SUMMARY
Background
The molecular mechanism of enhancement of the enzymatic activity of factor VIIa (FVIIa) by tissue factor (TF) is not fully understood, primarily due to the lack of atomic models for the membrane-bound form of the TF:FVIIa complex.
Objectives
To construct the first membrane-bound model of the TF:FVIIa complex and to investigate the dynamics of the complex in solution and on the surface of anionic membranes using large-scale molecular dynamics (MD) simulations in full atomic detail.
Methods
Membrane-bound models of the TF:FVIIa complex and the individual factors were constructed and subjected to MD simulations, in order to characterize protein-protein and protein-lipid interactions and to investigate the dynamics of TF and FVIIa.
Results
The MD trajectories reveal that isolated FVIIa undergoes large structural fluctuation primarily due to the hinge motions between its domains, while sTF is structurally stable. Upon complex formation, sTF restricts the motion of FVIIa significantly. The results also show that, in the membrane-bound form, sTF directly interacts with the lipid head groups, even in the absence of FVIIa.
Conclusion
The first atomic models of membrane-bound sTF:FVIIa, FVIIa, and sTF are presented, revealing sTF direct contacts with the lipids, both in the isolated form and in complex with FVIIa. The main effect of sTF binding to FVIIa is spatial stabilization of the catalytic site of FVIIa, which ensures optimal interaction with the substrate, factor X.
Keywords: coagulation cascade, tissue factor, factor VIIa, GLA domain, molecular dynamics simulation, protein-protein interaction
INTRODUCTION
The rate of the enzymatic activity of proteolytic reactions involved in the blood clotting cascade [1–3] is enhanced by several orders of magnitude upon binding of the coagulation factors to anionic regions of the cellular membrane, in particular, regions that are rich in phosphatidylserine (PS). For vitamin K-dependent coagulation factors, the key step of membrane binding hinges on the GLA domain, a major membrane-anchoring domain mediating the binding of coagulants to the surface of the membrane in a Ca2+-dependent manner (we use ’GLA’ for this Gla-rich domain and ’Gla’ to refer to γ-carboxyglutamic acid residues). Vitamin K-dependent coagulation proteins, namely, factor VII (FVII) [4–6], factor IX (FIX) [7–8], factor X (FX) [9,10], and prothrombin (PT) [11–13], as well as the anticoagulant protein C (PrC) [14] and its cofactor protein S (PrS) [15], and protein Z (PrZ) share this membrane-anchoring domain.
One of the initial steps along the extrinsic pathway is the activation of FX to FXa catalyzed through the formation of the ternary complex of activated FVII, FVIIa (the proteolytic enzyme), tissue factor, TF (the cofactor), and FX (the substrate). FVIIa is one of the essential serine proteases in the coagulation cascade. In order to gain full catalytic activity, FVIIa needs to bind to its cofactor, TF, forming the TF:FVIIa complex (the extrinsic FXase; Fig. 1). The catalytic efficiency (kcat/Km) of FX hydrolysis by FVIIa is increased by 106–107-fold when FVIIa is associated with TF [16]. TF is composed of four domains: a cytoplasmic domain which appears not to be needed for the coagulation function of this factor [17,18], a trans-membrane domain that anchors TF in the membrane (presumably a single α-helix for which currently no structure has been reported), a linker region connecting the trans-membrane and extracellular domains, and the extracellular domain (major functional domain). The extracellular domain of TF, often referred to as soluble TF (sTF) which is used throughout this article to distinguish this domain from the entire TF, has been expressed separately and shown to account for the activity of this factor [17].
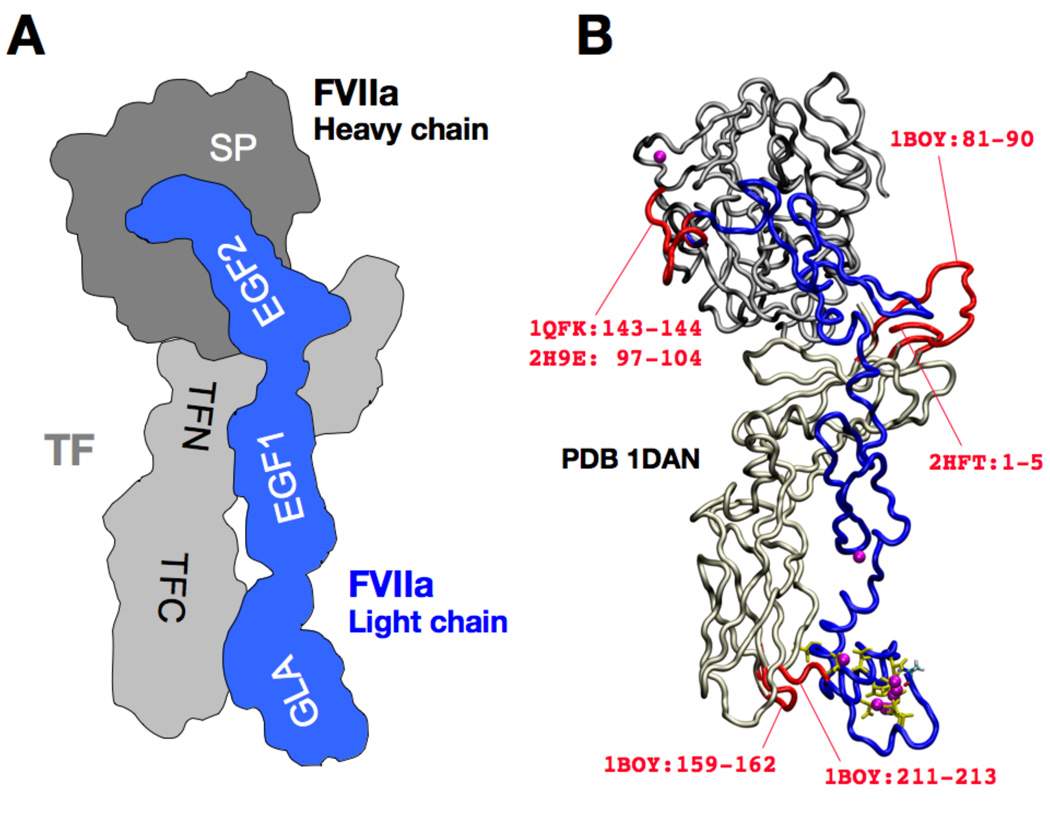
Figure 1. Architectural overview and structural modeling of the sTF:FVIIa complex.
(A) Schematic representation of the complex. sTF (shown in light gray) consists of N-terminal (residues 1–106) and C-terminal (residues 107–213) fibronectin type III domains (labeled as TFN and TFC, respectively). FVIIa consists of four domains: the serine protease (SP) domain which forms the heavy chain (dark gray), the GLA domain (GLA; residues 1–46), the EGF-like domain 1 (EGF1; residues 47–83), and the EGF-like domain 2 (EGF2; residues 87–128). The last three domains form the light chain (blue). (B) Construction of the complete model of the sTF:FVIIa complex starting from the PDB entry 1DAN [4]. The backbones of sTF (light gray), SP (dark gray), GLA, EGF1, and EGF2 (blue) are drawn in a tube representation. The Gla side chains of the GLA domain are drawn using a yellow stick representation, and FVIIa-bound Ca2+ ions are drawn as purple spheres. Missing loops (red) were constructed using other PDB entries, with the residue numbers specified.
