Abstract
Because Eurasian samples of Neoerysiphe collected on the Asteraceae were not identical in morphology, the molecular and morphological differences among these specimens were compared with those of the American N. cumminsiana. Neoerysiphe on Asteraceae was found to consist of at least four different species. Three of them are described as new species, viz. N. hiratae, N. joerstadii, and N. nevoi. Neoerysiphe hiratae is a Japanese species parasitizing hosts belonging to the genera Cacalia and Ligularia (tribe Senecioneae). Neoerysiphe joerstadii was found in Israel on Phagnalon rupestre (tribe Gnaphalieae). Neoerysiphe nevoi was recorded in Israel and Ukraine on a number of hosts in different genera but all belonging to tribe Cichorieae. Thus, Eurasian Neoerysiphe species infecting the Asteraceae are strongly specialised to particular tribes of this family. Phylogenetic analyses indicated that the three new species were not closely allied. Neoerysiphe hiratae is related to the American N. cumminsiana and species belonging to Oidium subg. Striatoidium. Neoerysiphe nevoi is sister to N. geranii, and N. joerstadii is allied to N. galii. In addition, Ukrainian Neoerysiphe samples on Geranium were phylogenetically and morphologically identical to Japanese samples of N. geranii, and this fungus seems to be an invasive species in Ukraine.
Keywords: 28S rDNA, Asteraceae, Erysiphales, Japan, Mediterranean region, new species, rDNA ITS region, systematics
INTRODUCTION
Based on the anamorph type, Heluta (1988) proposed to divide the genus Erysiphe into two separate genera, viz. Erysiphe s.str. and Golovinomyces. The former included all species with an anamorph of the Pseudoidium type (conidia formed singly on conidiophores), whereas the latter consisted of species with an anamorph of the Oidium s.str. type (= Euoidium; conidia catenate). Species belonging to two Erysiphe sections introduced by Braun (1978, 1981), namely Golovinomyces and Galeopsidis, were included in the genus Golovinomyces sensu Heluta (Heluta 1988). Only one species, E. galeopsidis, was contained in sect. Galeopsidis. This species differed from other Erysiphe representatives by its lobed appressoria and maturation of ascospores after wintering. It was clarified later that a few species very close to E. galeopsidis had to be included in sect. Galeopsidis, viz. E. chelones on Scrophulariaceae (USA), E. cumminsiana on Asteraceae (Asia, North America), E. galii on Rubiaceae (Europe, Asia), and E. geranii on Geraniaceae (Japan, New Zealand). However, molecular studies (Saenz & Taylor 1999, Mori et al. 2000) clearly indicated that sect. Golovinomyces did not group with sect. Galeopsidis. In addition, it was found that the conidium surface of species belonging to sect. Galeopsidis is unique among powdery mildew fungi enabling the creation of a new taxon for the anamorphs of this section, viz. Oidium subg. Striatoidium (Cook et al. 1997). Due to these morphological, biological, and molecular peculiarities of representatives of sect. Galeopsidis, Braun (1999) raised this section to genus rank and introduced the name Neoerysiphe. The five species in the section were transferred to this new genus with appropriate new taxonomic combinations. Later, another species, N. rubiae, was described on Rubia cf. tinctoria from Turkey (Bahcecioglu et al. 2006). In addition, Takamatsu et al. (2008) revealed that Oidium aloysiae on Aloysia citriodora, O. baccharidis on Baccharis linearis and B. racemosa, and O. maquii on Aristotelia chilensis are anamorphs of Neoerysiphe. Thus, at present this genus combines six teleomorph and three anamorph species, viz. N. chelones on the Scrophulariaceae, N. cumminsiana and O. baccharidis on the Asteraceae, N. galii and N. rubiae on the Rubiaceae, N. geranii on the Geraniaceae, O. aloysiae on the Verbenaceae, O. maquii on the Elaeocarpaceae, and N. galeopsidis parasitizing many hosts of Lamiaceae as the main host family but also a few species in Acanthaceae, Bignoniaceae, Dipsacaceae, and Malvaceae (Liu et al. 2005, Takamatsu et al. 2008). Each of these species has a quite different distribution. Neoerysiphe galeopsidis is nearly circumglobal, known from all Europe, Asia, Africa, North America, and New Zealand (Braun 1987). Distributions are rather limited for the remaining species. Neoerysiphe chelones is known from the USA, N. rubiae only from Turkey. Neoerysiphe galii is a Eurasian species. Neoerysiphe geranii was known from Japan and probably from New Zealand (Amano 1986, Nomura 1997). Heluta (2001) also reported this fungus from Ukraine. Some questions regarding the distributions of certain Neoerysiphe species have still to be answered. For instance, N. geranii seems to have a more disjunctive distribution. Furthermore, it is also possible that another species morphologically close to N. geranii is distributed in Ukraine. Braun (1983) described N. cumminsiana on Senecio seemannii from the USA and later reported it from North America and Japan on hosts belonging to Cacalia, Eupatorium, Heliopsis, and Ligularia (Braun 1987). Heluta (1989, 1999) first recorded a powdery mildew on Crepis and Taraxacum in Ukraine as Golovinomyces galii, and later changed it to G. cumminsianus. Voytyuk et al. (2004, 2006) reported N. cumminsiana from Israel on hosts of many genera of the Asteraceae, viz. Carthamus, Crepis, Filago, Hedypnois, Phagnalom, Rhagadiolus, Senecio, Thrincia, and Tolpis. According to Voytyuk et al. (2004), N. cumminsiana has a unique distribution being the only representative of the Erysiphales which must be classified as an American-African-Eurasian South Holarctic species. However, this does not correspond to the set of probable geographic and mycoflorogenetic units of powdery mildews proposed by Heluta (1993, 1995). These units consist of species having many factors in common, mainly their probable time and place of origin and current habitats. It is also not in accordance with Heluta’s (1992) hypothesis on the ways of powdery mildew migration. Therefore, Voytyuk et al. (2004) assumed that this hypothesis was either not fully correct or N. cumminsiana is a species complex with similar morphological characteristics. In the latter case ‘N. cumminsiana’ might be descended from an ancestor such as N. galeopsidis and might have emerged independently in several regions of North and South America, Africa, or Eurasia. In addition, specimens of Israeli ‘N. cumminsiana’ are morphologically not uniform. Voytyuk et al. (2004, 2006) reported that collections on Phagnalon rupestre had much larger chasmothecia and smaller peridial cells than those on other host plants. Furthermore, the taxonomic status of Eurasian ‘N. geranii’ and ‘N. cumminsiana’ was never examined with molecular methods. The goal of this study was to clarify the origin of the European populations of N. geranii and the Eurasian biotypes of ‘N. cumminsiana’, using mainly molecular methods.
MATERIALS AND METHODS
Molecular phylogenetic studies
The fungal species, host plants, location of collection, and accession numbers for the nucleotide sequence databases (DDBJ, EMBL and GenBank) are provided in Table 1. Isolation of whole-cell DNA was performed using the chelex method (Walsh et al. 1991) as described in Hirata & Takamatsu (1996). The 5′-end of the 28S rDNA, including the domains D1 and D2, and ITS region, including the 5.8S rDNA, were amplified by polymerase chain reaction (PCR) and then sequenced using direct sequencing as described in Takamatsu et al. (2006).
Table 1.
