNEUROSCI.11-12-03877.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueredo-Cardenas G, Anderson KD, Chen Q, Veenman CL, Reiner A. Relative survival of striatal projection neurons and interneurons after intrastriatal injection of quinolinic acid in rats. Exp Neurol. 1994;129:37–56. doi: 10.1006/exnr.1994.1145. [DOI] [PubMed] [Google Scholar]
- Fusco FR, Chen Q, Lamoreaux WJ, et al. Cellular localization of huntingtin in striatal and cortical neurons in rats: lack of correlation with neuronal vulnerability in Huntington's disease. J Neurosci. 1999;19:1189–1202. doi: 10.1523/JNEUROSCI.19-04-01189.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M, Charvin D, Caboche J. Expanded huntingtin activates the c-Jun terminal kinase/c-Jun pathway prior to aggregate formation in striatal neurons in culture. Neuroscience. 2004;127:859–870. doi: 10.1016/j.neuroscience.2004.05.054. [DOI] [PubMed] [Google Scholar]
- Garcia M, Vanhoutte P, Pages C, Besson MJ, Brouillet E, Caboche J. The mitochondrial toxin 3-nitropropionic acid induces striatal neurodegeneration via a c-Jun N-terminal kinase/c-Jun module. J Neurosci. 2002;22:2174–2184. doi: 10.1523/JNEUROSCI.22-06-02174.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasic GP, Hollmann M. Molecular neurobiology of glutamate receptors. Annu Rev Physiol. 1992;54:507–536. doi: 10.1146/annurev.ph.54.030192.002451. [DOI] [PubMed] [Google Scholar]
- Gauthier LR, Charrin BC, Borrell-Pages M, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004a;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Gauthier LR, Charrin BC, Borrell-Pages M, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004b;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Gerfen CR. Synaptic organization of the striatum. J Electron Microsc Tech. 1988;10:265–281. doi: 10.1002/jemt.1060100305. [DOI] [PubMed] [Google Scholar]
- Gerfen CR. The neostriatal mosaic: multiple levels of compartmental organization. Trends Neurosci. 1992;15:133–139. doi: 10.1016/0166-2236(92)90355-c. [DOI] [PubMed] [Google Scholar]
- Gerfen CR, Baimbridge KG, Miller JJ. The neostriatal mosaic: compartmental distribution of calcium-binding protein and parvalbumin in the basal ganglia of the rat and monkey. Proc Natl Acad Sci U S A. 1985;82:8780–8784. doi: 10.1073/pnas.82.24.8780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen CR, Engber TM, Mahan LC, Susel Z, Chase TN, Monsma FJ, Jr, Sibley DR. D1 and D2 dopamine receptor-regulated gene expression of striatonigral and striatopallidal neurons. Science. 1990;250:1429–1432. doi: 10.1126/science.2147780. [DOI] [PubMed] [Google Scholar]
- Gerfen CR, Young WS., 3rd Distribution of striatonigral and striatopallidal peptidergic neurons in both patch and matrix compartments: an in situ hybridization histochemistry and fluorescent retrograde tracing study. Brain Res. 1988;460:161–167. doi: 10.1016/0006-8993(88)91217-6. [DOI] [PubMed] [Google Scholar]
- Goebel DJ, Poosch MS. NMDA receptor subunit gene expression in the rat brain: a quantitative analysis of endogenous mRNA levels of NR1Com, NR2A, NR2B, NR2C, NR2D and NR3A. Brain Res Mol Brain Res. 1999;69:164–170. doi: 10.1016/s0169-328x(99)00100-x. [DOI] [PubMed] [Google Scholar]
- Gray M, Shirasaki DI, Cepeda C, et al. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci. 2008;28:6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybiel AM. Neurotransmitters and neuromodulators in the basal ganglia. Trends Neurosci. 1990;13:244–254. doi: 10.1016/0166-2236(90)90104-i. [DOI] [PubMed] [Google Scholar]
- Grunewald T, Beal MF. Bioenergetics in Huntington's disease. Ann N Y Acad Sci. 1999;893:203–213. doi: 10.1111/j.1749-6632.1999.tb07827.x. [DOI] [PubMed] [Google Scholar]
- Gu M, Gash MT, Mann VM, Javoy-Agid F, Cooper JM, Schapira AH. Mitochondrial defect in Huntington's disease caudate nucleus. Ann Neurol. 1996;39:385–389. doi: 10.1002/ana.410390317. [DOI] [PubMed] [Google Scholar]
- Gunawardena S, Her LS, Brusch RG, Laymon RA, Niesman IR, Gordesky-Gold B, Sintasath L, Bonini NM, Goldstein LS. Disruption of axonal transport by loss of huntingtin or expression of pathogenic polyQ proteins in Drosophila. Neuron. 2003;40:25–40. doi: 10.1016/s0896-6273(03)00594-4. [DOI] [PubMed] [Google Scholar]
- Gutekunst CA, Levey AI, Heilman CJ, Whaley WL, Yi H, Nash NR, Rees HD, Madden JJ, Hersch SM. Identification and localization of huntingtin in brain and human lymphoblastoid cell lines with anti-fusion protein antibodies. Proc Natl Acad Sci U S A. 1995;92:8710–8714. doi: 10.1073/pnas.92.19.8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutekunst CA, Li SH, Yi H, et al. Nuclear and neuropil aggregates in Huntington's disease: relationship to neuropathology. J Neurosci. 1999;19:2522–2534. doi: 10.1523/JNEUROSCI.19-07-02522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackam AS, Singaraja R, Wellington CL, Metzler M, McCutcheon K, Zhang T, Kalchman M, Hayden MR. The influence of huntingtin protein size on nuclear localization and cellular toxicity. J Cell Biol. 1998;141:1097–1105. doi: 10.1083/jcb.141.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafezparast M, Klocke R, Ruhrberg C, et al. Mutations in dynein link motor neuron degeneration to defects in retrograde transport. Science. 2003;300:808–812. doi: 10.1126/science.1083129. [DOI] [PubMed] [Google Scholar]