A number of studies have proposed different mechanisms by which the association with TF might affect the activity of FVIIa. Analysis of the binding of FVIIa and TF in solution indicated that the GLA domain of FVIIa might play roles other than membrane binding; it was found that the GLA domain is not only important for the formation of a more stable TF:FVIIa complex, but also contributes to an extended substrate recognition region [19,20]. Other biochemical studies found evidence that TF can also directly induce alteration of the FVIIa catalytic activity through an allosteric mechanism that is independent of the supporting membrane [16].
The structure of the sTF:FVIIa complex has been determined crystallographically [4]. Under physiological conditions, sTF is anchored to the membrane with a single trans-membrane segment, which is connected to sTF by a short (6 aa) linker. Due to the structural flexibility of the linker, the trans-membrane segment is most likely decoupled conformationally from sTF and, thus, is not expected to be important in controlling the relative orientation of sTF on the membrane [18]. Additionally, direct interaction between sTF and the membrane has been suspected to account for the observation that the removal of the GLA domain from FVIIa has little effect on its active site positioning relative to the membrane in the TF:FVII complex, suggesting that sTF is sufficiently well oriented on the membrane (even in the absence of its membrane-anchoring helix) to hold FVIIa in an optimal (vertical) orientation on the surface of the membrane [21,22]. However, the nature of such direct interactions between sTF (or TF) and the membrane, as well as that between (s)TF and FVIIa on the surface of the membrane, is completely unknown [23], although several sTF sites that are directly involved in the binding of the substrates (FIX, FX, and FVII itself) have been reported [24–28].
Several modeling and simulation studies have investigated various aspects of the sTF:FVIIa complex and how they might contribute to the increased enzymatic activity of FVIIa [29–33]. However, the role of the membrane was never addressed in any of these studies. Docking and MD studies of the sTF:FVIIa:FXa ternary complex in solution reported that the C-terminal region of the GLA domain of FVIIa interacts with that of FXa in the complex [32]. Another MD study corroborated by hydrogen exchange experiments reported that the TF-induced structural change in the serine protease domain of FVIIa [33]. The effect of lipid molecules, however, was again not taken into account in these studies, as the simulations were conducted in solution.
Here we seek to characterize the dynamical and structural consequences of complex formation between sTF and FVIIa, as well as the modes of interactions of these proteins, particularly sTF, with the membranes. We employ MD simulations of the sTF:FVIIa complex and individual monomers to investigate the changes in their dynamical behavior and to characterize protein-protein and protein-lipid interactions upon complex formation and how they might result in increased enzymatic activity of FVIIa.
Materials and Methods
Modeling of sTF and FVIIa
The coordinates of the complex of human sTF and FVIIa with seven bound Ca2+ ions were taken from Protein Data Bank (PDB) entry 1DAN [4]. Missing loop regions of sTF were modeled using the corresponding residues from other PDB entries of isolated sTF, i.e., 1BOY [34] and 2HFT [35] (Fig. 1B). The missing C-terminal residues 143 and 144 of chain L of FVIIa were modeled based on the corresponding residues from another PDB entry of isolated FVIIa, 1QFK [36], and residues 145–152 from a segment of FXa, PDB 2H9E [37] (res 97–104) which has the highest BLAST [38] score among the available structures in the PDB. Hydrogen atoms were added, and the N- and C-termini were modified so that they have charged amino and carboxy groups, respectively.
System preparation
The modeled proteins were solvated in a water box that provided a minimum padding of 12 Å on each side. The system was then neutralized by randomly replacing water molecules in the bulk with Na+ and Cl− ions to achieve a final ion concentration of 250 mM. To remove any steric clashes that might have been introduced during the modeling phase, the solvated, neutralized systems were subjected to several cycles of relaxation. Each cycle included an energy minimization step followed by a short (10 ps) MD simulation, which provides a more efficient way of removing possible steric clashes. In the first cycle only the modeled parts were allowed to move with the rest of the system constrained; in subsequent cycles, the constraints applied to the system were gradually removed with the last cycle treating the entire model without any constraints.
A more detailed description of the procedures used in system preparation is provided in Supplementary Material. In short, to construct membrane models, the sTF:FVIIa complex and isolated FVIIa were respectively superimposed using their GLA domains onto the membrane-bound model of the GLA domain of FVII [39,40]. The membrane-bound model of isolated sTF was prepared by removing FVIIa from the system of sTF:FVIIa on the DOPS (1,2-dioleoyl-sn-glycero-3-phosphoserine) membrane. A Pure DOPS lipid bilayer was employed in order to maximize the sampling of various modes of interaction between the proteins and PS head groups during the limited time scales accessible to MD simulations. These membrane-bound models were then solvated and neutralized in the same manner as the solution systems. All the systems were then simulated using equilibrium MD for 50–90 ns (Table 1).
Table 1.
The simulation systems reported in this study.
| System | Environment | time (ns) |
|---|---|---|
| sTF | Solution | 60 |
| FVIIa | Solution | 60 |
| sTF | Membrane | 50 |
| FVIIa | Membrane | 80 |
| sTF:FVIIa | Membrane | 90 |
Simulation procedures
All the simulations were performed using NAMD 2.6 [41] with the CHARMM27 force field parameters [42], and the TIP3P model for water [43]. Langevin dynamics with a damping coefficient of 1 ps−1 was used to maintain the temperature at 310 K. Langevin piston Nosé-Hoover method [44,45] was employed to maintain the pressure at 1 atm. The particle mesh Ewald (PME) method [46] with a grid density of slightly finer than 1 Å−3 was used to calculate long-range electrostatic forces without truncation. The cut-off for van der Waals interaction was set at 12 Å. Integration time steps were set at 1, 1, and 2 fs for bonded, nonbonded, and PME calculations, respectively. The solution systems were simulated in the NPT ensemble (constant pressure and temperature), and the membrane-bound systems in the NPnTA ensemble (constant membrane-normal pressure, Pn, temperature, and membrane area).
Results and Discussion
Locking effect of sTF on the rocking motion of FVIIa
The calculated RMSDs for sTF, FVIIa, and their complex in different simulations is provided in Table 2. In order to highlight the relative motion of the individual domains of these proteins and to contrast the high flexibility of the protein to the observed structural stability of the composing domains, we have also calculated RMSDs after superposition of the trajectories using individual domains (Table 3). The dynamics of FVIIa and sTF captured during the simulations of different systems is also displayed in Fig. 2 in which instantaneous conformations are superimposed using the Cα atoms of individual domains: GLA, EGF1, EGF2, or SP for FVIIa; TFN or TFC for sTF (see also the movie provided as Supplementary Material).
Table 2.
Average Cα-RMSDs and standard deviations (in parenthesis) for the entire protein(s) calculated from the simulations of individual proteins (sTF and FVIIa) and their complex (sTF:FVIIa).
| System | Environment | Cα-RMSD (Å) Mean (SD) |
|
|---|---|---|---|
| FVIIa | Solution | 12.34 (3.91) | |
| sTF | Solution | 3.03 (0.45) | |
| FVIIa | Membrane | 7.11 (2.01) | |
| sTF | Membrane | 2.37 (0.43) | |
| sTF:FVIIa | in sTF:FVIIa | Membrane | 2.94 (0.41) |
| FIIA | in sTF:FVIIa | Membrane | 2.88 (0.43) |
| sTF | in sTF:FVIIa | Membrane | 2.61 (0.45) |
Table 3. Cα-RMSD of sTF and FVIIa aligned using respective domains.