Sources of Neoerysiphe material used for molecular analyses and their accession numbers in DNA databases.
| Fungal species | Host | Location; year | Voucher no.1 | Accession no.2 |
|---|---|---|---|---|
| N. galeopsidis | Galeopsis sp. | Ukraine, Volhynian region; 2004 | KW 33697F / MUMH4657 | AB498940 |
| Lamium amplexicaule | Ukraine, Crimea; 2004 | KW 58375F / MUMH 4673 | AB498941 | |
| Lamium purpureum | Ukraine, Crimea; 2004 | KW 33682F / MUMH 4658 | AB498942 | |
| Marrubium praecox | Ukraine, Donetsk region; 2004 | KW 58376F / MUMH 4674 | AB498943 | |
| Phlomis pungens | Ukraine, Donetsk region; 2004 | KW 33698F / MUMH 4659 | AB498944 | |
| Phlomis pungens | Ukraine, Donetsk region; 2004 | KW 58377F / MUMH 4675 | AB498945 | |
| Phlomis tuberosa | Ukraine, Cherkasy region; 2005 | KW 33700F / MUMH 4660 | AB498946 | |
| Phlomis tuberosa | Ukraine, Donetsk region; 2004 | KW 33699F / MUMH 4661 | AB498947 | |
| Phlomis tuberosa | Ukraine, Donetsk region; 2004 | KW 58378F / MUMH 4676 | AB498948 | |
| Prasium majus | Israel, Tel-Aviv; 2004 | HAI 4322 / MUMH 4680 | AB498949 | |
| Stachys distans | Israel, Carmel Mt.; 2004 | HAI 4327 / MUMH 4681 | AB498950 | |
| N. galii | Galium aparine | Israel, Jordan Valley; 2004 | HAI 445 / MUMH 4682 | AB498951 |
| N. geranii | Geranium sibiricum var. popovii | Ukraine, Kyiv; 1998 | KW 28118F / MUMH 4662 | AB498952 |
| Geranium sibiricum var. popovii | Ukraine, Kyiv region; 1998 | KW 28121F / MUMH 4663 | AB498953 | |
| Geranium sibiricum var. popovii | Ukraine, Kyiv region; 1998 | KW 28123F / MUMH 4664 | AB498954 | |
| Geranium thunbergii | Japan, Hokkaido; 2004 | KW 34781F / MUMH 3555 | AB498955 | |
| Geranium sp. | Ukraine, Kyiv; 2007 | KW 33701F / MUMH 4665 | AB498956 | |
| N. hiratae | Cacalia delphiniifolia | Japan, Ehime; 1998 | KW 34784F / MUMH 567 | AB498957 |
| Cacalia hastata ssp. farfarifolia | Japan, Nagano; 2004 | KW 34785F / MUMH 3504 | AB498958 | |
| Ligularia fischeri | Japan, Okayama; 2006 | KW 34786F / MUMH 4471 | AB498959 | |
| Ligularia stenocephala | Japan, Mie; 2004 | KW 34789F / MUMH 3611 | AB498960 | |
| Ligularia stenocephala | Japan, Nagano; 2004 | KW 34788F / MUMH 3505 | AB498961 | |
| Ligularia stenocephala | Japan, Shiga; 2004 | KW 34787F / MUMH 3442 | AB498962 | |
| N. joerstadii | Phagnalon rupestre | Israel, Golan Heights; 2004 | HAI 4239 / MUMH 4668 | AB498976 |
| N. nevoi | Chondrilla sp. | Ukraine, Crimea; 2004 | KW 58373F / MUMH 4672 | AB498963 |
| Crepis aspera | Israel, Carmel Mt.; 2004 | HAI 4164 / MUMH 4667 | AB498964 | |
| Crepis aspera | Israel, Northern Negev; 2004 | KW 34790F / MUMH 4873 | AB498965 | |
| Crepis rhoeadifolia | Ukraine, Crimea; 1978 | KW 11753F / MUMH 4655 | AB498966 | |
| Crepis rhoeadifolia | Ukraine, Crimea; 1982 | KW 11755F / MUMH 4654 | AB498967 | |
| Hedypnois cretica | Israel, Northern Negev; 2004 | KW 34793F / MUMH 4875 | AB498968 | |
| Picris amalecitana | Israel, Mi’ilya; 2004 | HAI 4114 / MUMH 4669 | AB498969 | |
| Rhagadiolus stellatus | Israel, Northern Negev; 2004 | HAI 4329 / MUMH 4670 | AB498970 | |
| Rhagadiolus stellatus | Ukraine, Crimea; 2004 | KW 58374 / MUMH 4677 | AB498971 | |
| Taraxacum sp. | Ukraine, Crimea; 1981 | KW 11777F / MUMH 4656 | AB498972 | |
| Thrincia tuberosa | Israel, Lower Galilee; 2004 | HAI 4123 / MUMH 4678 | AB498973 | |
| Tolpis virgata | Israel, Upper Galilee; 2004 | HAI 4296 / MUMH 4679 | AB498974 | |
| N. nevoi var. scolymi | Scolymus hispanicus | Israel, Carmel Mt.; 2005 | HAI 5195 / MUMH 4671 | AB498975 |
1 Sources: HAI = Haifa University, Herbarium of the Institute of Evolution, Israel; KW = National Herbarium of the M.G. Kholodny Institute of Botany, Kiev, Ukraine; MUMH = Mie University, Mycological Herbarium, Japan.
2 The nucleotide sequence data will appear in the DDBJ, EMBL, and GenBank databases under the respective accession number.
The sequences were initially aligned using the Clustal X package (Thompson et al. 1997). The alignment was then visually refined with MacClade v4.08 (Maddison & Maddison 2005). The alignments were deposited in TreeBASE (www.treebase.org/). Phylogenetic trees were obtained from the data using the maximum parsimony (MP) method in PAUP* 4.0 (Swofford 2001) and Bayesian analysis in MrBayes 3.1.1 (Huelsenbeck & Ronquist 2001). MP analyses were performed with the heuristic search option using the ‘tree-bisection-reconstruction’ (TBR) algorithm with 100 random sequence additions to find the global optimum tree. All sites were treated as unordered and unweighted, with gaps treated as missing data. The strength of the internal branches of the resulting trees was tested with bootstrap (BS) analyses (Felsenstein 1985) using 1 000 replications with the stepwise addition option set as simple and maximum tree number as 100. BS values 70 % or higher are provided.
For Bayesian phylogenetic analyses, the best-fit evolutionary model was determined for each dataset by comparing different evolutionary models via the Akaike information criterion (AIC) using PAUP* and MrModeltest 2.2 (Nylander 2004). MrBayes was launched with random starting trees for 2 × 106 generations and the Markov chains were sampled every 100 generations, which resulted in 2 × 104 sampled trees. To ensure that the Markov chain did not become trapped in local optima, we used the MCMCMC algorithm, performing the estimation with four incrementally heated Markov chains. Bayesian posterior probability (PP) values 0.95 or higher are shown.
Morphological analysis
Powdery mildew specimens involved in the morphological analysis are listed after each description of new taxa. Morphological features of the specimens were examined and photographed using light microscopes MBI-6 (LOMO, Russia) with objectives ×16 and ×40 (Carl Zeiss, Germany) in phase contrast and Primo Star (Carl Zeiss, Germany) with objectives ×10 and ×40. Photographs were prepared by the digital cameras EOS 350D and PowerShot A640 (both Canon, Tokyo, Japan), accordingly. Shrivelled conidiophores, conidia and superficial hyphae were restored by heating to start of boiling in 40 % lactic acid. Only dry chasmothecia on host leaves were measured. For each morphological feature 30 structures were measured and the data processed statistically. Limits of variation were determined as M ± 1.96 σ, where M is a simple average and σ is a standard deviation. The SEM micrographs were obtained with a Jeol JSM–6060LA (Tokyo, Japan) SEM microscope. Dry pieces of leaf with mycelium, conidia, and ascomata were glued to metallic stubs and gold coated under vacuum. The specimens examined are deposited at HAI, HUJ, KW, MUMH, and TNS (abbreviations according to Holmgren et al. (1990)).