- Handley OJ, Naji JJ, Dunnett SB, Rosser AE. Pharmaceutical, cellular and genetic therapies for Huntington's disease. Clin Sci (Lond) 2006;110:73–88. doi: 10.1042/CS20050148. [DOI] [PubMed] [Google Scholar]
- HDCRG. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Hedreen JC, Folstein SE. Early loss of neostriatal striosome neurons in Huntington's disease. J Neuropathol Exp Neurol. 1995;54:105–120. doi: 10.1097/00005072-199501000-00013. [DOI] [PubMed] [Google Scholar]
- Hedreen JC, Peyser CE, Folstein SE, Ross CA. Neuronal loss in layers V and VI of cerebral cortex in Huntington's disease. Neurosci Lett. 1991;133:257–261. doi: 10.1016/0304-3940(91)90583-f. [DOI] [PubMed] [Google Scholar]
- Heinsen H, Rub U, Bauer M, et al. Nerve cell loss in the thalamic mediodorsal nucleus in Huntington's disease. Acta Neuropathol. 1999;97:613–622. doi: 10.1007/s004010051037. [DOI] [PubMed] [Google Scholar]
- Heinsen H, Strik M, Bauer M, Luther K, Ulmar G, Gangnus D, Jungkunz G, Eisenmenger W, Gotz M. Cortical and striatal neurone number in Huntington's disease. Acta Neuropathol. 1994;88:320–333. doi: 10.1007/BF00310376. [DOI] [PubMed] [Google Scholar]
- Her LS, Goldstein LS. Enhanced sensitivity of striatal neurons to axonal transport defects induced by mutant huntingtin. J Neurosci. 2008;28:13662–13672. doi: 10.1523/JNEUROSCI.4144-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt DJ, Graybiel AM, Saper CB. Neurochemical architecture of the human striatum. J Comp Neurol. 1997;384:1–25. doi: 10.1002/(sici)1096-9861(19970721)384:1<1::aid-cne1>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Humbert S, Bryson EA, Cordelieres FP, Connors NC, Datta SR, Finkbeiner S, Greenberg ME, Saudou F. The IGF-1/Akt pathway is neuroprotective in Huntington's disease and involves Huntingtin phosphorylation by Akt. Dev Cell. 2002;2:831–837. doi: 10.1016/s1534-5807(02)00188-0. [DOI] [PubMed] [Google Scholar]
- Ide K, Nukina N, Masuda N, Goto J, Kanazawa I. Abnormal gene product identified in Huntington's disease lymphocytes and brain. Biochem Biophys Res Commun. 1995;209:1119–1125. doi: 10.1006/bbrc.1995.1613. [DOI] [PubMed] [Google Scholar]
- Ishiguro H, Yamada K, Sawada H, et al. Age-dependent and tissue-specific CAG repeat instability occurs in mouse knock-in for a mutant Huntington's disease gene. J Neurosci Res. 2001;65:289–297. doi: 10.1002/jnr.1153. [DOI] [PubMed] [Google Scholar]
- Jakel RJ, Maragos WF. Neuronal cell death in Huntington's disease: a potential role for dopamine. Trends Neurosci. 2000;23:239–245. doi: 10.1016/s0166-2236(00)01568-x. [DOI] [PubMed] [Google Scholar]
- Jellinger KA. Challenges in neuronal apoptosis. Curr Alzheimer Res. 2006;3:377–391. doi: 10.2174/156720506778249434. [DOI] [PubMed] [Google Scholar]
- Jenkins BG, Koroshetz WJ, Beal MF, Rosen BR. Evidence for impairment of energy metabolism in vivo in Huntington's disease using localized 1H NMR spectroscopy. Neurology. 1993;43:2689–2695. doi: 10.1212/wnl.43.12.2689. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y. Neostriatal cell subtypes and their functional roles. Neurosci Res. 1997;27:1–8. doi: 10.1016/s0168-0102(96)01134-0. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Wilson CJ, Augood SJ, Emson PC. Striatal interneurones: chemical, physiological and morphological characterization. Trends Neurosci. 1995;18:527–535. doi: 10.1016/0166-2236(95)98374-8. [DOI] [PubMed] [Google Scholar]
- Kennedy L, Evans E, Chen CM, Craven L, Detloff PJ, Ennis M, Shelbourne PF. Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum Mol Genet. 2003;12:3359–3367. doi: 10.1093/hmg/ddg352. [DOI] [PubMed] [Google Scholar]
- Kennedy L, Shelbourne PF. Dramatic mutation instability in HD mouse striatum: does polyglutamine load contribute to cell-specific vulnerability in Huntington's disease? Hum Mol Genet. 2000;9:2539–2544. doi: 10.1093/hmg/9.17.2539. [DOI] [PubMed] [Google Scholar]
- Kieburtz K, Feigin A, McDermott M, et al. A controlled trial of remacemide hydrochloride in Huntington's disease. Mov Disord. 1996;11:273–277. doi: 10.1002/mds.870110310. [DOI] [PubMed] [Google Scholar]
- Kim AH, Yano H, Cho H, Meyer D, Monks B, Margolis B, Birnbaum MJ, Chao MV. Akt1 regulates a JNK scaffold during excitotoxic apoptosis. Neuron. 2002;35:697–709. doi: 10.1016/s0896-6273(02)00821-8. [DOI] [PubMed] [Google Scholar]