Average Cα-RMSDs and standard deviations (in parenthesis) calculated either for the entire molecule or for individual domains (GLA, EGF1, EGF2, SP, TFC, and TFN). In contrast to Table 2, the individual domains are used for the alignment of the snapshots in each trajectory.
| Aligned domain and RMSDs for all and the aligned domains (Å) | ||||||||
|---|---|---|---|---|---|---|---|---|
| System | Environment | Domain | GLA | EGF1 | EGF2 | SP | TFC | TFN |
| FVIIa | Solution | All | 48.88 (14.12) | 43.48 (14.11) | 24.75 (8.68) | 24.23 (8.55) | – | – |
| Aligned | 0.93 ( 0.17) | 1.84 ( 0.31) | 2.35 (0.47) | 2.27 (0.62) | – | – | ||
| sTF | Solution | All | – | – | – | – | 4.04 (0.86) | 4.60 (1.43) |
| Aligned | – | – | – | – | 1.83 (0.24) | 3.24 (0.57) | ||
| FVIIa | Membrane | All | 31.83 ( 8.51) | 35.62 (12.30) | 12.63 (4.59 | 12.64 (4.76) | – | – |
| Aligned | 0.73 ( 0.11) | 2.50 (0.34) | 2.04 (0.49) | 2.12 (0.53) | – | – | ||
| sTF | Membrane | All | – | – | – | – | 3.15 (0.77) | 3.19 (0.88) |
| Aligned | – | – | – | – | 1.85 (0.28) | 2.35 (0.57) | ||
| FVIIa of | Membrane | All | 9.30 ( 4.02) | 7.45 ( 2.39) | 5.94 (1.84) | 3.47 (0.66) | – | – |
| TF:FVIIa | Aligned | 1.04 ( 0.22) | 1.35 ( 0.24) | 1.55 (0.22) | 2.23 (0.42) | – | – | |
| sTF of | Membrane | All | – | – | – | – | 3.36 (0.77) | 3.46 (0.92) |
| TF:FVIIa | Aligned | – | – | – | – | 1.39 (0.25) | 3.04 (0.60) | |
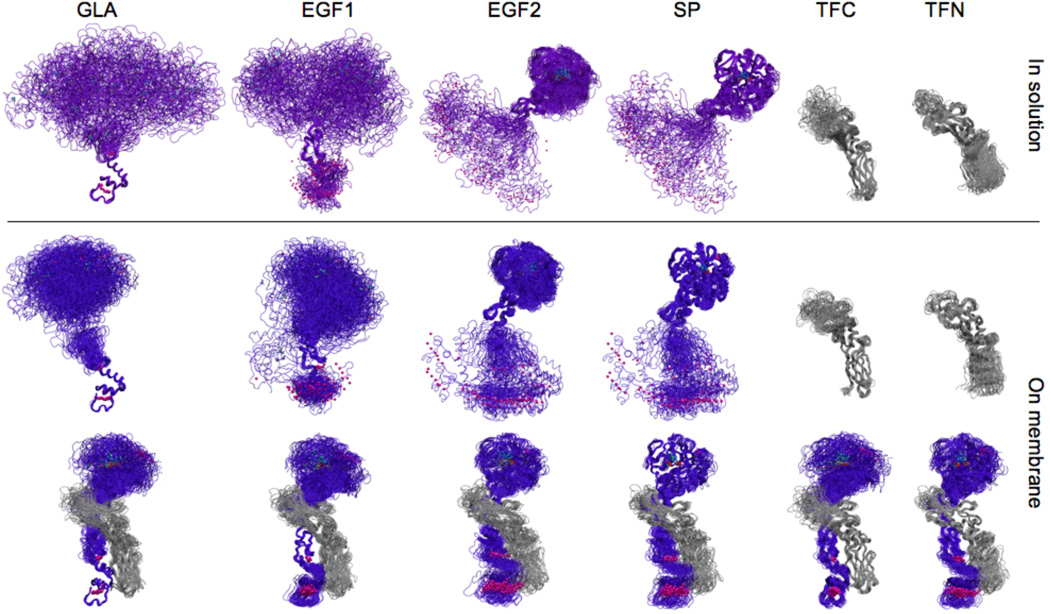
Figure 2. Dynamics of sTF and FVIIa.
Superimpose snapshots taken at 2-ns intervals from the simulations of individual sTF and FVIIa, as well as that of the sTF:FVIIa complex in solution or on the membrane are shown. sTF is colored gray, whereas FVIIa is colored purple when in solution, and blue when on the membrane. In each column a different domain (GLA, EGF1, EGF2, SP, TFC, or TFN) has been used for structural overlay of the snapshots, in order to depict relative movement of the domains and to emphasize that the individual domains are internally stable. FVIIa-bound Ca2+ ions are shown as purple spheres, and the side chains of the catalytic triad (CT) in the SP domain are shown in stick representations (see Fig. 3 for a more detailed view).
FVIIa exhibits significant flexibility and large fluctuations both in solution and on the surface of the membrane, while sTF presents a stable conformation regardless of the environment or configuration. The Cα-RMSDs of isolated FVIIa are over 30 Å when superimposed using the GLA or EGF1 domains, both in solution and on the membrane, while the corresponding RMSD values for FVIIa in the sTF:FVIIa complex are less than 10 Å. This trend, which is in good agreement with an earlier MD study of isolated FVIIa and sTF:FVIIa complex in solution [29], clearly indicates that TF binding has a significant confining effect on the internal motion of FVIIa (see also the movie provided as Supplementary Material). The observed phenomenon is supported by experimental results indicating that TF is able to restrict large-scale internal motion of FVIIa, although the nature of the movements involved could not be characterized [47].
Interestingly, the Cα-RMSDs within individual domains invariably remain small, at most 2.5 Å for EGF1 and around 1–2 Å for other domains (Table 3). These results demonstrate that the large motion of FVIIa primarily originates from the relative displacement of its individual domains which are each internally stable. This hinge-like (rocking) motion of FVIIa is significantly quenched upon complex formation with sTF. In other words, sTF rigidifies (or locks) FVIIa on the surface of the membrane. In comparison, the fluctuations of sTF are smaller. The Cα-RMSDs for the whole sTF or for its individual domains are around 2–3 Å. The RMSDs are slightly smaller for the membrane-bound sTF than those in solution, and for TFC than those of TFN.
Dynamics of FVIIa on the surface of the membrane
The simulated membrane-bound systems of isolated FVIIa and the sTF:FVIIa complex reveal an important role of sTF in rigidifying FVIIa (Table 3 and Fig. 2). Both the isolated FVIIa and the sTF:FVIIa complex remained on the surface of the DOPS membrane during the simulations exhibiting only slight changes in the orientation of the GLA domains against the membrane. However, FVIIa exhibits far larger fluctuation against the membrane in the isolated form (31.8 Å) than in complex with sTF (9.3 Å), as clearly demonstrated in Fig. 3A.
