Abstract
The development of leukemia as a consequence of vector-mediated genotoxicity in gene therapy trials for X-linked severe combined immunodeficiency (SCID-X1) has prompted substantial research effort into the design and safety testing of integrating vectors. An important element of vector design is the selection and evaluation of promoter-enhancer elements with sufficient strength to drive reliable immune reconstitution, but minimal propensity for enhancer-mediated insertional mutagenesis. In this study, we set out to explore the effect of promoter-enhancer selection on the efficacy and safety of human immunodeficiency virus-1-derived lentiviral vectors in γc-deficient mice. We observed incomplete or absent T- and B-cell development in mice transplanted with progenitors expressing γc from the phosphoglycerate kinase (PGK) and Wiscott–Aldrich syndrome (WAS) promoters, respectively. In contrast, functional T- and B-cell compartments were restored in mice receiving an equivalent vector containing the elongation factor-1-α (EF1α) promoter; however, 4 of 14 mice reconstituted with this vector subsequently developed lymphoma. Extensive analyses failed to implicate insertional mutagenesis or γc overexpression as the underlying mechanism. These findings highlight the need for detailed mechanistic analysis of tumor readouts in preclinical animal models assessing vector safety, and suggest the existence of other ill-defined risk factors for oncogenesis, including replicative stress, in gene therapy protocols targeting the hematopoietic compartment.
Introduction
X-linked severe combined immunodeficiency (SCID-X1) is caused by mutations in the IL2RG gene that encodes the common γ-chain (γc), a functionally indispensable subunit of interleukin (IL) receptors 2, 4, 7, 9, 15, and 21 (refs. 1,2). Affected infants have profound defects in cellular and humoral immunity, typically lack T and natural killer (NK) cells, and have normal or elevated numbers of B cells that are unable to undergo immunoglobulin class switching and antibody production.3,4 Failure of T- and NK-cell ontogeny is thought to result from loss of signaling through the receptors for IL-7 and IL-15, respectively.5,6 The treatment of choice is bone marrow transplantation from a human leukocyte antigen–identical sibling donor. Most infants, however, lack a suitable donor and conventionally undergo a human leukocyte antigen–mismatched transplant that is associated with an increased risk of morbidity and mortality.7,8 In many infants, immunological reconstitution remains incomplete, particularly B-cell function, with resultant lifelong requirement for immunoglobulin replacement therapy. Gene therapy offers these infants the potential for improved survival rates and more complete immunological reconstitution without the risk of graft-versus-host disease.
Gene therapy for SCID-X1 has been successfully employed in two related clinical trials with robust immune reconstitution observed in the majority of infants.9,10,11 In these trials, γc expression was driven by the viral long-terminal repeat (LTR) promoter of a MoMLV-based γ-retroviral vector. The profound proliferative capacity of lymphoid progenitors and the selective advantage of transduced γc-expressing progenitors in populating the T- and NK-cell compartments are hypothesized to be the major factors accounting for the success of gene therapy in this disease. The occurrence of vector-mediated insertional mutagenesis, however, in 5 of 20 infants treated12,13 has highlighted the previously underestimated risk of genotoxicity in gene therapy protocols targeting the hematopoietic compartment with integrating vectors. Interestingly, additional genetic abnormalities such as activating NOTCH1 mutations were observed in these patients.12,13 The in vivo expansion of clones containing activating insertions in growth promoting genes has also been observed in two adults in a clinical trial for X-linked chronic granulomatous disease.14 This has led to a major focus on the development of safer vector technology and the equally challenging task of developing preclinical assay systems capable of providing reliable and clinically meaningful readouts of the safety gains achieved.
Although avoidance of integrating vector systems is not currently a viable option for SCID-X1 gene therapy, the prospect of insertional mutagenesis can be significantly reduced by minimizing the risk associated with individual integration events. This is theoretically achievable by the use of vector systems with more favorable integration behavior, such as human immunodeficiency virus-1-derived lentiviral vectors,15 the use of self-inactivating LTRs, and improved expression cassette design, including the avoidance of strong promoter-enhancer elements.16,17 Selection of the latter must balance the need for sufficient γc expression to reliably achieve robust reconstitution of both the T- and NK-cell compartments18 while minimizing the risk of inadvertent enhancer-mediated gene activation at or near integration sites. Optimization of ex vivo transduction conditions may also improve protocol safety.19,20 Objectively quantifying the safety gains achieved by such measures, however, remains challenging, and available assay systems are particularly limited for evaluation of disease-specific risk. Challenges include the need to definitively link oncogenic events to insertional mutagenesis, background rates of tumor formation that limit sensitivity and specificity, and the inclusion of carefully considered and appropriate controls.17,21,22,23,24
In the current study, we set out to evaluate the efficacy and safety of lentiviral vectors containing the promoters from either the elongation factor-1-α (EF1α), phosphoglycerate kinase (PGK), or Wiscott–Aldrich syndrome (WAS) genes to drive expression of the human γc transgene in a murine model of SCID-X1. We observed restoration of lymphoid development and immune function in animals treated with progenitor cells transduced by the EF1α-containing vector. Immune reconstitution was poor in mice receiving cells modified with either the PGK or WAS-containing constructs. Long-term follow-up of mice reconstituted with either IL2RG−/− cells treated with the EF1α-γc vector or C57Bl/6 cells treated with EF1α-EGFP revealed a statistically significant incidence of lymphoma in the former group. In all instances, the malignant clones arose from γc-transduced IL2RG−/− progenitor cells, despite an approximately fourfold excess of untransduced IL2RG−/− cells in the reinfused population. Importantly, however, this correlation between receipt of gene therapy and tumor formation could not be attributed to insertional mutagenesis or γc overexpression, illustrating the significant limitations of murine models in the assessment of vector safety. Thus, these findings point to the existence of other ill-defined risk factors for oncogenesis, such as transduced progenitors being driven to undergo supraphysiological levels of replication (replicative stress), in gene therapy protocols targeting the hematopoietic compartment, and highlight the need for detailed mechanistic analysis of tumor readouts observed in preclinical animal models.
Results
Construction of a modular lentiviral vector expressing the human γc transgene
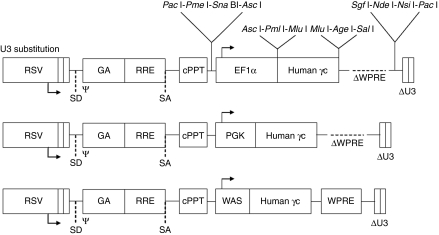
A self-inactivating lentiviral vector expressing the human IL2RG cDNA was constructed based on the previously described pRRLsin18.cPPT.WPRE vector backbone.25 In this construct, Pac I restriction endonuclease sites were introduced to flank the γc expression cassette (Figure 1). Polylinkers containing multiple rare unique sites were also introduced to facilitate sequential “retrofitting” of the construct with additional elements to optimize efficacy and safety in future applications. In this study, the human EF1α, human PGK, and human WAS promoters were selected to drive expression of γc based on their range of transcriptional activities in the ED-7R human T-cell line, with the promoter from the EF1α gene being approximately five- and ninefold more active in this line compared with the promoters from the PGK and WAS genes, respectively.18 In addition, a vector construct containing an EF1α-EGFP expression cassette was made and used as a control to transduce C57Bl/6 progenitor cells.
Figure 1.
Lentiviral vector constructs used in this study. Vectors contained the promoter elements from either the 1,177 base-pair (bp) human elongation factor-1-α (EF1α), 516 bp human phosphoglycerate kinase (PGK), or 481 bp human Wiskott–Aldrich syndrome protein (WAS) genes to drive expression of the human γc cDNA. ψ, packaging and dimerization signal; GA, fragment of the HIV-1 gag gene; cPPT, central polypurine tract; RRE, Rev responsive element; RSV, Rous sarcoma virus hybrid promoter; SD/SA, splice-donor and spice-acceptor sites; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element. The locations of unique and rare restriction endonuclease sites are indicated.