- Klapstein GJ, Fisher RS, Zanjani H, Cepeda C, Jokel ES, Chesselet MF, Levine MS. Electrophysiological and morphological changes in striatal spiny neurons in R6/2 Huntington's disease transgenic mice. J Neurophysiol. 2001;86:2667–2677. doi: 10.1152/jn.2001.86.6.2667. [DOI] [PubMed] [Google Scholar]
- Klivenyi P, Bende Z, Hartai Z, Penke Z, Nemeth H, Toldi J, Vecsei L. Behaviour changes in a transgenic model of Huntington's disease. Behav Brain Res. 2006;169:137–141. doi: 10.1016/j.bbr.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Kuemmerle S, Gutekunst CA, Klein AM, Li XJ, Li SH, Beal MF, Hersch SM, Ferrante RJ. Huntington aggregates may not predict neuronal death in Huntington's disease. Ann Neurol. 1999;46:842–849. [PubMed] [Google Scholar]
- Kuppenbender KD, Standaert DG, Feuerstein TJ, Penney JB, Jr, Young AB, Landwehrmeyer GB. Expression of NMDA receptor subunit mRNAs in neurochemically identified projection and interneurons in the human striatum. J Comp Neurol. 2000;419:407–421. doi: 10.1002/(sici)1096-9861(20000417)419:4<407::aid-cne1>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- La Spada AR, Taylor JP. Polyglutamines placed into context. Neuron. 2003;38:681–684. doi: 10.1016/s0896-6273(03)00328-3. [DOI] [PubMed] [Google Scholar]
- Laforet GA, Sapp E, Chase K, et al. Changes in cortical and striatal neurons predict behavioral and electrophysiological abnormalities in a transgenic murine model of Huntington's disease. J Neurosci. 2001;21:9112–9123. doi: 10.1523/JNEUROSCI.21-23-09112.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landwehrmeyer GB, McNeil SM, Dure LS, et al. Huntington's disease gene: regional and cellular expression in brain of normal and affected individuals. Ann Neurol. 1995a;37:218–230. doi: 10.1002/ana.410370213. [DOI] [PubMed] [Google Scholar]
- Landwehrmeyer GB, Standaert DG, Testa CM, Penney JB, Jr, Young AB. NMDA receptor subunit mRNA expression by projection neurons and interneurons in rat striatum. J Neurosci. 1995b;15:5297–5307. doi: 10.1523/JNEUROSCI.15-07-05297.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H, Thorner G, Hopf A, Schroder KF. Morphometric studies of the neuropathological changes in choreatic diseases. J Neurol Sci. 1976;28:401–425. doi: 10.1016/0022-510x(76)90114-3. [DOI] [PubMed] [Google Scholar]
- Laurie DJ, Bartke I, Schoepfer R, Naujoks K, Seeburg PH. Regional, developmental and interspecies expression of the four NMDAR2 subunits, examined using monoclonal antibodies. Brain Res Mol Brain Res. 1997;51:23–32. doi: 10.1016/s0169-328x(97)00206-4. [DOI] [PubMed] [Google Scholar]
- Lee JM, Ivanova EV, Seong IS, Cashorali T, Kohane I, Gusella JF, MacDonald ME. Unbiased gene expression analysis implicates the huntingtin polyglutamine tract in extra-mitochondrial energy metabolism. PLoS Genet. 2007;3:e135. doi: 10.1371/journal.pgen.0030135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WC, Yoshihara M, Littleton JT. Cytoplasmic aggregates trap polyglutamine-containing proteins and block axonal transport in a Drosophila model of Huntington's disease. Proc Natl Acad Sci U S A. 2004;101:3224–3229. doi: 10.1073/pnas.0400243101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MS, Cepeda C, Hickey MA, Fleming SM, Chesselet MF. Genetic mouse models of Huntington's and Parkinson's diseases: illuminating but imperfect. Trends Neurosci. 2004;27:691–697. doi: 10.1016/j.tins.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Li H, Li SH, Cheng AL, Mangiarini L, Bates GP, Li XJ. Ultrastructural localization and progressive formation of neuropil aggregates in Huntington's disease transgenic mice. Hum Mol Genet. 1999a;8:1227–1236. doi: 10.1093/hmg/8.7.1227. [DOI] [PubMed] [Google Scholar]
- Li H, Li SH, Yu ZX, Shelbourne P, Li XJ. Huntingtin aggregate-associated axonal degeneration is an early pathological event in Huntington's disease mice. J Neurosci. 2001;21:8473–8481. doi: 10.1523/JNEUROSCI.21-21-08473.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH, Cheng AL, Li H, Li XJ. Cellular defects and altered gene expression in PC12 cells stably expressing mutant huntingtin. J Neurosci. 1999b;19:5159–5172. doi: 10.1523/JNEUROSCI.19-13-05159.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH, Li XJ. Aggregation of N-terminal huntingtin is dependent on the length of its glutamine repeats. Hum Mol Genet. 1998;7:777–782. doi: 10.1093/hmg/7.5.777. [DOI] [PubMed] [Google Scholar]
- Li SH, Li XJ. Huntingtin and its role in neuronal degeneration. Neuroscientist. 2004;10:467–475. doi: 10.1177/1073858404266777. [DOI] [PubMed] [Google Scholar]
- Li SH, Schilling G, Young WS, III, et al. Huntington's disease gene (IT15) is widely expressed in human and rat tissues. Neuron. 1993;11:985–993. doi: 10.1016/0896-6273(93)90127-d. [DOI] [PubMed] [Google Scholar]