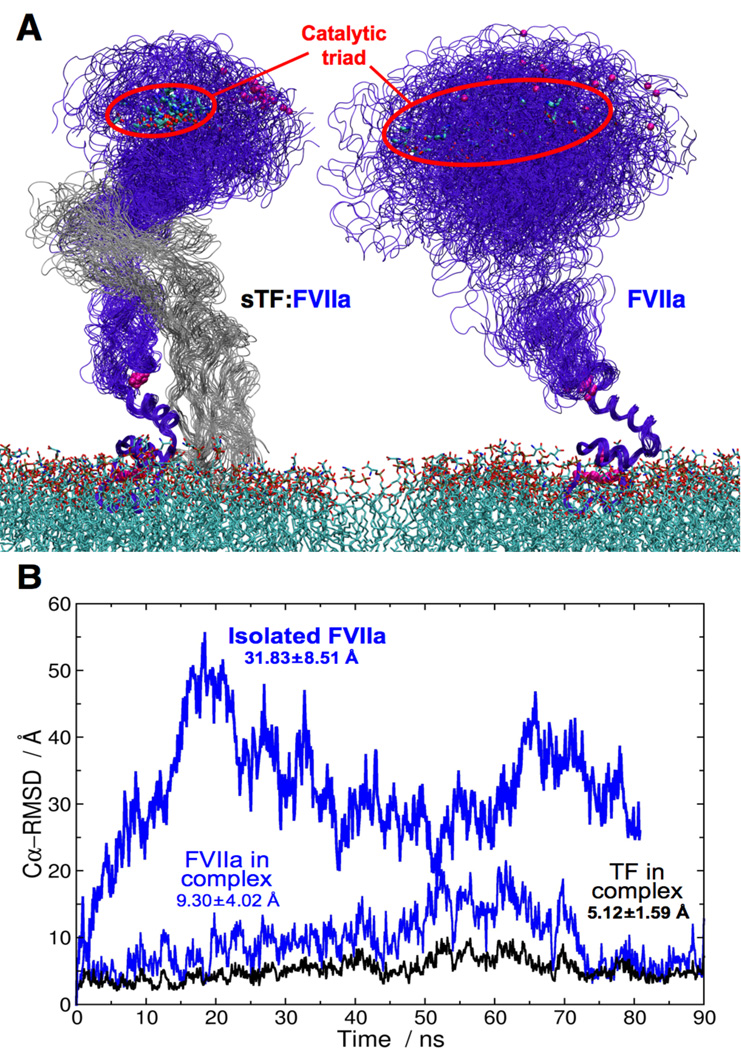
Figure 3. Impact of sTF on the dynamics of FVIIa on the membrane.
(A) Close-up views of the GLA-aligned snapshots taken from the simulations of sTF:FVIIa (left) and isolated FVIIa (right) on the membrane. The FVIIa-bound Ca2+ ions are drawn as purple spheres, and the CT and the DOPS lipids in stick representations. Red ovals are used to indicate the range of the CT fluctuation in the two systems. (B) Time series of the Cα-RMSDs calculated for isolated FVIIa on the membrane (thick blue line), FVIIa in the sTF:FVIIa complex on the membrane (blue line), and for sTF in the sTF:FVIIa complex on the membrane (black line). The average Cα-RMSDs and standard deviations calculated over the entire trajectory are also indicated for all systems.
The stabilizing role of sTF is further supported by the appearance of a large-scale, soft mode of fluctuation in the time series of Cα-RMSDs of FVIIa in its isolated form (Fig. 3B), although (about an order of magnitude) longer simulations would be required to fully validate this effect. The isolated FVIIa seems to exhibit Cα-RMSD fluctuations with a period of around 50 ns, whereas FVIIa in the complex exhibits only smaller, faster fluctuations on the order of a few nanoseconds, suggesting that the soft motion of FVIIa has vanished upon binding to sTF.
A major effect of the structural rigidification of FVIIa by sTF is the spatial confinement of the catalytic triad (CT, composed of His57, Asp102, and Ser195) in the SP domain, both in terms of its relative height and projection onto the surface of the membrane. This effect is visualized graphically by monitoring the positions of the side chains of the CT during the simulation, revealing a wide spatial scatter of this region, particularly with respect to the membrane projection of the CT, i.e., its position within the x-y plane parallel to the membrane (Fig. 3). The standard deviations of the x and y coordinates of the Ser195:Cα are ±11.2 Å and ±8.5 Å for isolated FVIIa, and only ±5.5 Å and ±6.3 Å for FVIIa in the complex (Fig. 4), indicating that the lateral fluctuation range of CT in the sTF:FVIIa complex is restricted to about one third of that in the isolated FVIIa. Furthermore, CT in the complex maintains a position directly above the GLA domain and sTF, whereas in the isolated FVIIa it scatters over a wide region, mostly distant from the position above the GLA domain (Fig. 4). Combined with the rotation of the SP domain, this results in a totally different position and orientation of CT in the absence of sTF. The relative position of the CT is of utmost importance for interaction with FX, which has been suggested to approach and bind the sTF:FVIIa complex from the putative exosite side of sTF (specified by a blue arrow in Fig. 4) [24–28].
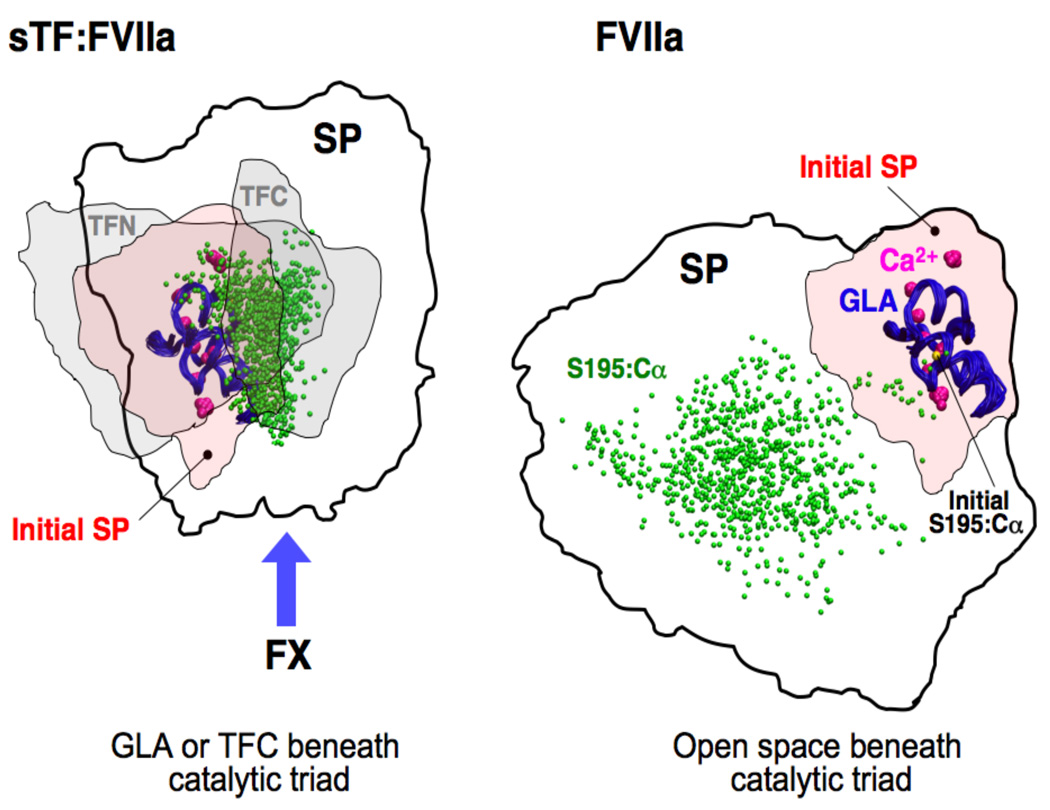
Figure 4. The effect of sTF on the fluctuation of the SP domain of the membrane-bound FVIIa.