The level of immune reconstitution correlates with promoter strength
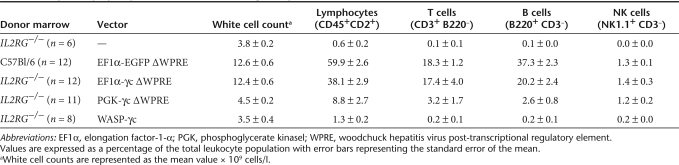
The EF1α-γc, PGK-γc, and WAS-γc lentiviral vectors described above were evaluated for their ability to reconstitute a murine model of SCID-X1. Using an ex vivo gene therapy approach, C57Bl/6 control or IL2RG−/− progenitor cells were vector-treated and transplanted into sublethally irradiated γc−/−Rag2−/−c5−/− recipients. To assess the transduction efficiency of the Sca1+ progenitors, cells were plated in semisolid medium and the presence of proviral DNA determined. For C57Bl/6 progenitors treated with the EF1α-EGFP vector, the transduction efficiency was determined to be 14.2 ± 3.5% (n = 3). This was similar to that for IL2RG−/− progenitors treated with the EF1α-γc vector, where 21.8 ± 2.4% (n = 3) of colonies were found to contain provirus. For Sca1+ progenitors treated with the PGK-γc and WAS-γc vectors, the level of transduction was comparable to that of the EF1α-γc vector and found to be 26.5 ± 3.6 (n = 3) and 32.4 ± 2.5 (n = 3), respectively. Lymphoid reconstitution was examined in peripheral blood samples between 12 and 20 weeks post-transplantation (Table 1). Reconstitution was consistently observed in mice receiving EF1α-EGFP C57Bl/6 control (n = 12) or EF1α-γc IL2RG−/− vector-treated cells (n = 12), although the percentage of B cells was reduced in the latter. Importantly, the levels of T- and NK-cell reconstitution in these cohorts were equivalent (P = 0.20 and 0.54, respectively). In contrast, reconstitution of the T-cell compartment in mice receiving IL2RG−/− cells treated with the PGK-γc construct (n = 11) was incomplete in most animals. Of interest, however, the NK-cell compartment was more reliably reconstituted in the same treatment cohort, achieving values comparable to mice receiving EF1α-EGFP C57Bl/6 control or EF1α-γc IL2RG−/− vector-treated cells (P = 0.83 and 0.81, respectively). Mice receiving WAS-γc-treated IL2RG−/− cells failed to reconstitute their lymphoid compartment (n = 8), and, as expected, lymphoid reconstitution was not observed in negative control mice receiving untransduced IL2RG−/− cells (n = 6).
Table 1.
Peripheral lymphocyte reconstitution following transplantation
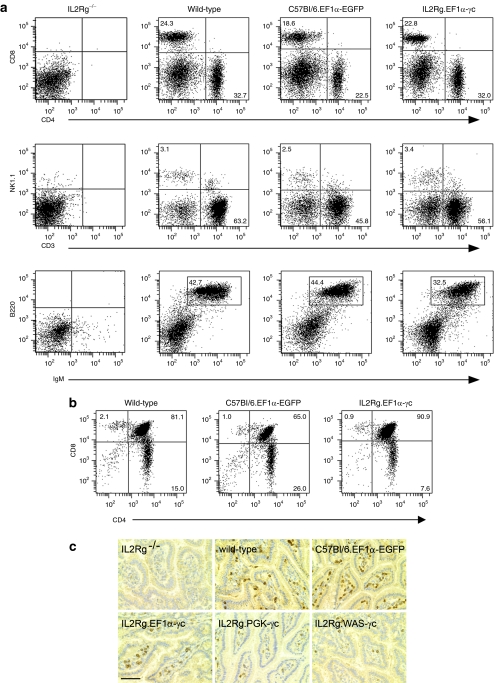
Engrafted mice (EF1α-EGFP C57Bl/6 control or EF1α-γc IL2RG−/− vector-treated cohorts) were killed, and the spleens, thymi, and small intestine assessed for immunological reconstitution. Flow cytometry analysis revealed the presence of T, B, and NK cells in the spleens of transplanted animals (Figure 2a and Table 2) and immature CD4+CD8+ double-positive T cells in the thymus (Figure 2b). In the bulk splenocyte populations from mice receiving EF1α-γc vector-treated IL2RG−/− cells, the vector copy number was low (0.2 ± 0.1 copies per cell, n = 9), consistent with the likelihood of single integration events in the majority of reconstituted lymphocytes. In addition to the correction of lymphoid defects in the spleen and thymus, expression of the γc transgene resulted in reconstitution of intraepithelial lymphocytes in the small intestine (Figure 2c).
Figure 2.
Restoration of lymphocyte populations following lentiviral vector–mediated gene transfer. (a) Splenocytes from transplant recipients were examined by flow cytometry using antibodies against murine B220, CD3, CD4, CD8, IgM, and NK1.1. (b) Thymopoiesis in transplant recipients receiving vector-treated progenitors was examined by flow cytometry using antibodies against CD3, CD4, and CD8. (c) Small intestinal samples were stained for CD3 and revealed intraepithelial lymphocyte development in wild-type mice and recipient mice following gene therapy. Bar = 100 µm.
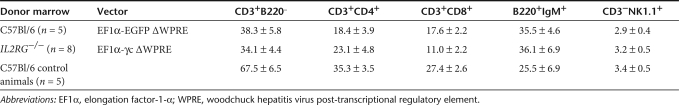
Table 2.
Lymphocyte subset determination in the spleens of mice following transplantation
Restoration of lymphocyte function
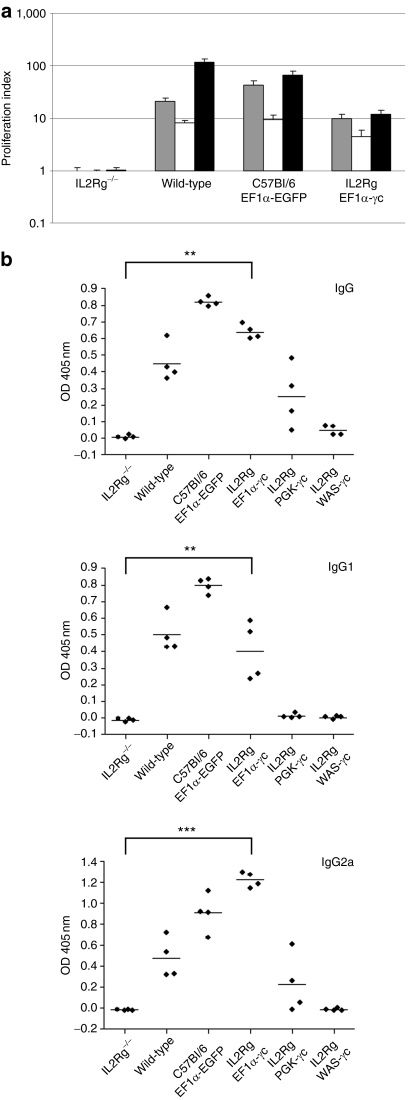
To assess restoration of lymphocyte function, splenocytes from mice transplanted with EF1α-γc vector-treated IL2RG−/− cells were stimulated under conditions to promote T-cell proliferation (Figure 3a). As expected, in the absence of γc expression, no proliferation above baseline values was detected. In mice that had received EF1α-γc vector-treated donor cells, T-cell function was confirmed by an increase in proliferative responses to mitogen stimulation. The level of proliferation, however, in wild-type control mice was higher and correlated with the greater percentage of CD3+ T cells in these animals (Table 2).
Figure 3.
Restoration of immune function following lentiviral vector–mediated gene transfer. (a) Splenocytes from transplant recipients or wild-type mice were stimulated under conditions to promote T-cell proliferation. Proliferating cells were evaluated by the incorporation of [3H] thymidine and expressed as the ratio of counts obtained for stimulated to unstimulated cells. Gray bars, conA alone; white bars, IL-2 alone; black bars, conA and IL-2. (b) Humoral immune responses, indicated by serum IgG, IgG1, and IgG2a levels, were examined in mice transplanted with vector-treated C57Bl/6 or IL2RG−/− progenitors, and compared to γc−/−Rag2−/−c5−/− mice transplanted with untransduced IL2RG−/− cells. Histograms represent the mean value for each group (n = 4) with error bars representing the standard error of the mean. *P < 0.05, **P < 0.01, ***P < 0.0001 (Wilcoxon rank-sum test). EF1α, elongation factor-1-α OD, optical density; PGK, phosphoglycerate kinase; WAS, Wiskott–Aldrich syndrome.