- Li XJ. The early cellular pathology of Huntington's disease. Mol Neurobiol. 1999;20:111–124. doi: 10.1007/BF02742437. [DOI] [PubMed] [Google Scholar]
- Li XJ, Orr AL, Li S. Impaired mitochondrial trafficking in Huntington's disease. Biochim Biophys Acta. 2010;1802:62–65. doi: 10.1016/j.bbadis.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievens JC, Woodman B, Mahal A, Bates GP. Abnormal phosphorylation of synapsin I predicts a neuronal transmission impairment in the R6/2 Huntington's disease transgenic mice. Mol Cell Neurosci. 2002;20:638–648. doi: 10.1006/mcne.2002.1152. [DOI] [PubMed] [Google Scholar]
- Lione LA, Carter RJ, Hunt MJ, Bates GP, Morton AJ, Dunnett SB. Selective discrimination learning impairments in mice expressing the human Huntington's disease mutation. J Neurosci. 1999;19:10428–10437. doi: 10.1523/JNEUROSCI.19-23-10428.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YF. Expression of polyglutamine-expanded Huntingtin activates the SEK1-JNK pathway and induces apoptosis in a hippocampal neuronal cell line. J Biol Chem. 1998;273:28873–28877. doi: 10.1074/jbc.273.44.28873. [DOI] [PubMed] [Google Scholar]
- Lopes-Cendes I, Maciel P, Kish S, et al. Somatic mosaicism in the central nervous system in spinocerebellar ataxia type 1 and Machado-Joseph disease. Ann Neurol. 1996;40:199–206. doi: 10.1002/ana.410400211. [DOI] [PubMed] [Google Scholar]
- Ludolph AC, He F, Spencer PS, Hammerstad J, Sabri M. 3-Nitropropionic acid-exogenous animal neurotoxin and possible human striatal toxin. Can J Neurol Sci. 1991;18:492–498. doi: 10.1017/s0317167100032212. [DOI] [PubMed] [Google Scholar]
- Luesse HG, Schiefer J, Spruenken A, Puls C, Block F, Kosinski CM. Evaluation of R6/2 HD transgenic mice for therapeutic studies in Huntington's disease: behavioral testing and impact of diabetes mellitus. Behav Brain Res. 2001;126:185–195. doi: 10.1016/s0166-4328(01)00261-3. [DOI] [PubMed] [Google Scholar]
- Lunkes A, Mandel JL. A cellular model that recapitulates major pathogenic steps of Huntington's disease. Hum Mol Genet. 1998;7:1355–1361. doi: 10.1093/hmg/7.9.1355. [DOI] [PubMed] [Google Scholar]
- Luo Y, Kokkonen GC, Wang X, Neve KA, Roth GS. D2 dopamine receptors stimulate mitogenesis through pertussis toxin-sensitive G proteins and Ras-involved ERK and SAP/JNK pathways in rat C6-D2L glioma cells. J Neurochem. 1998;71:980–990. doi: 10.1046/j.1471-4159.1998.71030980.x. [DOI] [PubMed] [Google Scholar]
- Luthi-Carter R, Strand A, Peters NL, et al. Decreased expression of striatal signaling genes in a mouse model of Huntington's disease. Hum Mol Genet. 2000;9:1259–1271. doi: 10.1093/hmg/9.9.1259. [DOI] [PubMed] [Google Scholar]
- MacDonald ME, Duyao M, Calzonetti T, et al. Targeted inactivation of the mouse Huntington's disease gene homolog Hdh. Cold Spring Harb Symp Quant Biol. 1996;61:627–638. [PubMed] [Google Scholar]
- Mann DM, Oliver R, Snowden JS. The topographic distribution of brain atrophy in Huntington's disease and progressive supranuclear palsy. Acta Neuropathol. 1993;85:553–559. doi: 10.1007/BF00230496. [DOI] [PubMed] [Google Scholar]
- Mattson MP, Magnus T. Ageing and neuronal vulnerability. Nat Rev Neurosci. 2006;7:278–294. doi: 10.1038/nrn1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006;16:R551–560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- McGeer EG, McGeer PL. Duplication of biochemical changes of Huntington's chorea by intrastriatal injections of glutamic and kainic acids. Nature. 1976;263:517–519. doi: 10.1038/263517a0. [DOI] [PubMed] [Google Scholar]
- McLin JP, Thompson LM, Steward O. Differential susceptibility to striatal neurodegeneration induced by quinolinic acid and kainate in inbred, outbred and hybrid mouse strains. Eur J Neurosci. 2006;24:3134–3140. doi: 10.1111/j.1460-9568.2006.05198.x. [DOI] [PubMed] [Google Scholar]
- Menalled LB. Knock-in mouse models of Huntington's disease. NeuroRx. 2005;2:465–470. doi: 10.1602/neurorx.2.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merienne K, Helmlinger D, Perkin GR, Devys D, Trottier Y. Polyglutamine expansion induces a protein-damaging stress connecting heat shock protein 70 to the JNK pathway. J Biol Chem. 2003;278:16957–16967. doi: 10.1074/jbc.M212049200. [DOI] [PubMed] [Google Scholar]
- Michalik A, Van BC. Pathogenesis of polyglutamine disorders: aggregation revisited. Hum Mol Genet. 2003;12:R173–R186. doi: 10.1093/hmg/ddg295. [DOI] [PubMed] [Google Scholar]
- Mitchell IJ, Cooper AJ, Griffiths MR. The selective vulnerability of striatopallidal neurons. Prog Neurobiol. 1999;59:691–719. doi: 10.1016/s0301-0082(99)00019-2. [DOI] [PubMed] [Google Scholar]
- Mitchell IJ, Griffiths MR. The differential susceptibility of specific neuronal populations: insights from Huntington's disease. IUBMB Life. 2003;55:293–298. doi: 10.1080/1521654031000153012. [DOI] [PubMed] [Google Scholar]