RESULTS
ITS phylogeny
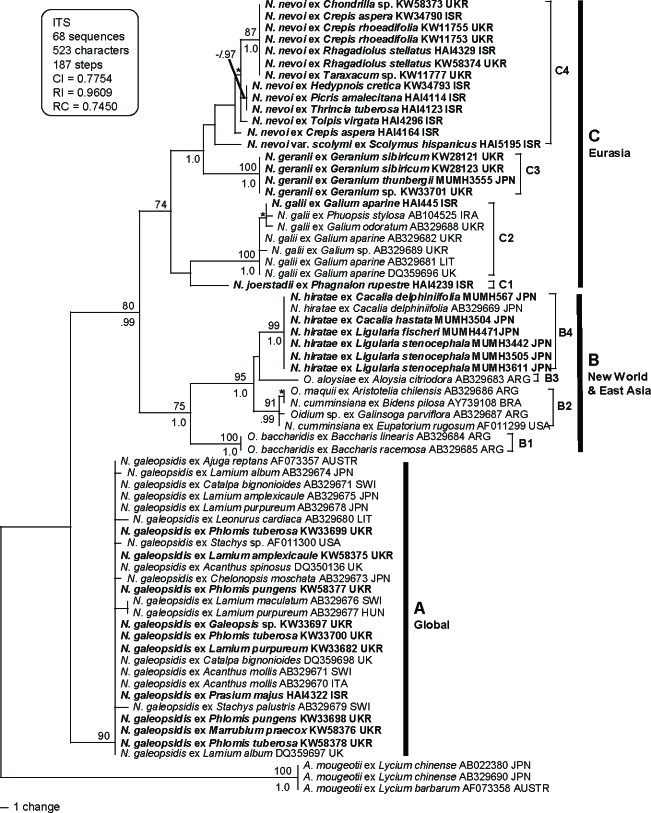
Thirty-five ITS sequences of Neoerysiphe spp. were newly determined in this study (Table 1). These sequences were aligned with 30 sequences of Neoerysiphe spp. and three sequences of Arthrocladiella mougeotii used as an outgroup taxon. The dataset consisted of 68 sequences and 523 characters. All characters were aligned unambiguously. Of the 523 characters, 119 were variable and 104 characters were phylogenetically informative for parsimony analysis. A total of 58 100 equally parsimonious trees with 187 steps (CI = 0.775, RI = 0.961, RC = 0.745) were generated by the MP analysis, when it had to be terminated due to the limit of memory size of the software. One of the trees is shown in Fig. 1. We also performed parsimony ratchet analysis (Nixon 1999) using PAUP* and PAUPRat v1 (Sikes & Lewis 2001) and confirmed the generation of almost identical tree topologies with the same tree length. Thus, we concluded that the tree shown in Fig. 1 is not the result of a local optimum. MrModeltest selected SYM+I+G model as the best for this dataset. Bayesian analysis was performed using this evolution model and yielded 2 × 104 trees. Of the trees, the first 14 130 were discarded (burn-in) because the average standard deviation of the split frequencies (ASDSF) dropped below 0.01. The remaining 5 870 trees were summarised in a majority-rule consensus tree, yielding the probability of each clade being monophyletic. The tree topology by the Bayesian analysis was almost identical to the MP tree, and thus the former tree is not shown.
Fig. 1.
Phylogenetic analysis of the nucleotide sequences of the ITS region including 5.8S rDNA for 68 sequences from Neoerysiphe with Arthrocladiella used as outgroup taxon. The tree is a phylogram of one of the 58 K MP trees with 178 steps obtained by a heuristic search employing the random stepwise addition option of PAUP*. Gaps were treated as missing data. Horizontal branch lengths are proportional to the number of nucleotide substitutions that were inferred to have occurred along a particular branch of the tree. Percentage BS support (1 000 replications; ≥ 70 %) and PP (≥ 0.95) are shown on and under branches, respectively. Nodes with asterisks (*) denote that the nodes collapsed in the strict consensus tree.
The 65 sequences of Neoerysiphe analyzed in this study were divided into three large clades (A, B and C) clearly defined by their geographical distributions and host plants. Clade A consisted of a single species, N. galeopsidis, and is supported strongly with both BS and PP values (BS = 90 %; PP = 0.90). Hosts of this species mostly belong to the Lamiaceae, but Acanthus (Acanthaceae) and Catalpa (Bignoniaceae) are also included in this clade as hosts. Maximum genetic divergence within this clade is only 0.8 %, which suggests that N. galeopsidis diverged on the Lamiaceae and sporadically infected other plant families recently. Clades B and C formed a larger clade (BS = 80 %; PP = 0.99). Clade B (BS = 75 %; PP = 1.0) consists of hosts of the Asteraceae, and one specimen from both Aloysia (Verbenaceae) and Aristotelia (Elaeocarpaceae) collected in North and South America and Japan. This clade is further divided into four subclades. B1 contains Oidium baccharidis on Baccharis (tribe Astereae, Asteraceae) and B3 contains O. aloysiae on Aloysia, both collected in Argentina. B2 consists of O. maquii on Aristotelia, N. cumminsiana on Bidens (tribe Heliantheae, Asteraceae) and Eupatorium (tribe Eupatorieae, Asteraceae), and Oidium sp. on Galinsoga (tribe Millerieae, Asteraceae) obtained from the USA and South America (BS = 91 %; PP = 0.99). B4 (BS = 99 %; PP = 1.0) comprises seven sequences of Neoerysiphe on Cacalia and Ligularia (tribe Senecioneae, Asteraceae) collected in Japan. These seven sequences are identical to each other. This fungus has been identified as N. cumminsiana (Nomura 1997, Takamatsu et al. 2008), but the present analysis indicates that the fungus forms an independent lineage different from N. cumminsiana collected in North and South America. Clade C (BS = 74 %; PP = 0.89) comprises Neoerysiphe spp. collected in Eurasia, especially in Mediterranean and circum Mediterranean areas like the north part of Israel and the south part of Ukraine, and is further divided into four subclades. C1 consists of a single sequence of a fungus on Phagnalon rupestre (tribe Gnaphalieae, Asteraceae) collected in Israel. The same sequence was obtained when the sequencing of the DNA extraction from this specimen was repeated. Subclades C2 and C3 consisted of N. galii on Galium spp. (Rubiaceae) and N. geranii on Geranium spp. (Geraniaceae), respectively. Both clades were strongly supported by BS and PP values (BS = 100 %; PP = 1.0 in both C2 and C3). All these specimens were collected in Europe, except for specimens of N. geranii collected in Japan and one sample of N. galii from Israel. C4 consisted of 13 sequences from fungi on tribe Cichorieae of the Asteraceae, collected in Israel and Ukraine. There was some genetic divergence within this clade. Subclades C3 and C4 formed a clade with strong support (BS = 56 %; PP = 1.0).
28S phylogeny
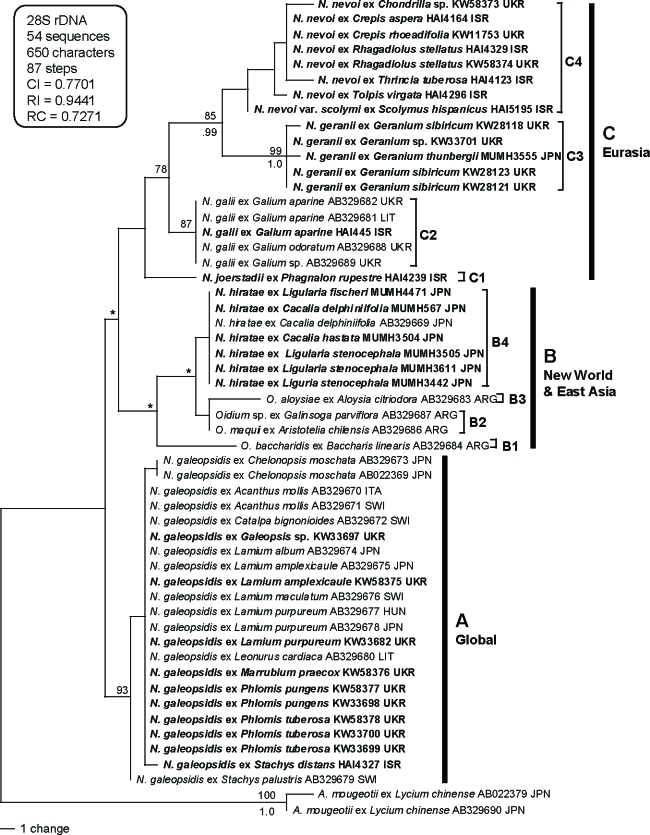
Thirty-one 28S rDNA sequences including D1/D2 domains of Neoerysiphe spp. were newly determined in this study (Table 1). These sequences were aligned with 21 sequences of Neoerysiphe spp. and two sequences of Arthrocladiella mougeotii used as an outgroup taxon. The dataset consisted of 54 sequences and 650 characters. All characters were aligned unambiguously. Of the 650 characters, 65 were variable and 51 characters were phylogenetically informative for parsimony analysis. A total of 14 equally parsimonious trees with 87 steps (CI = 0.770, RI = 0.944, RC = 0.727) were generated by the MP analysis. Of these 14 trees, a tree with the highest likelihood value is shown in Fig. 2. MrModeltest selected GTR+I model as the best for this dataset. Bayesian analysis using this evolution model yielded 2 × 104 trees. Of these, the first 7 020 were discarded (burn-in) because ASDSF dropped below 0.01. The remaining 12 980 trees were summarised in a majority-rule consensus tree, yielding the probability of each clade being monophyletic. The tree topology by the Bayesian analysis was almost identical to the MP tree, and thus the former tree is not shown.