Humoral immunity was examined by measuring the level of circulating plasma immunoglobulins in sera. Prior to transplantation, γc−/−Rag2−/−c5−/− recipient mice lacked detectable immunoglobulin, consistent with defective B-cell development in this model (Figure 3b). In contrast, IgG, IgG1, and IgG2a could be readily detected in sera from wild-type control mice and γc−/−Rag2−/−c5−/− recipients receiving EF1α-EGFP vector-treated C57Bl/6 progenitor cells. Following gene transfer, there was a significant increase in immunoglobulin levels for mice receiving cells treated with the EF1α-γc vector, indicating a restoration of B-cell function. For mice receiving PGK-γc vector-treated IL2RG−/− cells, the levels of circulating immunoglobulins were increased following treatment, although these were variable between individual mice and wild-type levels were not attained. Gene therapy using IL2RG−/− cells treated with the WAS-γc vector failed to restore humoral immunity. Taken together, the results obtained from these studies indicate that the level of γc expression delivered by the EF1α-γc vector is sufficient to restore both T- and B-lymphocyte function in a murine model of SCID-X1. Interestingly, as the level of γc expression is reduced, by using vectors containing weaker promoters, levels of reconstitution in the T- and B-cell compartments appear to be most affected with NK reconstitution retained at lower γc expression levels.
Reconstituted mice displayed a diverse T-cell repertoire
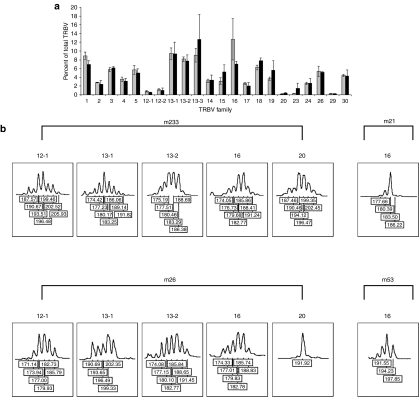
To examine the relative repertoire diversity of lymphocyte reconstitution between mice that had received either C57Bl/6 control or IL2RG−/− progenitor cells treated with the EF1α-EGFP or EF1α-γc vectors, respectively, T-cell receptor (TCR)Vβ repertoire and spectratype analyses on selected highly expressed TCRVβ families were performed on splenocytes between 6 and 18 months post-transplantation. Analysis of TCRVβ repertoire demonstrated that the percentage of each family was equivalent between the two groups (Figure 4a). Spectratype analysis revealed normal Gaussian distribution of peaks for the majority of highly expressed TCRVβ families investigated with an occasional family exhibiting a degree of oligoclonality with skewing and reduction in the number of peaks, irrespective of treatment group (Figure 4b).
Figure 4.
Analysis of T-cell receptor Vβ repertoire and CDR3 spectratyping in splenocytes from mice following lentiviral vector–mediated gene transfer. (a) Splenocytes were isolated from mice between 6 and 18 months post-transplantation, and Vβ repertoire analysis was performed by quantitative RT-PCR. The percentage of each Vβ family is indicated with results representing the mean of triplicate values for each group (n = 3) with error bars representing the SEM. Gray bars, C57Bl/6 control group; black bars, EF1α-γc treatment group. (b) CDR3 spectratypes of TCRVβ 12-1, 13-1, 13-2, 16, and 20 families in reconstituted mice receiving either C57Bl/6 control or IL2RG−/− progenitor cells treated with the EF1α-EGFP or EF1α-γc vectors (m233 and m26, respectively). Normal Gaussian distribution (6–11 peaks each separated by three nucleotides) was observed for TCRVβ 12-1, 13-1, 13-2, 16, and 20 in m223 and TCRVβ 12-1, 13-1, 13-2, and 16 in m26. Restricted CDR3 spectratyping of TCRVβ 16 in m21 (C57Bl/6 control progenitor cells treated with the EF1α-EGFP vector) and TCRVβ 20 in m26 (IL2RG−/− progenitor cells treated with the EF1α-γc vector) was observed. EF1α, elongation factor-1-α TRBV, T-cell receptor Vβ.
Hematopoietic malignancies were observed in transplanted mice
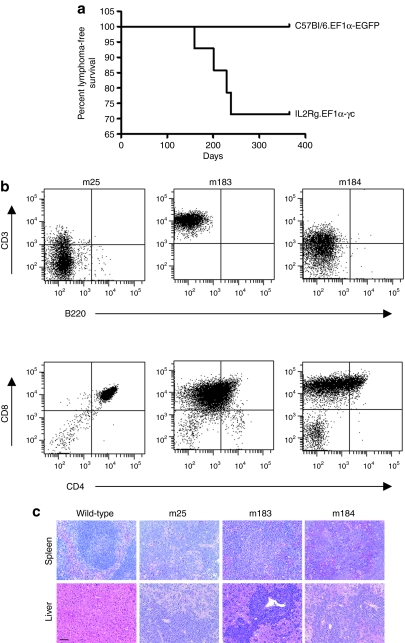
Cohorts of reconstituted mice that had received either C57Bl/6 control or IL2RG−/− progenitor cells, treated with the EF1α-EGFP or EF1α-γc vectors, respectively, were maintained for long-term observation. Between 5 and 9 months post-transplantation, lymphomas were detected exclusively in mice from the EF1α-γc treatment group. Of the 14 mice that had received EF1α-γc vector-treated IL2RG−/− cells, four developed malignancy (28.6%). This incidence was significantly different to the EF1α-EGFP vector-treated C57Bl/6 control group (n = 24) using a Fisher's exact test (P = 0.0273). In addition, there was a significant difference between the survival rates of these two groups (P = 0.0134, Figure 5a). FACS analysis revealed abnormal T-cell populations in three mice (m25, m183, and m184; Figure 5b) that were identified in the spleen, liver, thymus, kidney, bone marrow, and peripheral blood. White blood cell counts were also found to be elevated in these animals. Necropsy revealed hepatosplenomegaly with extensive blast infiltration, and histological analysis revealing marked disruption of tissue architecture in the liver and spleen (Figure 5c). The phenotype of the lymphoma from a fourth mouse (m45) could not be determined due to a lack of viable sample.
Figure 5.
Phenotypic characterization of lymphomas. (a) Kaplan–Meier survival analysis at 1 year post-transplantation. The percent lymphoma-free survival was significantly higher (P = 0.0134) for mice receiving C57Bl/6 progenitors treated with the EF1α-EGFP vector (n = 19) when compared to mice receiving EF1α-γc vector-treated IL2RG−/− cells (n = 14). (b) Malignant blasts, isolated from primary animals, were stained with antibodies against murine CD3, CD4, CD8, and B220, and analyzed by flow cytometry. (c) Hematoxylin and eosin staining revealed effacement of much of the normal splenic architecture by proliferating lymphoblastic cells (top panel) with marked hepatic infiltration by metastatic lymphoblasts (bottom panel) in mice from the EF1α-γc treatment group. Bar = 100 µm. EF1α, elongation factor-1-α.
The level of γc expression in malignant splenocytes was determined by quantitative RT-PCR using the comparative Ct method. In comparison to human CD3+ peripheral blood mononuclear cells, the level of γc expression was lower for all three samples. In addition, the level of γc expression was reduced in the malignant samples when compared to a mouse reconstituted with cells treated with the same vector (not showing signs of disease) being 0.86-, 0.66-, and 0.69-fold for m25, m183, and m184, respectively. This implies that transgene overexpression was not responsible for the observed hematopoietic malignancies. In addition, γc-dependent signal transduction, as determined by STAT5 phosphorylation, was not detected in malignant splenocytes following stimulation with cytokines IL-2, IL-7, or IL-15 (data not shown). Finally, no upregulation of JAK3 was observed by microarray analysis (http://pwbc.garvan.unsw.edu.au/caarray/project/ginn-00050) in the malignant cells when compared to CD3+ splenocytes isolated from a wild-type mouse. Collectively, these data suggest that the cellular expansions observed in at least three of the four mice were independent of γc overexpression.
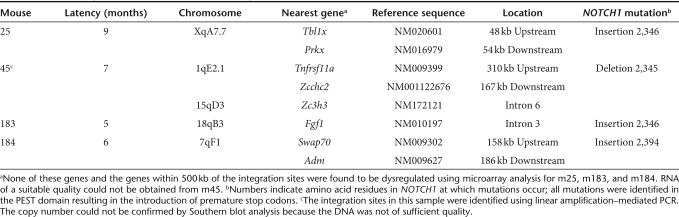
Molecular analysis revealed that integrated provirus was present in all four malignant clones, despite the significant numerical excess of unmarked IL2RG−/− progenitors in the vector-exposed cell populations transplanted into γc−/−Rag2−/−c5−/− recipients. Also of relevance, no tumors were seen in a small cohort of six mice transplanted with untransduced IL2RG−/− progenitor cells maintained for long-term observation (14.5 ± 2.0 months), further supporting transduction with the γc vector as a prerequisite for lymphoma development in the model system used. Southern blot analysis revealed single vector integrations in splenocytes from m25, m183, and m184 that were identified by linear amplification–mediated PCR (Table 3). Quantitative PCR confirmed the presence of single vector integration sites in the affected organs. The integration site for m183 was in reverse orientation and ~30.9 kb from the transcription start site of the fibroblast growth factor 1 gene (Fgf1), whereas the integration sites in the other two samples were not located near genes (>150 kb away). In addition, two sites were identified in genomic DNA from the spleen of m45. The copy number was again confirmed by quantitative PCR but could not be determined by Southern blot analysis due to the poor quality of the sample. Where viable splenocytes could be obtained from mice with the primary tumor, adoptive transfers were performed into nonirradiated γc−/−Rag2−/−c5−/− recipients. This consistently resulted in an equivalent disease phenotype in the recipients and recovery of identical integration sites. Finally, reverse transcriptase activity could not be detected in lysates from the malignant cells above background levels (data not shown), suggesting that the malignant transformation was not due to activation of an endogenous retrovirus in the recipient mice.