Top view of Fig. 3A showing the footprints (membrane projection) of various domains during the simulations of the sTF:FVIIa complex (left) and isolated FVIIa (right) on the membrane. Thick lines encompass the region in which the SP domain fluctuates in the x–y plane (parallel to the membrane). As a reference, the initial footprint of the SP domain (initial SP) is also shown using a rosé shading. In the left panel, the range of TFN and TFC fluctuations is marked with gray shadings. The GLA domains (used for structural alignment in both cases) are shown in blue tubes with the bound Ca2+ ions drawn as large purple spheres. As a measure of the spatial scatter of CT, the position of the Cα atom of Ser195 taken from multiple frames of the trajectories, is shown as small green spheres. The initial position of this residue is drawn as a small yellow sphere and labeled in the right panel. The putative FX binding interface is marked with a blue arrow in the left panel.
The distance between the membrane and CT (CT height), which has been experimentally estimated [21,22], can also play an important role in determining the optimal pose of the CT for interaction with FX. The CT height is defined here as the difference between the average z-coordinate of all carboxy oxygens in the serine head groups of the proximal lipid leaflet, which constitute the outermost layer of the DOPS bilayer [40,48], and that of the Cα atoms of the CT residues. The CT height of isolated FVIIa (85.1±3.8 Å) is slightly shorter than that of FVIIa in the complex (88.1±1.4 Å), and the standard deviation is larger for the former, indicating that sTF holds the SP domain in a more upright position, resulting in a slightly higher position of the CT than in the isolated FVIIa. However, the extent of the effect is much smaller compared to the confining effect of sTF on the projected position and orientation of the SP domain, which were described above. The CT heights calculated from the simulations are in close agreement with the distances derived from FRET measurements [21,22] which also concluded that FVIIa adopts a perpendicular orientation to the surface of the membrane.
Direct interaction of sTF with the membrane
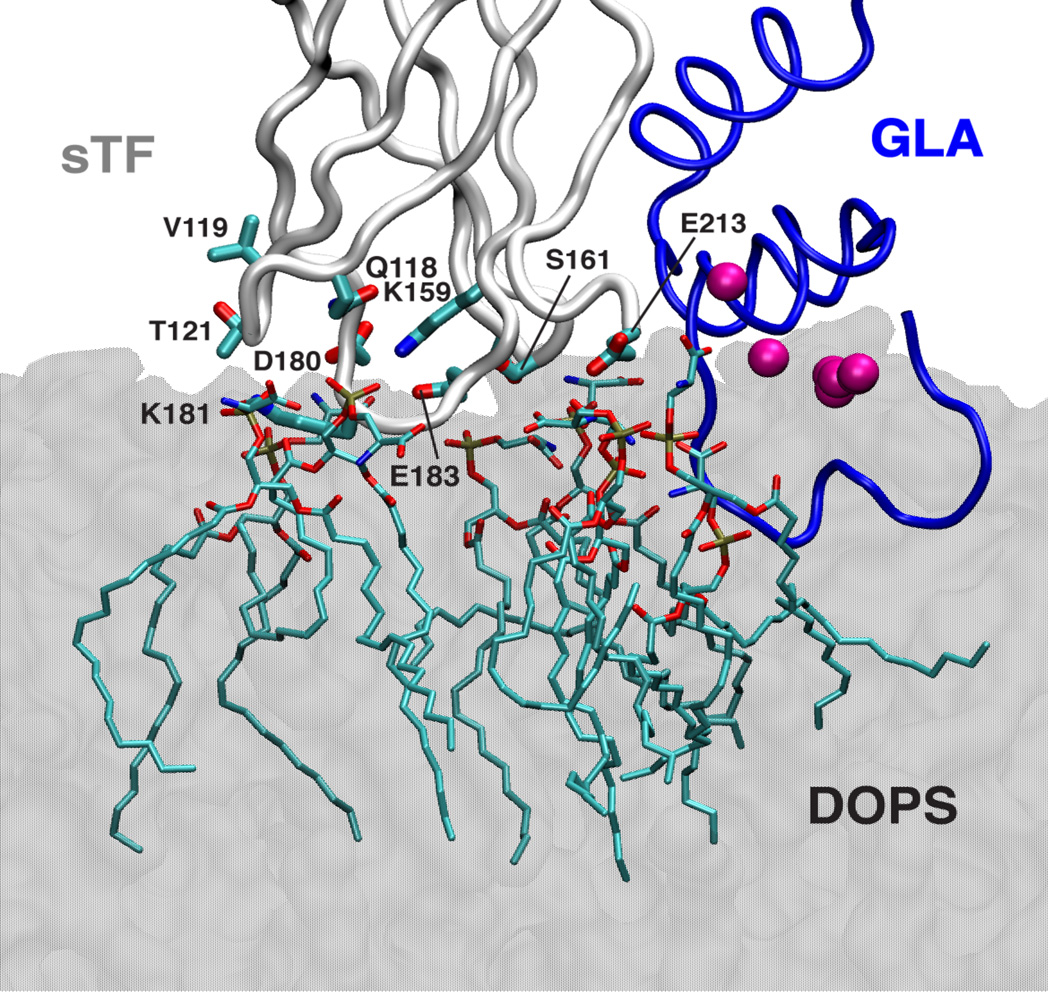
During the membrane simulations, sTF maintains its membrane-bound state both in its isolated form and in the sTF:FVIIa complex, establishing direct contacts with anionic DOPS molecules through several residues located at the bottom of TFC (Fig. 5). Based on these results we suggest that the trans-membrane and linker regions of TF are not required for establishing direct interactions between the membrane and TF and for keeping sTF on the surface of the membrane.
Figure 5. Direct interaction of sTF with the membrane.
A snapshot of the sTF:FVIIa complex on the membrane is shown in which direct contacts of sTF to several DOPS molecules in the membrane are highlighted. DOPS molecules and the side chains of sTF that are within 5 Å are shown in stick representations. The membrane is drawn using a transparent gray shading. The backbone of the GLA domain of FVIIa and sTF are shown in blue and light gray tube representations, respectively. The GLA domain-bound Ca2+ ions are drawn as purple spheres (not all seven ions are distinguishable in this view).