- Mizuno H, Shibayama H, Tanaka F, et al. An autopsy case with clinically and molecular genetically diagnosed Huntington's disease with only minimal non-specific neuropathological findings. Clin Neuropathol. 2000;19:94–103. [PubMed] [Google Scholar]
- Mootha VK, Bunkenborg J, Olsen JV, et al. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- Morfini G, Pigino G, Brady ST. Polyglutamine expansion diseases: failing to deliver. Trends Mol Med. 2005;11:64–70. doi: 10.1016/j.molmed.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Morfini G, Pigino G, Szebenyi G, You Y, Pollema S, Brady ST. JNK mediates pathogenic effects of polyglutamine-expanded androgen receptor on fast axonal transport. Nat Neurosci. 2006a;9:907–916. doi: 10.1038/nn1717. [DOI] [PubMed] [Google Scholar]
- Morfini G, Stenoien DL, Brady ST. Axonal Transport. In: Siegel GJ, Albers RW, Brady ST, Price DL, editors. Basic neurochemistry: molecular, cellular, and medical aspects. Elsevier Academic Press; San Diego, CA: 2006b. pp. 485–502. [Google Scholar]
- Morfini G, Szebenyi G, Richards B, Brady ST. Regulation of kinesin: implications for neuronal development. Dev Neurosci. 2001;23:364–376. doi: 10.1159/000048720. [DOI] [PubMed] [Google Scholar]
- Morfini GA, Burns M, Binder LI, et al. Axonal transport defects in neurodegenerative diseases. J Neurosci. 2009a;29:12776–12786. doi: 10.1523/JNEUROSCI.3463-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfini GA, You YM, Pollema SL, et al. Pathogenic huntingtin inhibits fast axonal transport by activating JNK3 and phosphorylating kinesin. Nat Neurosci. 2009b;12:864–871. doi: 10.1038/nn.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KP, Carter RJ, Lione LA, Mangiarini L, Mahal A, Bates GP, Dunnett SB, Morton AJ. Abnormal synaptic plasticity and impaired spatial cognition in mice transgenic for exon 1 of the human Huntington's disease mutation. J Neurosci. 2000;20:5115–5123. doi: 10.1523/JNEUROSCI.20-13-05115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir J, Floresco SB, O'Kusky JR, et al. Targeted disruption of the Huntington's disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- Nihei K, Kowall NW. Neurofilament and neural cell adhesion molecule immunocytochemistry of Huntington's disease striatum. Ann Neurol. 1992;31:59–63. doi: 10.1002/ana.410310111. [DOI] [PubMed] [Google Scholar]
- Oblinger MM, Brady ST, McQuarrie IG, Lasek RJ. Cytotypic differences in the protein composition of the axonally transported cytoskeleton in mammalian neurons. J Neurosci. 1987;7:453–462. doi: 10.1523/JNEUROSCI.07-02-00453.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okun MS. Huntington's disease: what we learned from the original essay. Neurologist. 2003;9:175–179. doi: 10.1097/01.nrl.0000080952.78533.1f. [DOI] [PubMed] [Google Scholar]
- Orr AL, Li S, Wang CE, et al. N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J Neurosci. 2008;28:2783–2792. doi: 10.1523/JNEUROSCI.0106-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- Oyanagi K, Takeda S, Takahashi H, Ohama E, Ikuta F. A quantitative investigation of the substantia nigra in Huntington's disease. Ann Neurol. 1989;26:13–19. doi: 10.1002/ana.410260103. [DOI] [PubMed] [Google Scholar]
- Palfi S, Ferrante RJ, Brouillet E, Beal MF, Dolan R, Guyot MC, Peschanski M, Hantraye P. Chronic 3-nitropropionic acid treatment in baboons replicates the cognitive and motor deficits of Huntington's disease. J Neurosci. 1996;16:3019–3025. doi: 10.1523/JNEUROSCI.16-09-03019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panov AV, Gutekunst CA, Leavitt BR, Hayden MR, Burke JR, Strittmatter WJ, Greenamyre JT. Early mitochondrial calcium defects in Huntington's disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- Parent A, Csonka C, Etienne P. The occurrence of large acetylcholinesterase-containing neurons in human neostriatum as disclosed in normal and Alzheimer-diseased brains. Brain Res. 1984;291:154–158. doi: 10.1016/0006-8993(84)90663-2. [DOI] [PubMed] [Google Scholar]
- Parent A, Hazrati LN. Functional anatomy of the basal ganglia. I. The cortico-basal ganglia-thalamo-cortical loop. Brain Res Brain Res Rev. 1995;20:91–127. doi: 10.1016/0165-0173(94)00007-c. [DOI] [PubMed] [Google Scholar]
- Parent A, O'Reilly-Fromentin J, Boucher R. Acetylcholinesterase-containing neurons in cat neostriatum: a morphological and quantitative analysis. Neurosci Lett. 1980;20:271–276. doi: 10.1016/0304-3940(80)90159-7. [DOI] [PubMed] [Google Scholar]
- Parent M, Parent A. Relationship between axonal collateralization and neuronal degeneration in basal ganglia. J Neural Transm Suppl. 2006:85–88. doi: 10.1007/978-3-211-45295-0_14. [DOI] [PubMed] [Google Scholar]