Table 3.
Vector integration sites recovered from malignant clones
To determine whether the pattern of gene expression around the lentiviral integration sites were altered, malignant splenocytes were compared to CD3+ splenocytes sorted from a wild-type animal. Importantly, microarray analysis revealed no significant changes in the pattern of gene expression between the normal CD3+ splenocytes and the leukemic clones, or between any of the leukemic samples for over 500 kb from the integration site in each direction (Table 3, http://pwbc.garvan.unsw.edu.au/caarray/project/ginn-00050). This suggests that the observed lymphomas were not the result of insertional mutagenesis and, furthermore, that the EF1α-containing lentiviral vector used did not alter nearby gene expression upon integration. Interestingly, the subset of genes exhibiting the highest levels of expression showed a high degree of commonality across the three malignant splenocyte samples examined, raising the possibility of a common transformation mechanism.
Finally, to investigate the presence of activating NOTCH1 mutations in the malignant clones, an occurrence previously observed in some SCID-X1 patients with insertional oncogenesis,12,13 DNA sequence analysis was performed across the HD, TAD, and PEST domains. Interestingly, activating NOTCH1 mutations were identified in the PEST domain from all four malignant clones (Table 3). These frameshift mutations are consistent with others identified in murine lymphomas and result in the introduction of premature stop codons with associated loss of the PEST domain.26,27 Microarray analysis also confirmed that the NOTCH1 downstream effector, HES1, was upregulated in splenocytes from m25, m83, and m184 in comparison to CD3+ splenocytes from a wild-type mouse by 6.1-, 6.6-, and 8.0-fold, respectively (http://pwbc.garvan.unsw.edu.au/caarray/project/ginn-00050).
Discussion
In two related clinical trials for SCID-X1, gene therapy successfully restored both cellular and humoral immunity in the majority of infants treated.9,10,11 However, the development of lymphoproliferative disease in 5 of 20 patients,12,13 that could be directly linked to the gene transfer technology used, highlighted the need for improved vector design. In particular, removal of promoter-enhancer elements located within the viral LTRs has been the focus of recent attention.16,17 In the current study, we sought to evaluate the relative efficacy and safety of self-inactivating lentiviral vectors expressing the human γc cDNA under the control of three different internal heterologous cellular promoters of varying transcriptional activity (human EF1α, PGK, or WAS) in a murine model of SCID-X1.
Our in vivo studies demonstrated that IL2RG−/− progenitor cells treated with a lentiviral vector using the promoter from the EF1α gene to drive expression of γc conferred robust and sustained correction of both cellular and humoral immunity. Importantly, the level of immune reconstitution observed was similar to that of mice receiving C57Bl/6 control cells. This result is consistent with other studies reporting high-level gene expression in hematopoietic stem cells using vectors containing this promoter.16,28,29 The promoter from the EF1α gene has also been reported to display a reduced propensity for insertional gene activation,30 further supporting the use of this promoter in future trials for diseases targeting the hematopoietic compartment. Similarly, the promoter from the PGK gene has been shown to display reduced levels of gene activation when compared to promoter sequences of γ-retroviral vectors.22,30,31 In this study, however, when an equivalent lentiviral vector containing the promoter from the PGK gene was used, levels of γc expression were insufficient to achieve consistently robust immune reconstitution in immunodeficient recipients. Similar results have been observed with retroviral vectors using the promoter from PGK gene to drive expression of O6-methylguanine DNA methyltransferase where selection of transduced cells could not be achieved with cells containing single integration events.32,33 Of note, the levels of reconstitution achieved in this study were highly variable between animals receiving PGK-γc-treated progenitors, possibly reflecting a greater dependence on favorable integration sites for γc expression. Interestingly, in mice receiving PGK-γc-treated progenitors, in comparison to the T- and B-cell compartments, there appeared to be preferential reconstitution of NK cells, a result consistent with limiting γc expression, as demonstrated in earlier in vitro studies performed in the ED-7R human T-cell line and SCID-X1 patient cells.18 Finally, mice transplanted with cells exposed to WAS-γc-treated progenitors failed to reconstitute, almost certainly the result of the low level of γc expression conferred by this vector.
Mice receiving either EF1α-γc IL2RG−/− or EF1α-EGFP C57Bl/6 vector-treated progenitor cells were maintained for long-term observation to monitor for signs of genotoxicity-related to lentiviral-mediated gene transfer. We observed a high incidence of T-cell lymphomas in mice receiving the EF1α-γc vector-treated progenitors (28.6%) that was significantly greater than the control group, where no cases of lymphoma were observed. The absence of lymphoma development in the control animals indicates that these events were not due to intrinsic properties of the model used, including the effects of irradiation or the background strain, both of which have been linked to malignancies observed previously in other murine models.24,34,35,36 In addition, the lack of lymphoma development in control mice would argue against the mechanistic involvement of reduced tumor immune surveillance prior to full immunological reconstitution. The observation of lymphoma development exclusively from the EF1α-γc transduced fraction of IL2RG−/− progenitors, and not the approximately fourfold more abundant untransduced fraction, strongly implies a requirement for transduction as a prerequisite for subsequent lymphoma development rather than malignant transformation being an intrinsic property of IL2RG−/− progenitor cells. Further supporting this conclusion, lymphoma development was not observed in a cohort of mice, albeit small, receiving untransduced IL2RG−/− progenitor cells. Finally, the T-lymphoid immunophenotype of the lymphomas examined implies that the progenitors from which they arose had progressed along a γc-dependent ontological pathway. This also supports a requirement for transduction.
Activation of endogenous retroviruses has also been proposed in recipient animals following irradiation conditioning.24 This possibility was precluded by the demonstrated lack of reverse transcriptase activity in the malignant clones. Given the occurrence of five cases of insertional mutagenesis in human SCID-X1 trials, we next established whether gene expression around the site of integration was altered in the malignant cells. From the four malignant samples, five integration sites were identified by linear amplification–mediated PCR; three were intergenic and not located near genes, with two being intragenic and located in intronic sequences. Importantly, in the three cases where samples of suitable quality could be obtained, microarray analysis demonstrated that there was no change in gene expression around the site of integration. This implies that, despite the high rate of lymphomagenesis observed, the lentiviral vector used in this study did not perturb expression of cellular genes upon integration, in contrast to a recent report using a lentiviral vector containing β-globin enhancer sequences.37
The possibility that the γc transgene may itself be oncogenic has also been suggested.21 In this study, the authors observed significantly more T-cell lymphomas in animals receiving cells treated with a lentiviral vector expressing γc from a strong hybrid CMV promoter containing the chicken actin enhancer (CAG) than immunodeficient animals receiving wild-type cells treated by mock transduction or an equivalent lentiviral vector expressing GFP. The authors concluded that, because no lymphomas developed in mice receiving cells treated with their GFP-expressing vector, constitutive overexpression of γc in hematopoietic cells, and not viral transduction, was likely to be the principal oncogenic determinant in the generation of T-cell lymphomas. Molecular analyses supporting this conclusion were not presented. We observed a similar rate of lymphomagenesis in our study, though in contrast, both quantitative RT-PCR and microarray analyses demonstrated that the level of γc expression was lower in the malignant clones when compared to both human CD3+ peripheral blood mononuclear cells or an animal reconstituted with cells treated with the same vector not showing signs of disease. This implies that the level of γc expression is unlikely to have resulted in the development of lymphoma, at least in our study. In support of the lack of γc involvement in our study, overexpression of the γc transgene has been shown to have no effect on T-cell development in a fetal thymus organ culture model.38 It seems more likely that the presence of gene marking in the malignant cells is indicative of T-cell development, without which there could be no T-cell lymphomas. In further support of the lack of γc involvement in mediating the oncogenic transformation, no upregulation of JAK3 was observed in the malignant splenocytes that were also unable to undergo signal transduction in response to the γc-dependent cytokines IL-2, IL-7, and IL-15. In support of our findings, transgenic mice expressing human γc under the control of the promoter from the CD2 gene and locus control region have been reported to show no increased predisposition to tumor development, even in tumor-prone models.39
Although it has been suggested that γc-deficient mice display an increased susceptibility to lymphomagenesis,23,39 tumor development has not been reported in the absence of manipulation. Using the tumor-prone Arf knockout mouse model, Shou and colleagues reported that the frequency of T-cell malignancy was significantly increased in γc-deficient mice reconstituted with Arf−/−/γc−/− compared to Arf−/−/γc+/+ donor cells transduced with a γc-encoding murine stem cell virus.23 Integration site analysis in a substantial proportion of the malignant clones arising provided evidence of insertional mutagenesis involving proto-oncogene loci. The authors concluded that the γc-deficient phenotype of the donor cells, transduction with a γc-encoding vector, and vector integration events near proto-oncogenes were critical elements of the increased incidence of tumorigenesis observed. Combined with the exclusion of γc overexpression, our observations support and extend these findings by clearly demonstrating that vector-mediated correction of the γc-deficient phenotype in mice can induce lymphoid malignancy by mechanisms that do not involve insertional mutagenesis. Although the possible contribution of replicative stress was recognized, its potential significance in tumorigenesis, at least in mice, is heightened by our observations.