Fig. 2.
Phylogenetic analysis of the divergent domains D1 and D2 sequences of the 28S rDNA for 54 sequences from Neoerysiphe with Arthrocladiella used as outgroup taxon. The tree is a phylogram of the tree with the highest likelihood score among the 14 MP trees with 87 steps, which was obtained and constructed as described for Fig. 1. Bold lines denote branches present in the strict consensus tree. Nodes with asterisks (*) denote that the nodes collapsed in the strict consensus tree.
The tree constructed by the 28S rDNA dataset strongly supported the phylogeny of Neoerysiphe shown in the ITS tree. Neoerysiphe sequences analysed in this study were divided into three large clades (A, B, and C), although clade B was collapsed in the strict consensus tree. Subclades B1, B2, B3, B4, C1, C2, C3, and C4 are also supported in the 28S tree, although BS and PP supports were lower than those in ITS tree. Subclades C3 and C4 formed a clade with strong support (BS = 85 %; PP = 0.99).
Morphology
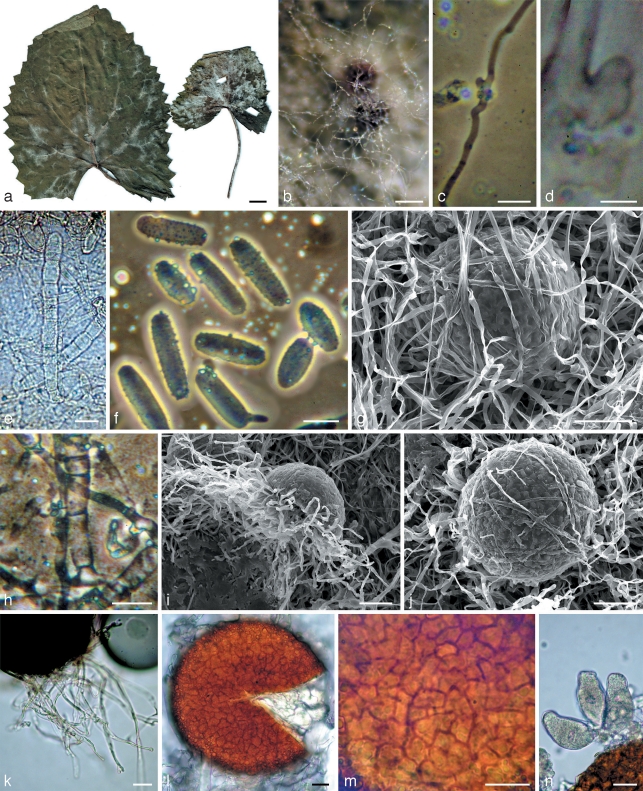
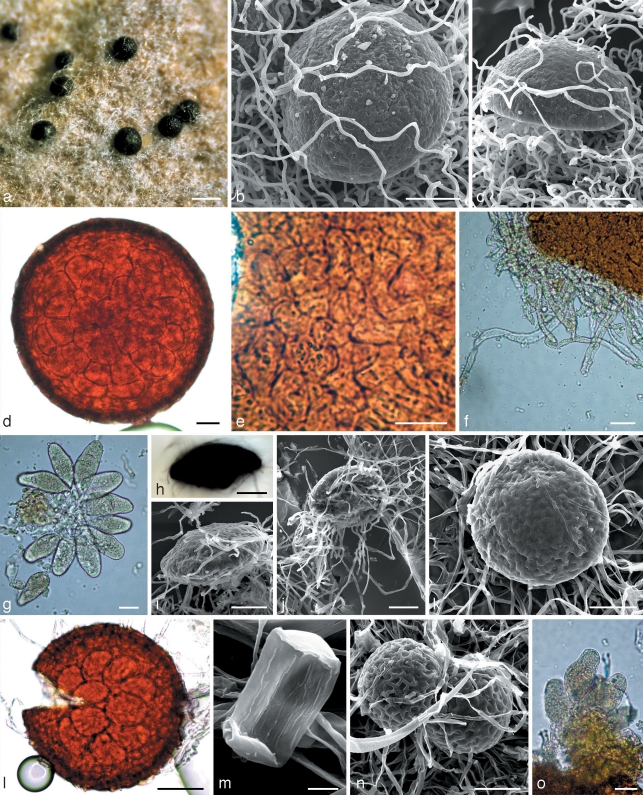
Mycelium of samples belonging to subclade B4 was well developed, especially along the veins of the leaves (Fig. 3a). Primary hyphae had distinct appressoria. The secondary mycelium appeared simultaneously with chasmothecium initials immediately following the sexual reproduction. This mycelium consisted of thin hyaline hyphae that later surrounded chasmothecia as an interlacing delicate or somewhat denser web (Fig. 3b, g). It should be noted that both mycelia are pure white without any yellowish or brownish tints. In contrast to these fungi, samples of subclade C4 had yellowish primary mycelium which gave rise to white secondary mycelium occasionally with somewhat yellowish hyphae. Mycelium of the fungus on Phagnalon rupestre (C1) was very weakly developed, almost invisible, greyish and only confined to primary mycelium. Hyphal appressoria in all groups were variable, of similar size, unlobed or somewhat lobate (Fig. 3c, d and 5a–c). Anamorphs were observed only in subclades B4 and C4. Conidiophores were very similar morphologically, with straight, cylindrical foot-cells, frequently increasing from base to top, 24–45.5 × 9.5–12.5 μm, followed by 1–2 shorter cells and conidium initials (Fig. 3e, h and 5e). However, in subclade C4 the foot-cells were occasionally very long, up to 100 μm. Conidia in subclade B4 were catenate, mainly cylindrical with rounded ends, often oblong ellipsoidal, long, up to 48 μm (Fig. 3f), whereas in C4 they were mainly ellipsoidal or short cylindrical with rounded ends, often almost limoniform, short, up to 36 μm (Fig. 5f); the length/width ratios were 1.9–3.5 (average 2.7) and 1.5–2.5 (average 1.9), respectively. Chasmothecia of all specimens studied were hemispherical, depressed or even concave in the lower part (Fig. 3i, 4c, and 5i) but on Phagnalon rupestre (C1) they were more flattened and distinctly larger, up to 200 μm diam. This contrasted with the other specimens, e.g. subclades B4 and C4 having chasmothecial diameters mainly up to 157 μm and 148 μm, respectively. The fungus on Phagnalon also had a rather transparent peridium, enabling the number of asci to be easily viewed and counted whilst still within the chasmothecium (Fig. 4d). Peridial cells of this fungus were obscure, polygonal or irregular in shape and small, 11–17(–33) × 10–14(–16) μm. The peridial surface was indistinctly close-meshed or knobbly (Fig. 4b, c), whilst in subclades B4 and C4 the peridium was less transparent with asci invisible within the chasmothecium. Peridial cells in subclade B4 were, however, more visible and more regular in shape than those of the Phagnalon parasite, but similarly small. Peridial cells in subclade C4 were distinguished from those in C1 and B4 by being distinct and perceptibly larger, up to 30 × 17 μm. The peridial surface also differed in being distinctly meshed and similar to the other samples in this group (Fig. 5i, j) with the exception of the fungus on Scolymus hispanicus. This fungus has chasmothecia with a deeply pitted peridium where cell junctions formed conspicuous ridges (Fig. 5k). Appendages of all specimens studied were well developed but very short and hyaline in chasmothecia on Phagnalon rupestre. In subclade B4 appendages were also hyaline and only in one specimen they were somewhat yellowish. All those in C4 were more or less pigmented. Asci in all three clades B4, C1, and C4 were similar in shape, mainly obpyriform, stipitate, and immature in the current season but they were more oblong on Phagnalon and more numerous, 16–32 per chasmothecium, in contrast to subclades B4 and C4 where the number of asci did not exceed 12 (compare Fig. 4g with Fig. 3n and 5n–q).
Fig. 3.