- Penney JB, Jr, Vonsattel JP, MacDonald ME, Gusella JF, Myers RH. CAG repeat number governs the development rate of pathology in Huntington's disease. Ann Neurol. 1997;41:689–692. doi: 10.1002/ana.410410521. [DOI] [PubMed] [Google Scholar]
- Perez-Navarro E, Canals JM, Gines S, Alberch J. Cellular and molecular mechanisms involved in the selective vulnerability of striatal projection neurons in Huntington's disease. Histol Histopathol. 2006;21:1217–1232. doi: 10.14670/HH-21.1217. [DOI] [PubMed] [Google Scholar]
- Perrin V, Dufour N, Raoul C, Hassig R, Brouillet E, Aebischer P, Luthi-Carter R, Deglon N. Implication of the JNK pathway in a rat model of Huntington's disease. Exp Neurol. 2009;215:191–200. doi: 10.1016/j.expneurol.2008.10.008. [DOI] [PubMed] [Google Scholar]
- Perutz MF. Glutamine repeats and inherited neurodegenerative diseases: molecular aspects. Curr Opin Struct Biol. 1996;6:848–858. doi: 10.1016/s0959-440x(96)80016-9. [DOI] [PubMed] [Google Scholar]
- Phelan DR, Price G, Liu YF, Dorow DS. Activated JNK phosphorylates the c-terminal domain of MLK2 that is required for MLK2-induced apoptosis. J Biol Chem. 2001;276:10801–10810. doi: 10.1074/jbc.M008237200. [DOI] [PubMed] [Google Scholar]
- Portera-Cailliau C, Hedreen JC, Price DL, Koliatsos VE. Evidence for apoptotic cell death in Huntington disease and excitotoxic animal models. J Neurosci. 1995;15:3775–3787. doi: 10.1523/JNEUROSCI.15-05-03775.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puls I, Jonnakuty C, LaMonte BH, et al. Mutant dynactin in motor neuron disease. Nat Genet. 2003;33:455–456. doi: 10.1038/ng1123. [DOI] [PubMed] [Google Scholar]
- Puls I, Oh SJ, Sumner CJ, et al. Distal spinal and bulbar muscular atrophy caused by dynactin mutation. Ann Neurol. 2005;57:687–694. doi: 10.1002/ana.20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, McBride JL, Herzog CD, Brandon E, Gasmi M, Bartus RT, Kordower JH. Neurturin gene therapy improves motor function and prevents death of striatal neurons in a 3-nitropropionic acid rat model of Huntington's disease. Neurobiol Dis. 2007a;26:375–384. doi: 10.1016/j.nbd.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, McBride JL, Kordower JH. Animal models of Huntington's disease. ILAR J. 2007b;48:356–373. doi: 10.1093/ilar.48.4.356. [DOI] [PubMed] [Google Scholar]
- Rauskolb S, Zagrebelsky M, Dreznjak A, et al. Global deprivation of brain-derived neurotrophic factor in the CNS reveals an area-specific requirement for dendritic growth. J Neurosci. 2010;30:1739–1749. doi: 10.1523/JNEUROSCI.5100-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading SA, Dziorny AC, Peroutka LA, et al. Functional brain changes in presymptomatic Huntington's disease. Ann Neurol. 2004;55:879–883. doi: 10.1002/ana.20121. [DOI] [PubMed] [Google Scholar]
- Reddy PH, Mao P, Manczak M. Mitochondrial structural and functional dynamics in Huntington's disease. Brain Res Rev. 2009;61:33–48. doi: 10.1016/j.brainresrev.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid E, Kloos M, Ashley-Koch A, et al. A Kinesin Heavy Chain (KIF5A) Mutation in Hereditary Spastic Paraplegia (SPG10) Am J Hum Genet. 2002;71:1189–1194. doi: 10.1086/344210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilmann R, Kirsten F, Quinn L, Henningsen H, Marder K, Gordon AM. Objective assessment of progression in Huntington's disease: a 3-year follow-up study. Neurology. 2001;57:920–924. doi: 10.1212/wnl.57.5.920. [DOI] [PubMed] [Google Scholar]
- Reiner A, Albin RL, Anderson KD, D'Amato CJ, Penney JB, Young AB. Differential loss of striatal projection neurons in Huntington disease. Proc Natl Acad Sci U S A. 1988;85:5733–5737. doi: 10.1073/pnas.85.15.5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richfield EK, Maguire-Zeiss KA, Vonkeman HE, Voorn P. Preferential loss of preproenkephalin versus preprotachykinin neurons from the striatum of Huntington's disease patients. Ann Neurol. 1995a;38:852–861. doi: 10.1002/ana.410380605. [DOI] [PubMed] [Google Scholar]
- Richfield EK, Maguire-Zeiss KA, Vonkeman HE, Voorn P. Preferential loss of preproenkephalin versus preprotachykinin neurons from the striatum of Huntington's disease patients. Ann Neurol. 1995b;38:852–861. doi: 10.1002/ana.410380605. [DOI] [PubMed] [Google Scholar]
- Rigby M, Le Bourdelles B, Heavens RP, et al. The messenger RNAs for the N-methyl-D-aspartate receptor subunits show region-specific expression of different subunit composition in the human brain. Neuroscience. 1996;73:429–447. doi: 10.1016/0306-4522(96)00089-9. [DOI] [PubMed] [Google Scholar]
- Rodda RA. Cerebellar atrophy in Huntington's disease. J Neurol Sci. 1981;50:147–157. doi: 10.1016/0022-510x(81)90049-6. [DOI] [PubMed] [Google Scholar]