The development of lymphoma is widely accepted as a “multistep” transformation process requiring the accumulation of multiple genetic changes. In mice, the estimated mutation rate is 4 × 10−8 mutations per cell division at the Hprt locus.40 Thus, it is plausible that immune reconstitution, when achieved from a reduced number of progenitor cells, necessitating higher rates of proliferation, could increase in the number of stochastic mutations acquired by individual cells during lymphoid ontogeny. A similar phenomenon has been observed clinically in patients undergoing hematopoietic regeneration where an increased risk of therapy-related myleodysplasia and acute myeloid leukemia has been observed following hematopoietic cell transplantation.41,42 It is interesting to speculate that, in studies attempting to define hematopoietic stem cell populations, the transplanted cells would have been under conditions of replicative stress. In the majority of cases, however, animals were either not maintained for long-term analysis or transplanted in the presence of carrier cells that included committed progenitors. In our study, every C57Bl/6 progenitor cell used to reconstitute control animals would have expressed wild-type levels of γc. In contrast, for mice receiving IL2RG−/− progenitors treated with the EF1α-γc vector, only the subset of cells successfully transduced, and with integration sites favoring sufficient γc expression, would have had the potential for lymphoid ontogeny. This subset of progenitor cells would therefore have been under increased proliferative pressure to achieve equivalent levels of lymphoid reconstitution. This could in turn have been associated with sufficiently high rates of stochastic mutation to drive lymphoma development. Long-term studies of γc-deficient mice reconstituted with limiting numbers of wild-type cells are underway to address this hypothesis. Such studies will also help elucidate the possible contributory role of abnormal lymphoid precursor populations described in γc-deficient mice.23
Extending the implications of replicative stress, any vector manipulation that reduces the efficiency of hematopoietic progenitor transduction and subsequent transgene expression, such as pseudotyping with a less efficient envelope protein or the use of a weaker promoter, could act as a risk factor for lymphoid malignancy. This in turn could result in misinterpretation of tumorigenic readouts in preclinical studies using mice intended to evaluate vector insertional mutagenic potential. A related consequence of reconstitution with limiting number of progenitor cells is oligoclonal reconstitution. Recently reported murine studies suggest that monoclonal T-cell populations develop malignancies with a greater propensity than their polyclonal counterparts, suggesting that polyclonal populations are more able to control the outgrowth of malignant T-cell clones.43 In our study, TCRVβ repertoire and spectratype analyses demonstrated a broad level of T-cell diversity between the two groups. Although this implies that the development of lymphoma was not due to the presence of a more clonal T-cell population in mice receiving IL2RG−/− progenitors treated with the EF1α-γc vector, it cannot be excluded in the mice that went on to develop malignancies.
In summary, we have demonstrated long-term efficacy and stability of lentiviral vector-mediated gene transfer of human γc, when expressed from the EF1α promoter, in a murine model of SCID-X1. Notably, functional T-and B-cell compartments were fully restored in the recipient animals, though the level of immune reconstitution diminished correspondingly when weaker promoters were used. In IL2RG−/− mice from the EF1α-γc treatment group maintained for long-term analysis, T-cell lymphomas developed following gene therapy. Importantly, the observation of malignancy appeared not to involve γc overexpression or insertional mutagenesis despite an absolute requirement for γc transduction. Although the underlying mechanism remains to be elucidated, there was a consistent correlation between mice receiving gene therapy and reconstitution from a substantially smaller number of progenitor cells. This raises the possibility of replicative stress as a risk factor for lymphomagenesis, and highlights the need for careful analysis and interpretation of tumorigenic readouts in preclinical animal models, which may not necessarily correlate with vector safety.
Materials and Methods
Lentiviral vector construction and virus production. The lentiviral vector constructs used in this study were derived from pRRLsin18.cPPT.CMV.EGFP.WPRE and have been described previously.18 These vectors contained the promoter elements from either the 1,177 base-pair (bp) human EF1α gene (Invitrogen, Carlsbad, CA), the 516 bp human PGK gene or the 487 bp primary WAS protein gene to drive expression of the human γc cDNA (GenBank accession number L19546) or EGFP (Clontech, Mountain View, CA). Vectors containing the promoter elements from either the EF1α or PGK genes lacked the woodchuck hepatitis virus post-transcriptional regulatory element (WPRE). Vector stocks were produced by using a four-plasmid transfection protocol as previously described44 and concentrated using two rounds of ultracentrifugation. Transduction titers, assigned by detection of integrated provirus, were determined on HEK293 cells in the presence of 8 µg/ml Polybrene (Sigma-Aldrich, St Louis, MO) by using quantitative PCR and previously published primer and probe sequences.45 Reactions were performed on a RotorGene 2000 (Corbett Research, Mortlake, Australia).
Animals. IL2RG−/− mice46 were obtained from The Jackson Laboratory (Bar Harbor, ME), γc−/−Rag2−/−c5−/− mice16 were kindly provided by Richard Lock (Children's Cancer Institute, Sydney, Australia), and C57Bl/6 mice were obtained from the Animal Resources Centre (ARC, Perth, Australia). All experimental procedures were approved by the Animal Care and Ethics Committee of the Children's Medical Research Institute and The Children's Hospital at Westmead.
Bone marrow isolation, lentiviral transduction and transplantation. Bone marrow was harvested from IL2RG−/− or C57Bl/6 mice, and Sca1+ progenitor cells isolated using the EasySep SCA1 positive selection kit (StemCell Technologies, Vancouver, British Columbia, Canada). Cells were plated in StemSpan SFEM serum-free media (StemCell Technologies) supplemented with 50 U/ml penicillin, 50 mg/ml streptomycin (Gibco, Grand Island, NY), 100 ng/ml mSCF (R&D Systems, Minneapolis, MN), 100 ng/ml hFlt-3 (R&D Systems), 100 ng/ml hIL-11 (R&D Systems), and 20 ng/ml mIL-3 (R&D Systems). Cells were transduced overnight with lentiviral vectors at a multiplicity of infection of 50. The following day, recipient γc−/−Rag2−/−c5−/− mice were exposed to sublethal (6 Gy) irradiation using a Gammacell 40 Exactor (MDS Nordion, Ottawa, Ontario, Canada). Vector-treated Sca1+ progenitor cells were washed, resuspended in RPMI 1640 medium (Gibco) and injected intravenously into recipient mice via the tail vein. To assess the transduction efficiency of vector-treated progenitors, cells were plated in MethoCult GF M3434 methylcellulose medium (StemCell Technologies) and incubated for 10–14 days. Colonies were harvested, washed in PBS−/− and lysed in 20 µl lysis buffer (50 mmol/l KCl, 10 mmol/l Tris-HCl, 2 mmol/l MgCl2, 0.45% (vol/vol) NP40, 0.45% (vol/vol) Tween-20, and 1 mg/ml proteinase K) at 37 °C. The following day, the lysate was heated to 95 °C for 10 minutes and 5 µl used in a PCR to confirm the presence of proviral sequence. The presence of amplifiable DNA was confirmed using primers to the titin gene (forward 5′-AAAACGAGCAGTGACGTGAGC-3′ and reverse 5′-TTCAGTCATGCTGCTAGCGC-3′).16
Flow cytometric analysis. Cells from the spleen, thymus, bone marrow, liver, and peripheral blood were stained and analyzed on a FACSCanto (BD Biosciences, San Jose, CA) running FACSDiva software (version 5.01). Lymphocytes were identified and gated based on their FSC and SSC profiles. Cell suspensions were treated with ACK lysis buffer (150 mmol/l NH4Cl, 10 mmol/l KHCO3, and 0.1 mmol/l disodium EDTA) and then stained with the following conjugated antibodies: B220-FITC (BD Biosciences), CD2-PE (Caltag Laboratories, Burlingame, CA), CD3ε-PerCP-Cy5.5 (BD Biosciences), CD4-FITC (Caltag Laboratories), CD8a-APC (Caltag Laboratories), CD11b-PE (BD Biosciences), CD45-Tri-Color (Caltag Laboratories), IgM-FITC (Caltag Laboratories), NK1.1-APC (Caltag Laboratories), and TCR γδ-PE (Caltag Laboratories).