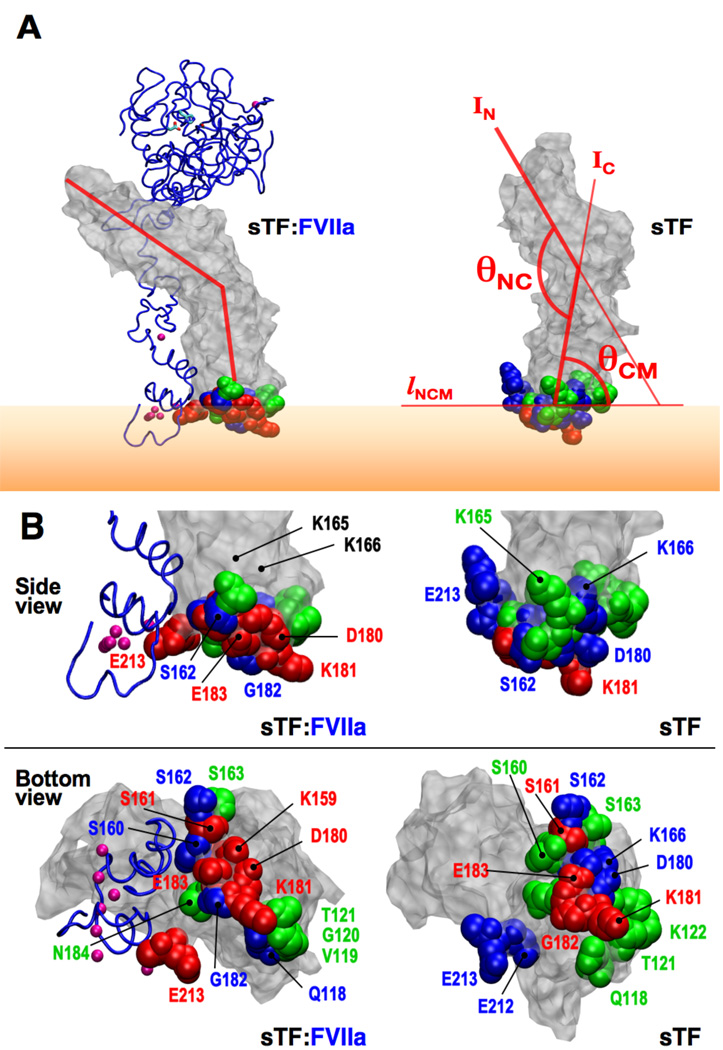
While the conformation of sTF remains mostly unchanged upon complex formation with FVIIa on the surface of the membrane, its orientation against the membrane changes significantly. Fig. 6A shows representative snapshots of sTF on the membrane with and without FVIIa. Two angles, θNC, the angle between TFN and TFC, and θCM, an angle between the membrane and sTF (see Fig. 6 for the exact definitions), are used to define the orientation of sTF on the surface of the membrane The average and standard deviation of θNC and θCM for isolated sTF and sTF in the complex are listed in Table 4. For both angles, the standard deviation decreases slightly in the complex. As discussed above, the effect of FVIIa on the internal motion of sTF seems to be small, thus resulting in a slight change in the average θNC, which is also evident from the overlay of the simulation snapshots shown in Fig. 2 and the Cα-RMSDs listed in Table 3. However, θCM in isolated sTF is approximately 20° smaller than that of sTF in the complex, reflecting the leaning motion of sTF toward FVIIa upon complex formation. This change in orientation results in a different pattern of membrane interaction for sTF, specifically the change in the contact probability of residues from sTF that directly interact with the lipid molecules. Fig. 6B shows these residues along with the fraction of the simulation time during which they maintained their contact to the lipids. Interestingly, some of the residues are proximal to the putative exosite of TF, the region that has been proposed to directly interact with the substrate (FX) [24–26,28]. Our results, therefore, suggest that complex formation and membrane binding might play a role in optimal presentation of the exosite region on TF, through modulating the conformation of specific regions and residues of sTF, as well as its orientation on the membrane. The direct interaction of PS head groups with specific residues in sTF most likely also affect the conformation of these residues. As such, one might expect that the lipids might have an active, direct participation in the decryption of sTF [49]. We are currently investigating this aspect in more detail. The GLA domain in the sTF:FVIIa complex did not change its structure or orientation against the membrane (Fig. 3A), preserving the initial Ca2+–PS interactions [40]. The N-terminal residue of the GLA domain (Ala1) remained “tucked” in the Ca2+-binding region, sustaining three hydrogen bonds with Gla20, Gln21, and Gla26 during all simulations (Fig. S1 in Supplementary Material).
Figure 6. Membrane orientation and lipid-interacting residues of sTF.
(A) Snapshots of membrane-bound sTF taken from the sTF:FVIIa complex (left) and isolated sTF (right) are shown. The backbone of FVIIa is drawn using a blue tube representation and sTF in gray surface representation with the membrane interacting residues in space-filling representations (green, blue, and red; see below for color coding). The GLA-bound Ca2+ ions are drawn as purple spheres (not all seven ions are distinguishable in this view). Membrane-interacting residues are defined using a cutoff distance of 3.5 Å from the lipid molecules, and colored based on the fraction of time they maintain contact with the lipid head groups during the simulations: green for <30 %, blue for 30–70 %, and red for >70 %. Orange shade represents the membrane. Two angles used to define the orientation of sTF (θNC and θCM) are also shown. IN and IC are the first two principal axes of TFN and TFC, respectively, and lNCM is the intersection between the plane of the membrane surface (whose normal is fixed along the z axis) and the plane spanned by the two principal axes IN and IC. θNC is the angle between IN and IC, and θCM is that between IC and lNCM. (B) Probability for sTF residues to directly interact with the membrane. Close-up side and bottom views of snapshots shown in (A) with individual lipid-interacting residues labeled. Same color coding as in (A) has been used. Residues K165 and K166, that are strongly suspected to mark the substrate-binding side of TF [24–26, 28], are also marked.
Table 4. Orientation of sTF on the membrane.
See Fig. 6 for the definition of the two angles, θNC and θCM, used to characterize the orientation of sTF on the membrane.
| System | θNC ( °) | θ CM( °) |
|---|---|---|
| sTF of sTF:FVIIa | 126.6±5.2 | 94.4±4.7 |
| sTF | 132.7±11.3 | 78.8±9.8 |
Concluding Remarks
Unraveling the molecular mechanism(s) by which complex formation between the coagulation factors and their respective cofactors results in an immense enhancement of their enzymatic activity, and the strong dependence of such reactions on anionic membranes hinges on atomic-resolution models of the membrane-bound forms of these complexes. In the present study, taking advantage of a crystal structure of the TF:FVIIa complex [4] and a recent model of the membrane-bound GLA domain of FVIIa [40], we have constructed the first membrane-bound model of the binary complex of TF and FVIIa at an atomic resolution. Extensive, large-scale MD simulations have been performed to establish optimal protein-protein and protein-lipid interactions, and to investigate functionally relevant dynamical aspects of TF and FVIIa, in their isolated form and as a complex, both in solution and on the surface of anionic membranes.
We show that FVIIa is highly dynamic in its isolated form and unlikely to provide a stable configuration for an optimal interaction with the substrate. Upon complex formation with TF, however, a significant reduction in fluctuation of FVIIa is observed in the simulations. We show that this effect results in spatial confinement, both in terms of the position and the orientation of the SP domain, and thereby the catalytic triad, not only with regard to the height of the site but also, and more dramatically, with respect to the projection of the SP domain on the membrane.
The equilibrated membrane-bound model of the sTF:FVIIa complex also provides the first view of the orientation and relative position of sTF on the surface of a membrane. The model suggests that sTF directly interacts with the lipid head groups, successfully accounting for experimental measurements indicating a stable orientation of sTF on the membrane [21], in a manner independent of the trans-membrane region of TF or its unstructured linker. Furthermore, the model provides a framework for new experiments, e.g., NMR measurements or mutagenesis studies, which will examine the involvement of the specific side chains of TF in interaction with the membrane that were identified by the simulations [50].