- Rosas HD, Lee SY, Bender A, et al. Altered white matter microstructure in the corpus callosum in Huntington's disease: Implications for cortical “disconnection”. Neuroimage. 2009 doi: 10.1016/j.neuroimage.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt A, Abbott MH, Gourley LM, Troncoso JC, Margolis RL, Brandt J, Ross CA. Predictors of neuropathological severity in 100 patients with Huntington's disease. Ann Neurol. 2003;54:488–493. doi: 10.1002/ana.10691. [DOI] [PubMed] [Google Scholar]
- Roy S, Zhang B, Lee VM, Trojanowski JQ. Axonal transport defects: a common theme in neurodegenerative diseases. Acta Neuropathol. 2005;109:5–13. doi: 10.1007/s00401-004-0952-x. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Carmichael J. Huntington's disease: molecular basis of neurodegeneration. Expert Rev Mol Med. 2003;5:1–21. doi: 10.1017/S1462399403006549. [DOI] [PubMed] [Google Scholar]
- Rudinskiy N, Kaneko YA, Beesen AA, Gokce O, Regulier E, Deglon N, Luthi-Carter R. Diminished hippocalcin expression in Huntington's disease brain does not account for increased striatal neuron vulnerability as assessed in primary neurons. J Neurochem. 2009;111:460–472. doi: 10.1111/j.1471-4159.2009.06344.x. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pernaute R, Garcia-Segura JM, del Barrio AA, Viano J, de Yebenes JG. Clinical correlation of striatal 1H MRS changes in Huntington's disease. Neurology. 1999;53:806–812. doi: 10.1212/wnl.53.4.806. [DOI] [PubMed] [Google Scholar]
- Sapp E, Penney J, Young A, Aronin N, Vonsattel JP, DiFiglia M. Axonal transport of N-terminal huntingtin suggests early pathology of corticostriatal projections in Huntington disease. J Neuropathol Exp Neurol. 1999;58:165–173. doi: 10.1097/00005072-199902000-00006. [DOI] [PubMed] [Google Scholar]
- Sassone J, Colciago C, Cislaghi G, Silani V, Ciammola A. Huntington's disease: the current state of research with peripheral tissues. Exp Neurol. 2009;219:385–397. doi: 10.1016/j.expneurol.2009.05.012. [DOI] [PubMed] [Google Scholar]
- Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- Schoepfer R, Monyer H, Sommer B, et al. Molecular biology of glutamate receptors. Prog Neurobiol. 1994;42:353–357. doi: 10.1016/0301-0082(94)90076-0. [DOI] [PubMed] [Google Scholar]
- Senut MC, Suhr ST, Kaspar B, Gage FH. Intraneuronal aggregate formation and cell death after viral expression of expanded polyglutamine tracts in the adult rat brain. J Neurosci. 2000;20:219–229. doi: 10.1523/JNEUROSCI.20-01-00219.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong IS, Ivanova E, Lee JM, et al. HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Hum Mol Genet. 2005;14:2871–2880. doi: 10.1093/hmg/ddi319. [DOI] [PubMed] [Google Scholar]
- Shelbourne PF, Keller-McGandy C, Bi WL, et al. Triplet repeat mutation length gains correlate with cell-type specific vulnerability in Huntington disease brain. Hum Mol Genet. 2007;16:1133–1142. doi: 10.1093/hmg/ddm054. [DOI] [PubMed] [Google Scholar]
- Sieradzan KA, Mann DM. The selective vulnerability of nerve cells in Huntington's disease. Neuropathol Appl Neurobiol. 2001;27:1–21. doi: 10.1046/j.0305-1846.2001.00299.x. [DOI] [PubMed] [Google Scholar]
- Sinadinos C, Burbidge-King T, Soh D, Thompson LM, Marsh JL, Wyttenbach A, Mudher AK. Live axonal transport disruption by mutant huntingtin fragments in Drosophila motor neuron axons. Neurobiol Dis. 2009;34:389–395. doi: 10.1016/j.nbd.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Smith Y, Bevan MD, Shink E, Bolam JP. Microcircuitry of the direct and indirect pathways of the basal ganglia. Neuroscience. 1998;86:353–387. doi: 10.1016/s0306-4522(98)00004-9. [DOI] [PubMed] [Google Scholar]
- Snell RG, MacMillan JC, Cheadle JP, et al. Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington's disease. Nat Genet. 1993;4:393–397. doi: 10.1038/ng0893-393. [DOI] [PubMed] [Google Scholar]
- Sotrel A, Paskevich PA, Kiely DK, Bird ED, Williams RS, Myers RH. Morphometric analysis of the prefrontal cortex in Huntington's disease. Neurology. 1991;41:1117–1123. doi: 10.1212/wnl.41.7.1117. [DOI] [PubMed] [Google Scholar]
- Storey E, Beal MF. Neurochemical substrates of rigidity and chorea in Huntington's disease. Brain. 1993;116(Pt 5):1201–1222. doi: 10.1093/brain/116.5.1201. [DOI] [PubMed] [Google Scholar]
- Szebenyi G, Morfini GA, Babcock A, et al. Neuropathogenic forms of huntingtin and androgen receptor inhibit fast axonal transport. Neuron. 2003a;40:41–52. doi: 10.1016/s0896-6273(03)00569-5. [DOI] [PubMed] [Google Scholar]
- Szebenyi G, Morfini GA, Babcock A, et al. Neuropathogenic Forms of Huntingtin and Androgen Receptor Inhibit Fast Axonal Transport. Neuron. 2003b;40:41–52. doi: 10.1016/s0896-6273(03)00569-5. [DOI] [PubMed] [Google Scholar]