Histology. Tissue was dissected from C57Bl/6 control or transplanted γc−/−Rag2−/−c5−/− recipient mice and fixed in 10% buffered formalin (Sigma-Aldrich). Paraffin-embedded sections (5 µm) were stained with hematoxylin and eosin, and evaluated by light microscopy. In addition, intestinal intraepithelial lymphocytes were identified by positive CD3 staining with slides independently scored by the Institute of Medical and Veterinary Science (IMVS, Adelaide, Australia). Images were captured from a Zeiss Axio Imager.A1 Microscope (Carl Zeiss MicroImaging, Göttingen, Germany) using a Spot enhanced camera (Diagnostic Instruments, SciTech, Preston, Australia) running Spot Software version 4.0.1.
Splenocyte proliferation. Splenocytes were seeded in triplicate (3 × 105 cells/well) in 96-well U-bottom plates (BD Biosciences) with RPMI 1640 medium containing 20% (vol/vol) FCS (HyClone, South Logan, UT) and 50 µmol/l 2-mercaptoethanol (Sigma-Aldrich). Splenocytes were stimulated for 48 hours with or without the addition of 4 µg/ml concanavalin A (Sigma-Aldrich), murine IL-2 (R&D Systems), or both. Splenocytes were then pulsed with 1 µCi/well [3H]-thymidine (PerkinElmer, Waltham, MA) for 18 hours, harvested, and incorporated radioactivity determined using a TopCount NXT scintillation counter (PerkinElmer). Results are expressed as a proliferative index, the ratio of counts obtained for simulated to unstimulated cells.
Enzyme-linked immunosorbent assay detection of serum Ig. Serum was obtained from mice at the time of killing by cardiac puncture. Serum IgG, IgG1, or IgG2a levels were measured using an enzyme-linked immunosorbent assay as described previously.16 Briefly, nonsaturating serum dilutions were added to 96-well Maxisorp Nunc-Immuno plates (Nunc, Roskilde, Denmark) precoated with IgG (AbD Serotec, Kidlington, UK), IgG1 (BD Biosciences), or IgG2a (BD Biosciences) capture antibodies. Bound IgGs were quantified using horseradish peroxidase–conjugated antibodies and the enzyme-linked immunosorbent assay DuoSet substrate reagent pack (R&D Systems). The absorbance of each plate was read at 405 nm using a VERSAmax microplate reader (Molecular Devices, Sunnyvale, CA) running SoftMax Pro 4.7.1.
Quantitative PCR. Genomic DNA was isolated using the Puregene genomic DNA purification kit (Gentra Systems, Minneapolis, MN) and proviral copy number determined by quantitative PCR using previously published primer and probe sequences.47 Reactions were performed on a RotorGene 2000 (Corbett Research) and standardized using primers to the murine titin gene (probe 5′-HEX-TGCACGGAAGCGTCTCGTCTCAGTC-BHQ-3′) and the comparative Ct method. Alternatively, to determine the level of γc expression, RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA), and quantitative RT-PCR performed using the SuperScript III Platinum One-Step qRT-PCR System (Invitrogen).
Analysis of TCR repertoire and CDR3 spectratyping. Individual TCRVβ families in splenocytes from transplanted mice were detected using quantitative RT-PCR and primers specific for the TCR β-chain.48 All analyses were performed in triplicate as previously described.49 The TCRVβ repertoire diversity was subsequently analyzed by CDR3 spectratyping as outlined previously.48
Analysis of viral integration sites. To identify the number and location of viral integration sites, genomic DNA was isolated from splenocytes using the Puregene genomic DNA purification kit. The number of viral integration sites in malignant clones was determined by Southern blot analysis using standard protocols. Genomic DNA (10 µg) was digested with the restriction endonuclease XbaI, separated on a 0.8% (wt/vol) agarose gel and transferred to a Biodyne B nylon membrane (Pall, Pensacola, FL). Blots were hybridized at 65 °C with an 826 bp α [32P]-labeled probe to the γc transgene.
Linear amplification–mediated PCR was performed as described previously50 using 100 ng starting template. The biotinylated primer LTR1 (5′-AGGGTCTGAGGGATCTCTAGTTAC-3′) was used for the linear preamplification of the vector-genome junctions. Following magnetic capture and random hexanucleotide priming, the products were digested with the restriction endonucleases CviA II, Aci I, or HinP1 I, and a linker cassette (5′-GTGTGACCCGGGAGATCTGAATTCAGTGGCACAGCAGTTAGGC-3′ and 5′-ATGCCTAACTGCTGTGCCACTGAATTCAGATCTCCCGGGTCAC-3′) ligated at the 5′ termini of the genomic sequence. Two rounds of exponential PCR were then performed (LTR2 5′-CAGTGGGTTCCCTAGTTAGC-3′ and LTR3 5′-GCAAAAAGCAGATCTTGTCTTC-3′) and linear amplification–mediated PCR amplicons cloned into the TOPO TA vector (Invitrogen) and sequenced (ABI PRISM 3100 Genetic Analyzer; Applied Biosystems, Foster City, CA). Integration sites were aligned to the murine genome using the University of California–Santa Cruz (UCSC) BLAT genome browser (http://genome.ucsc.edu).
Detection of reverse transcriptase activity. Samples were tested for the presence of reverse transcriptase activity using the EnzChek Reverse Transcriptase Assay Kit (Molecular Probes, Eugene, OR) containing the PicoGreen dsDNA quantitation reagent. Cell pellets were resuspended in disruption buffer,51 and a standard curve using a dilution series of M-MLV reverse transcriptase (Promega, Madison, WI) was prepared in the same buffer with the limit of detection determined to be 0.001 U. At the completion of the reaction, the level of fluorescence in each sample was determined by the Victor3V Multilabel Counter (PerkinElmer) using standard fluorescein wavelengths.
Analysis of STAT5 phosphorylation in malignant splenocytes. Detection of STAT5 phosphorylation was used to assess γc-dependent signal transduction as previously described.18 Briefly, cells were washed in RPMI 1640 medium and incubated overnight. The following day, cells were incubated with IL-2, IL-7, or IL-15 (all at 10 nmol/l) (R&D Systems) for 20 minutes followed by chilling to 4 °C to halt cytokine activation. Cells were then cytospun and fixed onto prepared slides and labeled with rabbit anti-pSTAT5 (Cell Signaling, Genesearch, Arundel, Australia). Nuclei were counterstained with 4-6-diamidino-2-phenylindole (Sigma-Aldrich). Cells, positive or negative for pSTAT5, were identified using a BX51 Olympus Microscope (Olympus, Mount Waverley, Australia).
Microarray analysis of gene expression. RNA for expression analysis was isolated using the RNeasy Mini Kit (Qiagen), and whole genome expression profiling was performed by the Australian Genome Research Facility (The Walter and Eliza Hall Institute of Medical Research, Parkville, Australia). Briefly, total RNA was labeled using the Total Prep RNA amplification kit (Ambion, Austin, TX), and the quantity of labeled product determined using a Bioanalyser 2100 and the NanoChip protocol (Agilent Technologies, Forest Hill, Australia). A total of 1.5 µg of labeled cRNA was then prepared for hybridization to the Mouse Sentrix-6 V1.1 BeadChip (Illumina, San Diego, CA) array by preparing a probe cocktail (cRNA at 0.05 µg/µl) that includes GEX-HYB Hybridization Buffer (Illumina). A total hybridization volume of 30 µl was prepared for each sample and loaded into a single array on the Beadchip. Hybridization was performed at 58 °C for 16 hours in an oven with a rocking platform. After hybridization, the chip was washed, coupled with Cy3 and scanned in the BeadArray reader operating BeadStudio software (Illumina). The level of gene expression for each malignant sample was analyzed within a 500 kb window on either side of the vector integration site using microarray data, where it was anticipated that insertional dysregulation was most likely to occur.37 Gene networks were identified using the Ingenuity Pathway Analysis (IPA) software package (Ingenuity Systems, Redwood City, CA).