Supplementary Material
Acknowledgements
This work was supported by NIH grants R01-GM086749, R01-HL47014 and P41-RR05969. All the simulations have been performed using the computational resources provided by TeraGrid allocation (grant number MCA06N060), primarily on the Big Red cluster at Indiana University. The authors thank Chad M. Rienstra, Mark A. McLean, and Steve Sligar for insightful discussion and Giray Enkavi for technical assistance.
Footnotes
Disclosure of Conflict of Interests
The authors state that they have no conflict of interest.
References
- 1.Furie B, Furie BC. The molecular basis of blood coagulation. Cell. 1988;53:505–518. doi: 10.1016/0092-8674(88)90567-3. [DOI] [PubMed] [Google Scholar]
- 2.Stace CL, Ktistakis NT. Phosphatidic acid- and phosphatidylserine-binding proteins. Biochim Biophys Acta. 2006;176:913–926. doi: 10.1016/j.bbalip.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Zwaal RFA, Comfurius P, Bevers EM. Lipid-protein interactions in blood coagulation. Biochim Biophys Acta. 1998;1376:433–453. doi: 10.1016/s0304-4157(98)00018-5. [DOI] [PubMed] [Google Scholar]
- 4.Banner DW, D’Arcy A, Chène C, Winkler FK, Guha A, Konigsberg WH, Nemerson Y. The crystal structure of the complex of blood coagulation factor VIIa with soluble tissue factor. Nature. 1996;380:41–46. doi: 10.1038/380041a0. [DOI] [PubMed] [Google Scholar]
- 5.Harvey SB, Martinez MDSMB, Nelsestuen GL. Mutagenesis of the γ-carboxyglutamic acid domain of human factore VII to generate maximum enhancement of the membrane contact site. J Biol Chem. 2003;278:8363–8369. doi: 10.1074/jbc.M211629200. [DOI] [PubMed] [Google Scholar]
- 6.Nelsestuen GL. Enhancement of vitamin-K-dependent protein function by modification of the γ-carboxyglutamic acid domain: Studies of protein C and factor VII. Trends Cardiovasc Med. 1999;9:162–167. doi: 10.1016/s1050-1738(99)00024-9. [DOI] [PubMed] [Google Scholar]
- 7.Huang M, Furie BC, Furie B. Crystal structure of the calcium-stabilized human factor IX Gla domain bound to a conformation-specific anti-factor IX antibody. J Biol Chem. 2004;279:14338–14346. doi: 10.1074/jbc.M314011200. [DOI] [PubMed] [Google Scholar]
- 8.Shikamoto Y, Morita T, Fujimoto Z, Mizuno H. Crystal structure of Mg2+- and Ca2+-bound Gla domain of factor IX complexed with binding protein. J Biol Chem. 2003;278:24090–24094. doi: 10.1074/jbc.M300650200. [DOI] [PubMed] [Google Scholar]
- 9.Mizuno H, Fujimoto Z, Atoda H, Morita T. Crystal structure of an anticoagulant protein in complex with the Gla domain of factor X. Proc Natl Acad Sci USA. 2001;98:7230–7234. doi: 10.1073/pnas.131179698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabharwal AK, Padmanabhan K, Tulinsky A, Mathur A, Gorka J, Bajaj SP. Interaction of calcium with native and decarboxylated human factor X. J Biol Chem. 1997;272:22037–22045. doi: 10.1074/jbc.272.35.22037. [DOI] [PubMed] [Google Scholar]
- 11.Falls LA, Furie BC, Jacobs M, Furie B, Rigby AC. The ω-loop region of the human prothrombin γ-carboxyglutamic acid domain penetrates anionic phospholipid membranes. J Biol Chem. 2001;276:23895–23902. doi: 10.1074/jbc.M008332200. [DOI] [PubMed] [Google Scholar]
- 12.Huang M, Rigby AC, Morelli X, Grant MA, Huang G, Furie B, Seaton B, Furie BC. Structural basis of membrane binding by Gla domains of vitamin K-dependent proteins. Nat Struct Biol. 2003;10:751–756. doi: 10.1038/nsb971. [DOI] [PubMed] [Google Scholar]
- 13.Ratcliffe JV, Furie B, Furie BC. The importance of specific γ-carboxyglutamic acid residues in prothrombin - evaluation by site-specific mutagenesis. J Biol Chem. 1993;268:24339–24345. [PubMed] [Google Scholar]
- 14.Oganesyan V, Oganesyan N, Terzyan S, Qu D, Dauter Z, Esmon NL, Esmon CT. The crystal structure of the endothelial protein C receptor and a bound phospholipid. J Biol Chem. 2002;277:24851–24854. doi: 10.1074/jbc.C200163200. [DOI] [PubMed] [Google Scholar]
- 15.Rezende SM, Simmonds RE, Lane DA. Coagulation, inflammation, and apoptosis: different roles for protein S and the protein S-C4b binding protein complex. Blood. 2004;103:1192–1201. doi: 10.1182/blood-2003-05-1551. [DOI] [PubMed] [Google Scholar]
- 16.Lawson JH, Butenas S, Mann KG. The evaluation of complex-dependent alterations in human factor VIIa. J Biol Chem. 1992;267:4834–4843. [PubMed] [Google Scholar]
- 17.Ruf W, Rehemtullas A, Edgington TS. Phospholipid-independent and -dependent interactions required for tissue factor receptor and cofactor function. J Biol Chem. 1991;266:2158–2166. [PubMed] [Google Scholar]
- 18.Paborsky LR, Carasli IW, Fisher KL, Gorman CM. Lipid association, but not transmembrane domain, is required the for tissue factor activity — substitution of the transmembrane domain with a phosphatidylinositol anchor. J Biol Chem. 1991;266:21911–21916. [PubMed] [Google Scholar]
- 19.Ruf W, Kalnik MW, Lund-Hansen T, Edgington TS. Characterization of factor VII association with tissue factor in solution — high and low affinity calcium binding sites in factor VII contribute to functionally distinct interactions. J Biol Chem. 1991;266:15719–15725. [PubMed] [Google Scholar]
- 20.Neuenschwander PF, Morrissey JH. Roles of the membrane-interactive regions of factor VIIa and tissue factor - the factor VIIa Gla domain is dispensable for binding to tissue factor but important for activation of factor X. J Biol Chem. 1994;269:8007–8013. [PubMed] [Google Scholar]
- 21.McCallum CD, Su B, Neuenschwander PF, Morrissey JH, Johnson AE. Tissue factor positions and maintains the factor VIIa active site far above the membrane surface even in the absence of the factor VIIa Gla domain – a fluorescence resonance energy transfer study. J Biol Chem. 1997;272:30160–30166. doi: 10.1074/jbc.272.48.30160. [DOI] [PubMed] [Google Scholar]
- 22.McCallum CD, Hapak RC, Neuenschwander PF, Morrissey JH, Johnson AE. The location of the active site of blood coagulation factor VIIa above the membrane surface and its reorientation upon association with tissue factor. J Biol Chem. 1996;271:28168–28175. doi: 10.1074/jbc.271.45.28168. [DOI] [PubMed] [Google Scholar]
- 23.Morrissey JH, Neuenschwander PF, Huang Q, McCallum CD, Su B, Johnson AE. Factor VIIa-tissue factor: functional importance of protein-membrane interactions. Thromb Haemost. 1997;78:112–116. [PubMed] [Google Scholar]