- Tang TS, Chen X, Liu J, Bezprozvanny I. Dopaminergic signaling and striatal neurodegeneration in Huntington's disease. J Neurosci. 2007;27:7899–7910. doi: 10.1523/JNEUROSCI.1396-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas EA. Striatal specificity of gene expression dysregulation in Huntington's disease. J Neurosci Res. 2006;84:1151–1164. doi: 10.1002/jnr.21046. [DOI] [PubMed] [Google Scholar]
- Tobin AJ, Signer ER. Huntington's disease: the challenge for cell biologists. Trends Cell Biol. 2000;10:531–536. doi: 10.1016/s0962-8924(00)01853-5. [DOI] [PubMed] [Google Scholar]
- Trottier Y, Devys D, Imbert G, et al. Cellular localization of the Huntington's disease protein and discrimination of the normal and mutated form. Nat Genet. 1995;10:104–110. doi: 10.1038/ng0595-104. [DOI] [PubMed] [Google Scholar]
- Truant R, Atwal RS, Desmond C, Munsie L, Tran T. Huntington's disease: revisiting the aggregation hypothesis in polyglutamine neurodegenerative diseases. FEBS J. 2008;275:4252–4262. doi: 10.1111/j.1742-4658.2008.06561.x. [DOI] [PubMed] [Google Scholar]
- Trushina E, Dyer RB, Badger JD, et al. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol Cell Biol. 2004;24:8195–8209. doi: 10.1128/MCB.24.18.8195-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila M, Przedborski S. Targeting programmed cell death in neurodegenerative diseases. Nat Rev Neurosci. 2003;4:365–375. doi: 10.1038/nrn1100. [DOI] [PubMed] [Google Scholar]
- Vis JC, Schipper E, de Boer-van Huizen RT, Verbeek MM, de Waal RM, Wesseling P, ten Donkelaar HJ, Kremer B. Expression pattern of apoptosis-related markers in Huntington's disease. Acta Neuropathol. 2005;109:321–328. doi: 10.1007/s00401-004-0957-5. [DOI] [PubMed] [Google Scholar]
- von Horsten S, Schmitt I, Nguyen HP, et al. Transgenic rat model of Huntington's disease. Hum Mol Genet. 2003;12:617–624. doi: 10.1093/hmg/ddg075. [DOI] [PubMed] [Google Scholar]
- Vonsattel JP. Huntington disease models and human neuropathology: similarities and differences. Acta Neuropathol. 2008;115:55–69. doi: 10.1007/s00401-007-0306-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP., Jr Neuropathological classification of Huntington's disease. J Neuropathol Exp Neurol. 1985a;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP., Jr Neuropathological classification of Huntington's disease. J Neuropathol Exp Neurol. 1985b;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- Wade A, Jacobs P, Morton AJ. Atrophy and degeneration in sciatic nerve of presymptomatic mice carrying the Huntington's disease mutation. Brain Res. 2008;1188:61–68. doi: 10.1016/j.brainres.2007.06.059. [DOI] [PubMed] [Google Scholar]
- Walker FO. Huntington's Disease. Semin Neurol. 2007;27:143–150. doi: 10.1055/s-2007-971176. [DOI] [PubMed] [Google Scholar]
- Wang CE, Zhou H, McGuire JR, Cerullo V, Lee B, Li SH, Li XJ. Suppression of neuropil aggregates and neurological symptoms by an intracellular antibody implicates the cytoplasmic toxicity of mutant huntingtin. J Cell Biol. 2008;181:803–816. doi: 10.1083/jcb.200710158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watase K, Venken KJ, Sun Y, Orr HT, Zoghbi HY. Regional differences of somatic CAG repeat instability do not account for selective neuronal vulnerability in a knock-in mouse model of SCA1. Hum Mol Genet. 2003;12:2789–2795. doi: 10.1093/hmg/ddg300. [DOI] [PubMed] [Google Scholar]
- Weaver KE, Richards TL, Liang O, Laurino MY, Sami A, Aylward EH. Longitudinal diffusion tensor imaging in Huntington's Disease. Exp Neurol. 2009 doi: 10.1016/j.expneurol.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CJ, Kawaguchi Y. The origins of two-state spontaneous membrane potential fluctuations of neostriatal spiny neurons. J Neurosci. 1996;16:2397–2410. doi: 10.1523/JNEUROSCI.16-07-02397.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DD, Kuan CY, Whitmarsh AJ, Rincon M, Zheng TS, Davis RJ, Rakic P, Flavell RA. Absence of excitotoxicity-induced apoptosis in the hippocampus of mice lacking the Jnk3 gene. Nature. 1997;389:865–870. doi: 10.1038/39899. [DOI] [PubMed] [Google Scholar]
- Ye W, Qiang G, Jingmin W, Yanling Y, Xiru W, Yuwu J. Clinical and genetic study in Chinese patients with Alexander disease. J Child Neurol. 2008;23:173–177. doi: 10.1177/0883073807308691. [DOI] [PubMed] [Google Scholar]
- Zhen X, Uryu K, Wang HY, Friedman E. D1 dopamine receptor agonists mediate activation of p38 mitogen-activated protein kinase and c-Jun amino-terminal kinase by a protein kinase A-dependent mechanism in SK-N-MC human neuroblastoma cells. Mol Pharmacol. 1998;54:453–458. doi: 10.1124/mol.54.3.453. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Cattaneo E. Role of brain-derived neurotrophic factor in Huntington's disease. Prog Neurobiol. 2007;81:294–330. doi: 10.1016/j.pneurobio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Ciammola A, Rigamonti D, et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]