Identification of NOTCH1 gene mutations. Mutations within the proline, glutamate, serine, and threonine (PEST), transactivation (TAD), or heterodimerization (HD) domains of NOTCH1 were identified by sequencing PCR products amplified using Pfu DNA polymerase (Promega). Primers covering exons 26, 27, and 34, where the HD, TAD, and PEST domains are located, are described elsewhere.26
Statistical analysis. Results were analyzed by the Wilcoxon rank-sum (Mann–Whitney) (http://elegans.swmed.edu/~leon/stats/utest.html) or Fisher's exact test (http://www.exactoid.com/fisher/index.php). For comparison of population survival, cohorts of mice were compared using Kaplan–Meier analysis. Results considered significant when P < 0.05 and values represent the mean ± SEM.
Acknowledgments
We thank Inder Verma (Salk Institute, San Diego, CA) for providing lentiviral vector reagents and Jeff Crosbie (Centenary Institute, Sydney, Australia) for assistance with the Gammacell 40 Exactor. We also thank Christine Smyth for performing STAT5 phosphorylation assays. Finally, we thank Daniel Catchpoole (The Children's Hospital at Westmead, Sydney, Australia) and Warren Kaplan (Peter Wills Bioinformatics Centre, Garvan Institute of Medical Research, Sydney, Australia) for help with MIAME compliance and Grant Logan (Children's Medical Research Institute, Sydney, Australia) for critical review of the manuscript. A.J.T. is a Wellcome Trust Senior Clinical Fellow and S.L.G. is the recipient of a fellowship honoring the memory of Noel Dowling.
REFERENCES
- Sugamura K, Asao H, Kondo M, Tanaka N, Ishii N, Nakamura M, et al. The common gamma-chain for multiple cytokine receptors. Adv Immunol. 1995;59:225–277. doi: 10.1016/s0065-2776(08)60632-x. [DOI] [PubMed] [Google Scholar]
- Asao H, Okuyama C, Kumaki S, Ishii N, Tsuchiya S, Foster D, et al. Cutting edge: the common gamma-chain is an indispensable subunit of the IL-21 receptor complex. J Immunol. 2001;167:1–5. doi: 10.4049/jimmunol.167.1.1. [DOI] [PubMed] [Google Scholar]
- Noguchi M, Nakamura Y, Russell SM, Ziegler SF, Tsang M, Cao X, et al. Interleukin-2 receptor gamma chain: a functional component of the interleukin-7 receptor. Science. 1993;262:1877–1880. doi: 10.1126/science.8266077. [DOI] [PubMed] [Google Scholar]
- Buckley RH, Schiff RI, Schiff SE, Markert ML, Williams LW, Harville TO, et al. Human severe combined immunodeficiency: genetic, phenotypic, and functional diversity in one hundred eight infants. J Pediatr. 1997;130:378–387. doi: 10.1016/s0022-3476(97)70199-9. [DOI] [PubMed] [Google Scholar]
- Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, De Coene C, Selz F, Le Deist F, et al. Role of interleukin-2 (IL-2), IL-7, and IL-15 in natural killer cell differentiation from cord blood hematopoietic progenitor cells and from gamma c transduced severe combined immunodeficiency X1 bone marrow cells. Blood. 1996;88:3901–3909. [PubMed] [Google Scholar]
- Puel A, Ziegler SF, Buckley RH., and , Leonard WJ. Defective IL7R expression in T(-)B(+)NK(+) severe combined immunodeficiency. Nat Genet. 1998;20:394–397. doi: 10.1038/3877. [DOI] [PubMed] [Google Scholar]
- Buckley RH, Schiff SE, Schiff RI, Markert L, Williams LW, Roberts JL, et al. Hematopoietic stem-cell transplantation for the treatment of severe combined immunodeficiency. N Engl J Med. 1999;340:508–516. doi: 10.1056/NEJM199902183400703. [DOI] [PubMed] [Google Scholar]
- Antoine C, Müller S, Cant A, Cavazzana-Calvo M, Veys P, Vossen J, European Group for Blood and Marrow Transplantation; European Society for Immunodeficiency et al. Long-term survival and transplantation of haemopoietic stem cells for immunodeficiencies: report of the European experience 1968-99. Lancet. 2003;361:553–560. doi: 10.1016/s0140-6736(03)12513-5. [DOI] [PubMed] [Google Scholar]
- Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, Gross F, Yvon E, Nusbaum P, et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288:669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Le Deist F, Carlier F, Bouneaud C, Hue C, De Villartay JP, et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N Engl J Med. 2002;346:1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- Gaspar HB, Parsley KL, Howe S, King D, Gilmour KC, Sinclair J, et al. Gene therapy of X-linked severe combined immunodeficiency by use of a pseudotyped gammaretroviral vector. Lancet. 2004;364:2181–2187. doi: 10.1016/S0140-6736(04)17590-9. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- De Palma M, Montini E, Santoni de Sio FR, Benedicenti F, Gentile A, Medico E, et al. Promoter trapping reveals significant differences in integration site selection between MLV and HIV vectors in primary hematopoietic cells. Blood. 2005;105:2307–2315. doi: 10.1182/blood-2004-03-0798. [DOI] [PubMed] [Google Scholar]
- Thornhill SI, Schambach A, Howe SJ, Ulaganathan M, Grassman E, Williams D, et al. Self-inactivating gammaretroviral vectors for gene therapy of X-linked severe combined immunodeficiency. Mol Ther. 2008;16:590–598. doi: 10.1038/sj.mt.6300393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montini E, Cesana D, Schmidt M, Sanvito F, Bartholomae CC, Ranzani M, et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J Clin Invest. 2009;119:964–975. doi: 10.1172/JCI37630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth CM, Ginn SL, Deakin CT, Logan GJ., and , Alexander IE. Limiting {gamma}c expression differentially affects signaling via the interleukin (IL)-7 and IL-15 receptors. Blood. 2007;110:91–98. doi: 10.1182/blood-2006-11-055442. [DOI] [PubMed] [Google Scholar]
- Li Z, Schwieger M, Lange C, Kraunus J, Sun H, van den Akker E, et al. Predictable and efficient retroviral gene transfer into murine bone marrow repopulating cells using a defined vector dose. Exp Hematol. 2003;31:1206–1214. doi: 10.1016/j.exphem.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Kustikova OS, Wahlers A, Kuhlcke K, Stahle B, Zander AR, Baum C, et al. Dose finding with retroviral vectors: correlation of retroviral vector copy numbers in single cells with gene transfer efficiency in a cell population. Blood. 2003;102:3934–3937. doi: 10.1182/blood-2003-05-1424. [DOI] [PubMed] [Google Scholar]
- Woods NB, Bottero V, Schmidt M, von Kalle C., and , Verma IM. Gene therapy: therapeutic gene causing lymphoma. Nature. 2006;440:1123. doi: 10.1038/4401123a. [DOI] [PubMed] [Google Scholar]
- Montini E, Cesana D, Schmidt M, Sanvito F, Ponzoni M, Bartholomae C, et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat Biotechnol. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- Shou Y, Ma Z, Lu T., and , Sorrentino BP. Unique risk factors for insertional mutagenesis in a mouse model of XSCID gene therapy. Proc Natl Acad Sci USA. 2006;103:11730–11735. doi: 10.1073/pnas.0603635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siapati EK, Bigger BW, Kashofer K, Themis M, Thrasher AJ., and , Bonnet D. Murine leukemia following irradiation conditioning for transplantation of lentivirally-modified hematopoietic stem cells. Eur J Haematol. 2007;78:303–313. doi: 10.1111/j.1600-0609.2006.00813.x. [DOI] [PubMed] [Google Scholar]
- Follenzi A, Ailles LE, Bakovic S, Geuna M., and , Naldini L. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat Genet. 2000;25:217–222. doi: 10.1038/76095. [DOI] [PubMed] [Google Scholar]