Morphology within phylogenetic subclade B4. a, b, g, i–n: Neoerysiphe hiratae (isotype, KW 34787F) on Ligularia stenocephala; c–f, h: N. hiratae (KW 34783F) on L. delphiniifolia. a. The infected host; b. chasmothecia in reflected light covered by the secondary mycelium; c, d. hyphae of the primary mycelium with appressoria; e. conidiophores; f. conidia; g, i, j. chasmothecia viewed by scanning electron microscope: g, j – covered by hyphae of the secondary mycelium, i – side view; h. basal part of conidiophore; k. chasmothecial appendages; l. chasmothecium viewed by light microscope; m. peridial cells; n. asci. — Scale bars: a = 1 cm; b = 100 μm; c, e, f, h, k–n = 20 μm; d = 5 μm; g, i, j = 50 μm.
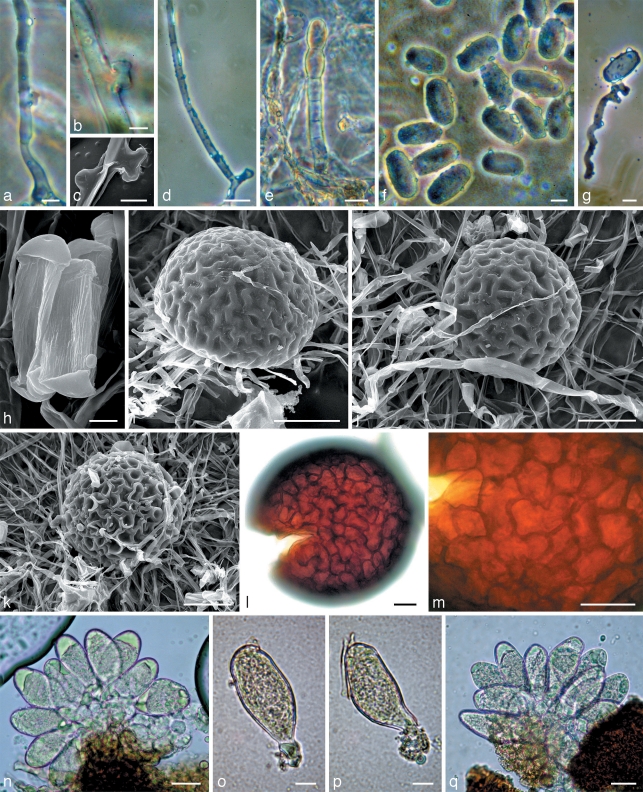
Fig. 5.
Morphology within phylogenetic subclade C4. a, b, d–g, q: Neoerysiphe nevoi (KW 35726F) on Thrincia tuberosa; c, h–j, l–p: N. nevoi (holotype, KW 34802F) on Tolpis virgata; k: N. nevoi var. scolymi (holotype, KW 34800F) on Scolymus hispanicus. a–c. Hyphae of the primary mycelium with appressoria; d. secondary hypha arisen from the primary hypha; e. conidiophore; f–h. conidia; g. germinated conidium with a hypha extending from a lobed appressorium of the Striatoidium type; i–k. chasmothecia viewed by scanning electron microscope; l. chasmothecium viewed by light microscope; m. peridial cells; n–q. asci. — Scale bars: a, f, g, o, p = 10 μm; b, c, h = 5 μm; d, e, l–n, q = 20 μm; i–k = 50 μm.
Fig. 4.
Morphology within phylogenetic subclades C1, B2 and C3. a–g: Neoerysiphe joerstadii (holotype, KW 35717F, subclade C1) on Phagnalon rupestre; h–l: N. cumminsiana (isotype, HAL 1462F, subclade B2) on Senecio seemannii; m–o: N. geranii (KW 34782F, subclade C3) on Geranium sp. a. Chasmothecia in reflected light; b, c. chasmothecia viewed by scanning electron microscope: c – side view; d. chasmothecium in transmitted light; e. peridial cells; f. chasmothecial appendages; g. asci; h. chasmothecium with evagination on the lower side, side view; i–k. chasmothecia viewed by scanning electron microscope: i – side view, j – bottom view; l. chasmothecium in transmitted light; m. conidium with longitudinal ridges; n. chasmothecia; o. asci. — Scale bars: a = 200 μm; b–d, h–l, n = 50 μm; e–g, o = 20 μm; m = 5 μm.
Morphological analysis indicated that the fungi belonging to subclades B4, C1 and C4 had obvious differences and must be treated as separate species. This conclusion fully agreed with the results of our phylogenetic analysis. The type specimen of N. cumminsiana, another Neoerysiphe species parasitizing the Asteraceae, was also included in the morphological analysis. In this specimen secondary mycelium was also formed, but it is barely visible and appressed to the substrate. Chasmothecia were large like those on Phagnalon but differed in having a unique structure, characterised by a clearly visible basal evagination up to 27 μm height (Fig. 4h–j). Such a feature is unknown in any other Neoerysiphe species. Although the peridium of N. cumminsiana was also transparent and the chasmothecia were large like those on Phagnalon, the asci were fewer, mainly 10, and notably larger, 50–57 × 31–35.5 μm. Thus, all the studied samples of Eurasian Neoerysiphe parasitizing Asteraceae differed from N. cumminsiana morphologically and so do undoubtedly not belong to this species. An attempt to sequence the type specimen of N. cumminsiana failed.
Results of the phylogenetic analysis indicated that subclades C4 and C3 (N. geranii) were sister groups. Their propinquity was confirmed by morphological examinations of these fungi, including the type specimen of N. geranii. However, subclade C4 had a more developed secondary mycelium, a conidial surface with larger number of lengthwise striations, a less transparent peridium, and asci more ellipsoidal than obpyriform (compare Fig. 5h–q with 4l–o).
Neoerysiphe on Phagnalon (C1) differs strongly from all known Neoerysiphe species, first of all, by its large chasmothecia with numerous somewhat elongated asci. Powdery mildews in subclade B4 are allied to American N. cumminsiana but they differ in having smaller fruiting bodies, chasmothecium being concave in the lower half, without evagination and aerial secondary mycelium, not appressed to the substrate. Morphologically, this group seems to be closer to subclade C4 but is distinguished by long cylindrical conidia, white secondary mycelium, and a less sculptured chasmothecial surface.
Taxonomy
Phylogenetic and morphological analyses have indicated that all three groups of Eurasian Neoerysiphe specimens parasitizing Asteraceae do not correspond to N. cumminsiana or any other known species of this genus, i.e. they have to be considered separate, new species, which are described as N. hiratae from Japan, N. joerstadii from Israel, and N. nevoi, including its variety scolymi, mainly from the Mediterranean region.
Neoerysiphe hiratae Heluta & S. Takam., sp. nov. — MycoBank MB513278; Fig. 3
Anamorph. Oidium subgenus Striatoidium.
Species nostra Neoerysiphe cumminsianae affinis est tamen mycelio secundario albo et tomentoso, conidiis longis, chasmotheciis minoribus et basi chasmothecii protuberatione carens bene differt.
Etymology. Named in honour of the famous Japanese mycologist Koji Amano (Hirata).
Mycelium amphigenous, often more developed along the veins of leaves, also caulicolous and on petioles, at first forming patches, then confluent. Primary mycelium thin, greyish, hyphal diam 5–6(–11) μm. Secondary mycelium arising from primary hyphae, pure white, hyphae smooth, without appressoria, 5–6 μm diam, forming a delicate or thick and tomentose web around ascomata. Hyphal appressoria very variable in shape and size, distinct, unlobed or slightly lobate, frequently in pairs, 7–10 × 4.5–6.5 μm. Conidiophores straight or sometimes arcuate, 112–154 μm, foot-cells cylindrical, 24–41 × 11–12 μm, frequently increasing in width towards the tip, followed by one shorter cell and conidial initials. Conidia catenate, mainly cylindrical with rounded ends, often oblong ellipsoidal, 27–48 × 11–17.5 μm, length/width ratio 1.9–3.5 (average 2.7). Chasmothecia scattered, hemispherical, depressed in the lower part, with an indistinct close-meshed or often knobbly peridial surface, (102–)105–153(–157) μm diam. Peridial cells polygonal or irregular, small, 9–20 × 5–12 μm. Appendages numerous, in the basal part of the chasmothecium, mycelioid, well developed, 0.5–2 times as long as the chasmothecial diam, 5–6 μm wide, hyaline, rarely somewhat brownish. Asci 7–12 per chasmothecium, immature, oblong ellipsoid, obpyriform, with an irregular outline, wide in the lower part and abruptly narrowed in the upper part, 46–57 × 21–30 μm, short-stalked, ascospores not developed before overwintering.