- 24.Huang Q, Neuenschwander PF, Rezaie AR, Morrissey JH. Substrate recognition by tissue factor-factor VIIa – evidence for interaction of residues Lys165 and Lys166 of tissue factor with the 4-carboxyglutamate-rich domain of factor X. J Biol Chem. 1996;271:21752–21757. doi: 10.1074/jbc.271.36.21752. [DOI] [PubMed] [Google Scholar]
- 25.Carlsson K, Freskgård P, Persson E, Carlsson U, Svensson M. Probing the interface between factor Xa and tissue factor in the quaternary complex tissue factor-factor VIIa-factor Xa-tissue factor pathway inhibitor. Eur J Biochem. 2003;270:2576–2582. doi: 10.1046/j.1432-1033.2003.03625.x. [DOI] [PubMed] [Google Scholar]
- 26.Kirchhofer D, Eigenbrot C, Lipari MT, Moran P, Peek M, Kelley RF. The tissue factor region that interacts with factor Xa in the activation of factor VII. Biochemistry. 2001;40:675–682. doi: 10.1021/bi002013v. [DOI] [PubMed] [Google Scholar]
- 27.Kirchhofer D, Lipari MT, Moran P, Eigenbrot C, Kelley RF. The tissue factor region that interacts with substrates factor IX and factor X. Biochemistry. 2000;39:7380–7387. doi: 10.1021/bi000182+. [DOI] [PubMed] [Google Scholar]
- 28.Manithody C, Yang L, Rezaie AR. Identification of a basic region on tissue factor that interacts with the first epidermal growth factor-like domain of factor X. Biochemistry. 2007;46:3193–3199. doi: 10.1021/bi6025193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colina CM, Venkateswarlu D, Duke R, Perera L, Pedersen LG. What causes the enhancement of activity of factor VIIa by tissue factor? J Thromb Haem. 2006;4:2726–2729. doi: 10.1111/j.1538-7836.2006.02222.x. [DOI] [PubMed] [Google Scholar]
- 30.Hoffman M, Colina CM, McDonald AG, Arepally GM, Pedersen L, Monroe DM. Tissue factor around dermal vessels has bound factor VII in the absence of injury. J Thromb Haem. 2007;5:1403–1408. doi: 10.1111/j.1538-7836.2007.02576.x. [DOI] [PubMed] [Google Scholar]
- 31.Norledge BV, Petrovan RJ, Ruf W, Olson AJ. The tissue factor/factor VIIa/factor Xa complex: A model built by docking and site-directed mutagenesis. Proteins. 2003;53:640–648. doi: 10.1002/prot.10445. [DOI] [PubMed] [Google Scholar]
- 32.Venkateswarlu D, Duke R, Perera L, Darden TA, Pedersen LG. An all-atom solution-equilibrated model for human extrinsic blood coagulation complex (sTF-VIIa-Xa): a protein-protein docking and molecular dynamics refinement study. J Thromb Haem. 2003;1:2577–2588. doi: 10.1111/j.1538-7836.2003.00421.x. [DOI] [PubMed] [Google Scholar]
- 33.Olsen OE, Rand KD, Ostergaard H, Persson E. A combined structural dynamics approach identifies a putative switch in factor VIIa employed by tissue factor to initiate blood coagulation. Prot Sci. 2007;16:671–682. doi: 10.1110/ps.062504907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harlos K, Martin DMA, O’Brien DP, Jones EY, Stuart DI, Polikarpov I, Miller A, Tuddenham EGD, Boys CWG. Crystal structure of the extracellular region of human tissue factor. Nature. 1994;370:662–666. doi: 10.1038/370662a0. [DOI] [PubMed] [Google Scholar]
- 35.Yves A, Muller MHU, de Vos AM. The crystal structure of the extracellular domain of human tissue factor refined to 1.7 Å resolution. J Mol Biol. 1999;289:103–112. doi: 10.1006/jmbi.1996.0073. [DOI] [PubMed] [Google Scholar]
- 36.Pike ACW, Brzozowski AM, Roberts SM, Olsen OH, Persson E. Structure of human factor VIIa and its implications for the triggering of blood coagulation. Proc Natl Acad Sci USA. 1999;96:8925–8930. doi: 10.1073/pnas.96.16.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murakami MT, Rios-Steiner J, Weaver SE, Tulinsky A, Geiger JH, Arni RK. Intermolecular interactions and characterization of the novel factor Xa exosite involved in macromolecular recognition and inhibition: Crystal structure of human Gla-domainless factor Xa complexed with the anticoagulant protein NAPc2 from the hematophagous nematode Ancylostoma caninum. J Mol Biol. 2007;366:602–610. doi: 10.1016/j.jmb.2006.11.040. [DOI] [PubMed] [Google Scholar]
- 38.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 39.Morrissey JH, Pureza V, Davis-Harrison RL, Sligar SG, Ohkubo YZ, Tajkhorshid E. Blood clotting reactions on nanoscale phospholipid bilayers. Trombosis Research. 2008;122:S23–S26. doi: 10.1016/S0049-3848(08)70014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohkubo YZ, Tajkhorshid E. Distinct structural and adhesive roles of Ca2+ in membrane binding of blood coagulation factors. Structure. 2008;16:72–81. doi: 10.1016/j.str.2007.10.021. [DOI] [PubMed] [Google Scholar]
- 41.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comp Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck J, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher IWE, Roux B, Schlenkrich M, Smith J, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 43.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 44.Feller SE, Zhang YH, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation — the Langevin piston method. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- 45.Martyna GJ, Tobias DJ, Klein ML. Constant pressure molecular dynamics algorithms. J Chem Phys. 1994;101:4177–4189. [Google Scholar]
- 46.Darden T, York D, Pedersen L. Particle mesh Ewald. An N·log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 47.Waxman E, Laws WR, Laue TM, Nemerson Y, Ross JBA. Human factor VIIa and its complex with soluble tissue factor: Evaluation of asymmetry and conformational dynamics by ultracentrifugation and fluorescence anisotropy decay methods. Biochemistry. 1993;32:3005–3012. doi: 10.1021/bi00063a011. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Ohkubo YZ, Tajkhorshid E. Gas conduction of lipid bilayers and membrane channels. chap. 60. In: Feller S, editor. Current Topics in Membranes: Computational Modeling of Membrane Bilayers. Elsevier; 2008. pp. 343–367. [Google Scholar]
- 49.Bach RR. Tissue factor encryption. Arter Thromb Vasc Biol. 2006;26:456–461. doi: 10.1161/01.ATV.0000202656.53964.04. [DOI] [PubMed] [Google Scholar]
- 50.Morrissey JH, Pureza V, Davis-Harrison RL, Sligar SG, Rienstra CM, Kijak AZ, Ohkubo YZ, Tajkhorshid E. Protein-membrane interactions: blood clotting on nanoscale bilayers. J Thromb Haem. 2009;7(S1):169–172. doi: 10.1111/j.1538-7836.2009.03390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.