- Karlsson A, Ungerbäck J, Rasmussen A, French JE., and , Söderkvist P. Notch1 is a frequent mutational target in chemically induced lymphoma in mouse. Int J Cancer. 2008;123:2720–2724. doi: 10.1002/ijc.23832. [DOI] [PubMed] [Google Scholar]
- O'Neil J, Calvo J, McKenna K, Krishnamoorthy V, Aster JC, Bassing CH, et al. Activating Notch1 mutations in mouse models of T-ALL. Blood. 2006;107:781–785. doi: 10.1182/blood-2005-06-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramezani A, Hawley TS., and , Hawley RG. Lentiviral vectors for enhanced gene expression in human hematopoietic cells. Mol Ther. 2000;2:458–469. doi: 10.1006/mthe.2000.0190. [DOI] [PubMed] [Google Scholar]
- Benhamida S, Pflumio F, Dubart-Kupperschmitt A, Zhao-Emonet JC, Cavazzana-Calvo M, Rocchiccioli F, et al. Transduced CD34+ cells from adrenoleukodystrophy patients with HIV-derived vector mediate long-term engraftment of NOD/SCID mice. Mol Ther. 2003;7:317–324. doi: 10.1016/s1525-0016(03)00002-9. [DOI] [PubMed] [Google Scholar]
- Zychlinski D, Schambach A, Modlich U, Maetzig T, Meyer J, Grassman E, et al. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Mol Ther. 2008;16:718–725. doi: 10.1038/mt.2008.5. [DOI] [PubMed] [Google Scholar]
- Hendrie PC, Huo Y, Stolitenko RB., and , Russell DW. A rapid and quantitative assay for measuring neighboring gene activation by vector proviruses. Mol Ther. 2008;16:534–540. doi: 10.1038/sj.mt.6300398. [DOI] [PubMed] [Google Scholar]
- Robert-Richard E, Richard E, Malik P, Ged C, de Verneuil H., and , Moreau-Gaudry F. Murine retroviral but not human cellular promoters induce in vivo erythroid-specific deregulation that can be partially prevented by insulators. Mol Ther. 2007;15:173–182. doi: 10.1038/sj.mt.6300030. [DOI] [PubMed] [Google Scholar]
- Schambach A, Bohne J, Chandra S, Will E, Margison GP, Williams DA, et al. Equal potency of gammaretroviral and lentiviral SIN vectors for expression of O6-methylguanine-DNA methyltransferase in hematopoietic cells. Mol Ther. 2006;13:391–400. doi: 10.1016/j.ymthe.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Benjelloun F, Garrigue A, Demerens-de Chappedelaine C, Soulas-Sprauel P, Malassis-Séris M, Stockholm D, et al. Stable and functional lymphoid reconstitution in artemis-deficient mice following lentiviral artemis gene transfer into hematopoietic stem cells. Mol Ther. 2008;16:1490–1499. doi: 10.1038/mt.2008.118. [DOI] [PubMed] [Google Scholar]
- Will E, Bailey J, Schuesler T, Modlich U, Balcik B, Burzynski B, et al. Importance of murine study design for testing toxicity of retroviral vectors in support of phase I trials. Mol Ther. 2007;15:782–791. doi: 10.1038/sj.mt.6300083. [DOI] [PubMed] [Google Scholar]
- Mortellaro A, Hernandez RJ, Guerrini MM, Carlucci F, Tabucchi A, Ponzoni M, et al. Ex vivo gene therapy with lentiviral vectors rescues adenosine deaminase (ADA)-deficient mice and corrects their immune and metabolic defects. Blood. 2006;108:2979–2988. doi: 10.1182/blood-2006-05-023507. [DOI] [PubMed] [Google Scholar]
- Hargrove PW, Kepes S, Hanawa H, Obenauer JC, Pei D, Cheng C, et al. Globin lentiviral vector insertions can perturb the expression of endogenous genes in beta-thalassemic hematopoietic cells. Mol Ther. 2008;16:525–533. doi: 10.1038/sj.mt.6300394. [DOI] [PubMed] [Google Scholar]
- Pike-Overzet K, de Ridder D, Weerkamp F, Baert MR, Verstegen MM, Brugman MH, et al. Ectopic retroviral expression of LMO2, but not IL2Rgamma, blocks human T-cell development from CD34+ cells: implications for leukemogenesis in gene therapy. Leukemia. 2007;21:754–763. doi: 10.1038/sj.leu.2404563. [DOI] [PubMed] [Google Scholar]
- Scobie L, Hector RD, Grant L, Bell M, Nielsen AA, Meikle S, et al. A novel model of SCID-X1 reconstitution reveals predisposition to retrovirus-induced lymphoma but no evidence of gammaC gene oncogenicity. Mol Ther. 2009;17:1031–1038. doi: 10.1038/mt.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan RN, Treuting PM, Fuller ED, Goldsby RE, Norwood TH, Gooley TA, et al. Mutation at the polymerase active site of mouse DNA polymerase delta increases genomic instability and accelerates tumorigenesis. Mol Cell Biol. 2007;27:7669–7682. doi: 10.1128/MCB.00002-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fern L, Pallis M, Ian Carter G, Seedhouse C, Russell N., and , Byrne J. Clonal haemopoiesis may occur after conventional chemotherapy and is associated with accelerated telomere shortening and defects in the NQO1 pathway; possible mechanisms leading to an increased risk of t-AML/MDS. Br J Haematol. 2004;126:63–71. doi: 10.1111/j.1365-2141.2004.05006.x. [DOI] [PubMed] [Google Scholar]
- Bhatia R, Van Heijzen K, Palmer A, Komiya A, Slovak ML, Chang KL, et al. Longitudinal assessment of hematopoietic abnormalities after autologous hematopoietic cell transplantation for lymphoma. J Clin Oncol. 2005;23:6699–6711. doi: 10.1200/JCO.2005.10.330. [DOI] [PubMed] [Google Scholar]
- Newrzela S, Al-Ghaili N, Heinrich T, Cornils K, Li Z, Baum C, et al. Control mechanisms in mature T cell leukemia/lymphoma. Kyoto T Cell Conference 5th International Workshop, 1–4 June. 2009.
- Ginn SL, Fleming J, Rowe PB., and , Alexander IE. Promoter interference mediated by the U3 region in early-generation HIV-1-derived lentivirus vectors can influence detection of transgene expression in a cell-type and species-specific manner. Hum Gene Ther. 2003;14:1127–1137. doi: 10.1089/104303403322167975. [DOI] [PubMed] [Google Scholar]
- Sastry L, Johnson T, Hobson MJ, Smucker B., and , Cornetta K. Titering lentiviral vectors: comparison of DNA, RNA and marker expression methods. Gene Ther. 2002;9:1155–1162. doi: 10.1038/sj.gt.3301731. [DOI] [PubMed] [Google Scholar]
- Lo M, Bloom ML, Imada K, Berg M, Bollenbacher JM, Bloom ET, et al. Restoration of lymphoid populations in a murine model of X-linked severe combined immunodeficiency by a gene-therapy approach. Blood. 1999;94:3027–3036. [PubMed] [Google Scholar]
- Zhang F, Thornhill SI, Howe SJ, Ulaganathan M, Schambach A, Sinclair J, et al. Lentiviral vectors containing an enhancer-less ubiquitously acting chromatin opening element (UCOE) provide highly reproducible and stable transgene expression in hematopoietic cells. Blood. 2007;110:1448–1457. doi: 10.1182/blood-2006-12-060814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Watson D, Zhang GY, Graf N, Wang YM, Sartor M, et al. Long-term cardiac allograft survival across an MHC mismatch after “pruning” of alloreactive CD4 T cells. J Immunol. 2008;180:6593–6603. doi: 10.4049/jimmunol.180.10.6593. [DOI] [PubMed] [Google Scholar]
- Walters G., and , Alexander SI. T cell receptor BV repertoires using real time PCR: a comparison of SYBR green and a dual-labelled HuTrec fluorescent probe. J Immunol Methods. 2004;294:43–52. doi: 10.1016/j.jim.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Schwarzwaelder K, Bartholomae C, Zaoui K, Ball C, Pilz I, et al. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR) Nat Methods. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- Lovatt A, Black J, Galbraith D, Doherty I, Moran MW, Shepherd AJ, et al. High throughput detection of retrovirus-associated reverse transcriptase using an improved fluorescent product enhanced reverse transcriptase assay and its comparison to conventional detection methods. J Virol Methods. 1999;82:185–200. doi: 10.1016/s0166-0934(99)00111-1. [DOI] [PubMed] [Google Scholar]