Specimens examined. Japan, Shiga, Mt Ibuki, on Ligularia stenocephala (Maxim.) Matsum. & Koidz. (Asteraceae), 7 Nov. 2004, S. Takamatsu, holotype TNS F-25684, isotype KW 34787F, MUMH 3442, rDNA sequence ex-type AB498962; Echime, Mt Ishiduchi, on Cacalia delphiniifolia Siebold & Zucc., 9 Nov. 1998, S. Takamatsu, MUMH 552, KW 34783F; Mt Bingamori, on C. delphiniifolia, 10 Nov. 1998, S. Takamatsu, MUMH 567, KW 34784F; Nagano, Kamikouchi, on C. hastata L. ssp. farfaraefolia, 3 Sept. 2004, S. Takamatsu, MUMH 3504, KW 34785F; Okayama, Kagamino Town, Forest Park, on Ligularia fischeri Turcz., 2 Nov. 2006, S. Takamatsu, MUMH 4471, KW 34786F; Mie, Mt Nonobori, on L. stenocephala, 21 Nov. 2004, S. Takamatsu, MUMH 3611, KW 34789F; Nagano, Kamikouchi, on L. stenocephala, 4 Sept. 2004, S. Takamatsu, MUMH 3505, KW 34788F.
Neoerysiphe joerstadii Heluta & S. Takam., sp. nov. — MycoBank MB513279; Fig. 4a–g
Anamorph. Not observed.
Species nostra Neoerysiphe cumminsianae similis est tamen absentia mycelii secundarii, numero ascorum majoribus, 16–32 in chasmothecio, ascis longioribus et basi chasmothecii protuberatione carens bene differt.
Etymology. Named in honour of the famous Norwegian mycologist Ivar Jørstad.
Primary mycelium amphigenous, very sparse, inconspicuous. Secondary mycelium absent. Appressoria obscure, unlobed or slightly lobate. Anamorph not observed. Chasmothecia scattered, hemispherical, very depressed in the lower part, with an indistinct close-meshed or knobbly peridial surface, (118–)125–171(–200) μm diam and 92–97 μm high. Peridial cells obscure, polygonal or irregular, small, 11–17(–33) × 10–14(–16) μm. Appendages numerous, in the basal part of the chasmothecium, mycelioid, 0.5–1 times as long as the chasmothecial diam, 4–6 μm wide, always hyaline, somewhat rough, interlaced with host fibres. Asci numerous, 16–32 per chasmothecium, oblong ellipsoid, with irregular outline when immature, increased in the lower part and narrowed in the upper part, 42–60 × 20–31 μm, stalked, ascospores not developed before overwintering.
Specimens examined. Israel, Golan Heights, Yehudiyya, 32°56′N, 35°41′E, on Phagnalon rupestre (L.) DC. (Asteraceae), 17 May 2004, S. Voytyuk, holotype KW 35717F, isotypes HAI 4239, 4245, KW 34794F, 34795F, MUMH 4668, rDNA sequence ex-type AB498976; Upper Galilee, Mt Meron, Nahal Keziv, on Phagnalon rupestre, 18 Mar. 2002, T. Andrianova, KW 35716F; Zefat (= Safed), 22 Aug. 1953, T. Rayss, HUJ 301/111 147S.
Neoerysiphe nevoi Heluta & S. Takam., sp. nov. — MycoBank MB513280; Fig. 5
Anamorph. Oidium subgenus Striatoidium.
Species nostra Neoerysiphe geranii affinis est tamen mycelio secundario bene evoluto, conidiis magis striatulis, peridio minus translucenti, ascis magis ellipsoideis et minus pyriformibus differt.
Etymology. Named in honour of the famous Israeli biologist Eviatar Nevo.
Mycelium amphigenous, also caulicolous, effuse. Primary mycelium thin, yellowish, hyphal diam 4.5–7.5 μm. Secondary mycelium arising from primary hyphae, white to brownish, hyphae smooth, without appressoria, 4.5–6.5 μm diam, surrounding ascomata mainly as a delicate web, persistent or evanescent. Hyphal appressoria on primary mycelium variable in shape and size, distinct, unlobed or slightly lobate, 6.5–8.5 × 4–5 μm. Conidiophores straight or somewhat arcuate, 125–165 μm, foot-cells cylindrical, 28.5–45.5(–100) × 9.5–12.5 μm, frequently increasing in width towards the tip, followed by 1–2 shorter cells and conidial initials. Conidia catenate, mainly ellipsoidal or short cylindrical with rounded ends, often almost limoniform, 23.5–36 × 11–17.5 μm, length/width ratio 1.5–2.5 (average 1.9). Conidial germ tube with a lobed appressorium typical of the Striatoidium type as defined by Cook & Braun (2009). Chasmothecia scattered, hemispherical, depressed in the lower part, with a distinctly meshed peridial surface, (90–)94–138(–148) μm diam. Peridial cells rather distinct, polygonal, rounded or irregular in shape, 11–30 × 11–17 μm. Appendages numerous, in the basal part of the chasmothecium, mycelioid, well developed or occasionally poorly developed, 0.5–2 times as long as the chasmothecial diam, 5.5–6.5(–8) μm wide, somewhat rough, brownish, yellow, rarely hyaline. Asci 6–12 per chasmothecium, immature, oblong ovoid to obpyriform, with irregular outline, wide in the lower part and narrowed in the upper part, 40.5–51.5 × 21–25 μm, stalked, ascospores not developed before overwintering.
Specimens examined. Israel, Upper Galilee, near ‘En Kamonnim, 32°54′N, 35°26′E, on Tolpis virgata (Desf.) Bertol. (Asteraceae), 2 May 2004, S. Voytyuk, holotype KW 34802F; isotypes KW 34803F, MUMH 4679, rDNA sequence ex-type AB498974; Haifa, Mt Carmel, near Institute of Evolution, on Crepis aspera L., 22 Apr. 2004, S. Voytyuk, HAI 4164, KW 34791F, MUMH 4667; Atlit near Haifa, on C. aspera, 5 Apr. 2004, S. Voytyuk, HAI 4235, KW 34790F, MUMH 4873; Lower Halilee, Mi’ilya near Ma’a lot, on C. sancta (L.) Babc., 25 Mar. 2004, S. Voytyuk, HAI 4238, KW 34792F, MUMH 4874; Carmel Coast, near Atlit, on Crepis sp., 19 Mar. 2002, V. Heluta, anamorph, KW 35721F; Haifa, on Crepis sp., 15 Mar. 2002, V. Heluta, KW 35722F; Pardes Hana, on Crepis sp., 12 Apr. 2002, E. Nevo, KW 35723F; Carmel Coast, near Atlit, on Hedypnois cretica (L.) Willd., 27 Apr. 2004, S. Voytyuk, HAI 4101, 4102, KW 34793F, 35724F, MUMH 4875; Golan Heights, Avné Etan near Ramat Magshimim, on Picris altissima Ledeb. ex Rchb., 2 Mar. 2004, S. Voytyuk, anamorph, HAI 4110, 4112, KW 34796F, 34797F; Mi’ilya, on P. amalecitana (Boiss.) Eig, 25 Mar. 2004, S. Voytyuk, HAI 4113, 4114, KW 34798F, 35725F, MUMH 4669; Northern Negev, near Lahav, on Rhagadiolus stellatus Gaertn., 21 Mar. 2004, S. Voytyuk, KW 34799F, MUMH 4670; Lower Galilee, Alloné Abba near Qirat Tiv’on, on Thrincia tuberosa DC., 18 Apr. 2004, S. Voytyuk, HAI 4123, KW 34801F, MUMH 4678; Mt Carmel, Muchraka, on T. tuberosa, 17 Mar. 2004, S. Voytyuk, HAI 4124, KW 35726F; Upper Galilee, near ‘En Kamonnim, on Tolpis virgata, 2 May 2004, S. Voytyuk, KW 34802F, 34803F, MUMH 4679; Lower Galilee, near Alloné Abba near Qirat Tiv’on, on T. virgata, 18 Apr. 2004, S. Voytyuk, HAI 4182, KW 35727F. – Ukraine, Autonomous Republic of Crimea, Yalta, Livadia, on Chondrilla sp., 22 Aug. 2004, V. Heluta, KW 58373F, MUMH 4672; Alupka, on Crepis micrantha Czerep., 17 July 1982, V. Heluta, KW 11752F; Miskhor, on C. micrantha, 8 July 1959, M. Sokolova, KW 35720F; Chornomorske urban village, on C. rhoeadifolia M. Bieb., 13 July 1982, V. Heluta, anamorph, KW 11754F; Chornomorske distr., Daleke village, on C. rhoeadifolia, 12 July 1982, V. Heluta, KW 11755F; Chornomorske distr., Olenivka village, on C. rhoeadifolia, 16 July 1982, V. Heluta, KW 11756F; Saky, on C. rhoeadifolia, 15 July 1978, V. Heluta, anamorph, KW 11753F; Kherson region, Hola Prystan distr., Black Sea Biosphere reserve, on C. rhoeadifolia, 5 July 1978, V. Heluta, anamorph, KW 11757F; Odessa region, Mykolayiv distr., Nastasiyivka village, on C. rhoeadifolia, 26 Sept. 1977, V. Heluta, anamorph, KW 11758F; Autonomous Republic of Crimea, Yalta Nature reserve, Mt Ai-Petri, on Taraxacum sp., 27 July 1981, V. Heluta, KW 11777, MUMH 4656.
Neoerysiphe nevoi var. scolymi Heluta & S. Takam., var. nov. — MycoBank MB513281; Fig. 5k
Anamorph. Oidium subgenus Striatoidium.
A typo peridio profunde scrobiculato differt.
Etymology. Named from host plant genus.
This variety differs from the type by a deeply pitted peridial surface, i.e. a clearly visible mesh is formed by high ridges surrounding cell margins.
Specimen examined. Israel, Haifa, Mt Carmel, Nahal Oren, on Scolymus hispanicus L. (Asteraceae), 31 May 2005, S. Voytyuk, holotype KW 34800F, isotype MUMH 4671, rDNA sequence ex-type AB498975.
KEY FOR IDENTIFICATION OF NEOERYSIPHE SPECIES PARASITIZING ASTERACEAE
-
1.
Mycelium inconspicuous, secondary mycelium absent; chasmothecia rather large, mainly 125–170 μm, up to 200 μm; asci 16–32 per chasmothecium; on Phagnalon; in the Mediterranean region……………N. joerstadii
-
1.
Mycelium well developed, secondary mycelium present; chasmothecia smaller, usually 95–150 μm, if larger then only about 10 asci per chasmothecium; on other hosts……………2
-
2.
Chasmothecia large, mainly 125–165 μm, with visible evagination on the lower side; on Senecio and probably other hosts; in North and South America……………N. cumminsiana
-
2.
Chasmothecia smaller, concave in the lower part……………3
-
3.
Both primary and secondary mycelia pure white; conidia mainly cylindrical with rounded ends, often oblong ellipsoidal, average length/width ratio of 2.7; chasmothecial appendages hyaline, occasionally somewhat brownish; peridial surface with indistinct meshes or even knobbly; on Cacalia and Ligularia; in Japan……………N. hiratae
-
3.
Primary mycelium yellowish, even somewhat brownish, secondary mycelium mainly white or sometimes faintly pigmented; conidia ellipsoidal or short cylindrical with rounded ends, often somewhat limoniform, average length/width ratio 1.9; appendages brownish, occasionally hyaline; peridial surface with a clear meshes or even deeply pitted; on different members of the Asteraceae; mainly in the Mediterranean region……………N. nevoi
DISCUSSION
Based on the ITS sequences as well as the 28S rDNA sequences, Takamatsu et al. (2008) reported that Neoerysiphe specimens of hosts belonging to the Asteraceae are divided into three subgroups, each of which corresponds to different host tribes, viz. Heliantheae and Eupatorieae (Group 2a), Senecioneae (2c), and Astereae (2d) (see Fig. 2 in Takamatsu et al. (2008) and Table 2). American and Japanese samples of ‘N. cumminsiana’ appeared in different subgroups. Consequently, the authors concluded that ‘N. cumminsiana’ could be divided into two different species. The present study confirmed this assumption. Morphological analysis demonstrated that Japanese samples are uniform, differ clearly from the true N. cumminsiana, and belong to a separate species here named N. hiratae. This species parasitizes Cacalia and Ligularia (tribe Senecioneae, subfamily Asteroideae) and is known only from Japan (East Asia). Other new species analysed here were N. joerstadii collected on Phagnalon rupestre (tribe Gnaphalieae in Asteroideae) in Israel (West Asia) and N. nevoi infecting several hosts belonging to different genera in the tribe Cichorieae (in Cichorioideae), viz. Chondrilla, Crepis, Hedypnois, Picris, Rhagadiolus, Scolymus, Taraxacum, Thrincia (= Leontodon), and Tolpis (Table 2). Thus, Eurasian Neoerysiphe species are clearly connected with specific tribes of the Asteraceae. Similar close relationships between powdery mildews and their host tribes of the Asteraceae were also reported in Golovinomyces (Matsuda & Takamatsu 2003).
Table 2.
Neoerysiphe and Oidium subgenus Striatoidium species aligned with their hosts in the Asteraceae and their geographical regions.
| Fungal species | Subfamily | Tribe | Genus | Geographical region |
|---|---|---|---|---|
| Neoerysiphe | ||||
| cumminsiana | Asteroideae | Coreopsideae | Bidens | South America |
| Eupatorieae | Eupatorium | Norh America | ||
| Senecioneae | Senecio | North America | ||
| hiratae | Asteroideae | Senecioneae | Cacalia (=Parasenecio), | Eastern Asia |
| Ligularia | ||||
| joerstadii | Asteroideae | Gnaphalieae | Phagnalon | Mediterranean |
| nevoi | Cichorioideae | Cichorieae | Chondrilla, Crepis, | Mediterranean |
| Hedypnois, Picris, | ||||
| Rhagadiolus, Scolymus, | ||||
| Taraxacum, Thrincia | ||||
| (= Leontodon), Tolpis | ||||
| Oidium | ||||
| baccharidis sp. | Asteroideae | Astereae | Baccharis | South America |
| Millerieae | Galinsoga | South America | ||
As mentioned above, Voytyuk et al. (2004) expressed doubt on the correctness of Heluta’s (1992) hypothesis about migrations of powdery mildew fungi because the natural distribution of N. cumminsiana sensu Heluta was not in accordance with this hypothesis. However, it is now clear that these authors dealt with a species complex composed of four different species, namely the American genuine N. cumminsiana, the Japanese N. hiratae, and the Mediterranean taxa N. joerstadii and N. nevoi. We should note that the description of N. cumminsiana in the monograph of Braun (1987) combined characteristics of N. cumminsiana and N. hiratae. Therefore, only the original description published by Braun (1983) refers to N. cumminsiana s.str. Neoerysiphe nevoi is currently only known from Israel and Ukraine but this species has probably a much wider distribution. It is very likely that all collections formerly reported as Erysiphe cumminsiana, E. galeopsidis or E. galii on Asteraceae from European countries and Africa (Amano 1986, Braun 1987, Gorter 1987) belong to this species. A few years ago we examined all specimens of powdery mildews from the herbarium of Jerusalem University (HUJ, Israel). Many specimens originally identified as E. cichoracearum actually belonged to N. nevoi. It is possible that numerous records of ‘E. cichoracearum’ on hosts belonging to the genera Crepis, Filago, Hedypnois, Hypochaeris, Picris, and Rhagadiolus on the Canary and the Balearic Islands (Jørstad 1962a, b), in Portugal (de Mendonça & de Sequeira 1963, de Sequeira & de Mendonça 1965, de Sequeira 1969, 1975, 1978, 1981), Romania (Sandu-Ville 1967, Bontea 1986), Spain (Durrieu & Mercé 1972), and central Europe (Blumer 1967) also belong to N. nevoi. De Sequeira (1978) reported that fruiting bodies of ‘Erysiphe cichoracearum’ on Hedypnois cretica, Picris echioides, P. hieracioides, and Tolpis barbata collected in Portugal were immature, and the descriptions of other characters of these fungi agree well with those of N. nevoi.
We collected N. joerstadii only on Phagnalon rupestre, but according to Amano (1986), E. cichoracearum was recorded on this host and Ph. saxatile in France, on the Balearic and the Canary Islands, and in the Spanish Sahara. These specimens very likely belong to N. joerstadii. Furthermore, the chasmothecia of this fungus on Ph. saxatile from the Balearic and the Canary Islands measured 130–200 μm diam (Jørstad 1962a, b). Such a size range fully conforms to N. joerstadii.
As explained above, a powdery mildew on hosts belonging to Geranium in Ukraine was identified as N. geranii by Heluta (2001). However, since this species was known only from Japan and New Zealand (Amano 1986, Nomura 1997), we compared Japanese and Ukrainian samples including the type specimen. Phylogenetically and morphologically, all specimens were found to be uniform. Thus, the true N. geranii was correctly recorded in Ukraine as an invasive species.
In conclusion, molecular and morphological evidence revealed that at least four Neoerysiphe species, viz. N. cumminsiana, N. hiratae, N. joerstadii, and N. nevoi, are able to infect Asteraceae. Some of these fungi, above all N. joerstadii and N. nevoi, are probably common in the Mediterranean region but have been formerly identified and reported mainly as E. cichoracearum. Thus, the identity of powdery mildews collected on the Asteraceae in the Mediterranean and adjacent regions needs re-examination in the light of these findings.
Acknowledgments
We are indebted to Tetiana V. Andrianova (Ukraine) for kindly donating a specimen of N. joerstadii, Nadiya and Sergej Mosyakin (Ukraine) for specimens of N. geranii, Uwe Braun (Germany) for providing the isotype of N. cumminsiana, Eviatar Nevo and Solomon P. Wasser (Israel) for appreciable support of our field work in Israel. We thank Roger Cook (Great Britain) for help with the English and valuable comments on the manuscript. We also gratefully acknowledge the late Mr. D. Diomenko, for his help with scanning electron microscopy.
REFERENCES
- Amano K. 1986. Host range and geographical distribution of the powdery mildew fungi Japan Scientific Societies Press, Tokyo, Japan: . [Google Scholar]
- Bahcecioglu Z, Braun U, Kabaktepe S. 2006. Neoërysiphe rubiae – a new powdery mildew species on Rubia cf. tinctoria from Turkey. Nova Hedwigia 83: 489 – 492 . [Google Scholar]
- Blumer S. 1967. Echte Mehltaupilze (Erysiphaceae). Ein Bestimmungsbuch für die in Europa vorkommenden Arten Fischer Verlag, Jena, Germany: . [Google Scholar]
- Bontea V. 1986. Ciuperci parazite si saprofite din România. Vol. 2 Editura Academici Republicii Socialiste România, Bucuresti, Romania: . [Google Scholar]
- Braun U. 1978. Beitrag zur Systematik und Nomenklatur der Erysiphales. Feddes Repertorium 88: 655 – 665 . [Google Scholar]
- Braun U. 1981. Taxonomic studies in the genus Erysiphe. I. Generic delimitation and position in the system of the Erysiphaceae. Nova Hedwigia 34: 679 – 719 . [Google Scholar]
- Braun U. 1983. Descriptions of new species and combinations in Microsphaera and Erysiphe IV. Mycotaxon 18: 113 – 129 . [Google Scholar]
- Braun U. 1987. A monograph of the Erysiphales (powdery mildews). Beihefte zur Nova Hedwigia 89: 1 – 700 . [Google Scholar]
- Braun U. 1999. Some critical notes on the classification and the generic concept of the Erysiphaceae. Schlechtendalia 3: 48 – 54 . [Google Scholar]
- Cook RTA, Braun U. 2009. Conidial germination patterns in powdery mildews. Mycological Research 113: 616 – 636 . [DOI] [PubMed] [Google Scholar]
- Cook RTA, Inman AJ, Billings C. 1997. Identification and classification of powdery mildew anamorphs using light and scanning electron microscopy and host range data. Mycological Research 101: 975 – 1002 . [Google Scholar]
- Durrieu G, Mercé J . 1972. Erysiphacées du sud-est de l’Espagne. Bulletin de la Société Mycologique de France 38: 175 – 191 . [Google Scholar]
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783 – 791 . [DOI] [PubMed] [Google Scholar]
- Gorter GJMA. 1987. Relevance of conidial surface structure for identifying Erysiphe galeopsidis DC. South African Journal of Science 83: 112 – 114 . [Google Scholar]
- Heluta VP. 1988. Filogeneticheskie vzaimosvyazi mezhdu rodami erizifalnykh gribov i nekotorye voprosy sistematiki poryadka Erysiphales. Biologicheskii Zhurnal Armenii 41: 351 – 358 . [Google Scholar]
- Heluta VP. [as Geluta WP]. 1989. Flora gribov Ukrainy. Muchnistorosyanye griby Naukova Dumka, Kiev, Ukraine: . [Google Scholar]
- Heluta VP. 1992. Hypotesa pro pokhodzhennya ta migratsii grybiv poryadku Erysiphales. Ukrainskyi Botanichnyi Zhurnal 49, 5: 5 – 14 . [Google Scholar]
- Heluta VP. 1993. Geografichnyi analiz boroshnystorosyanykh grybiv Ukrainy. Ukrainskyi Botanichnyi Zhurnal 50, 2: 79 – 85 . [Google Scholar]
- Heluta VP. 1995. Micoflorogenetychnyi analiz vydovogo skladu boroshnystorosyanykh grybiv Ukrainy. Ukrainskyi Botanichnyi Zhurnal 52: 200 – 206 . [Google Scholar]
- Heluta VP. 1999. Poshyrennya v Ukraini Golovinomyces cumminsianus (U. Braun) Heluta (Erysiphales). Ukrainskyi Botanichnyi Zhurnal 56: 431 – 433 . [Google Scholar]
- Heluta VP. 2001. Neoerysiphe geranii (Y. Nomura) U. Braun – novyi dlya Ukrainy vyd boroshnystorosyanogo gryba. Ukrainskyi Botanichnyi Zhurnal 58: 239 – 242 . [Google Scholar]
- Hirata T, Takamatsu S. 1996. Nucleotide sequence diversity of rDNA internal transcribed spacers extracted from conidia and cleistothecia of several powdery mildew fungi. Mycoscience 37: 283 – 288 . [Google Scholar]
- Holmgren PK, Holmgren NH, Barbett LC. 1990. Index herbariorum, Part. 1: The Herbaria of the World. 8th edn Regnum vegetabile 120: 1 – 163 . [Google Scholar]
- Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754 – 755 . [DOI] [PubMed] [Google Scholar]
- Jørstad I . 1962a. Investigations on the Uredinales and other parasitic fungi in Mallorca and Menorca. Skrifter utgitt av Det Norske Videnskaps-Akademi i Oslo. I. Matematisk-Naturvidenskapelig Klasse. Ny Serie 2: 1 – 73 . [Google Scholar]
- Jørstad I . 1962b. Parasitic micromycetes from the Canary Islands. Skrifter utgitt av Det Norske Videnskaps-Akademi i Oslo. I. Matematisk-Naturvidenskapelig Klasse. Ny Serie 7: 1 – 71 . [Google Scholar]
- Liu SY, Takamatsu S, Yang LL, Wang XM, Lu D, Luo L. 2005. First report of Neoerysiphe galeopsidis on Althaea rosea. Plant Pathology 55: 297 . [Google Scholar]
- Maddison DR, Maddison WP. 2005. MacClade 4: Analysis of phylogeny and character evolution. Version 4.08 Sinauer, Sunderland, MA: . [DOI] [PubMed] [Google Scholar]
- Matsuda S, Takamatsu S. 2003. Evolution of host-parasite relationship of Golovinomyces (Ascomycete: Erysiphales) inferred from nuclear rDNA sequences. Molecular Phylogenetics and Evolution 27: 314 – 327 . [DOI] [PubMed] [Google Scholar]
- Mendonça AA de, Sequeira MPS de . 1963. Erysiphaceae Lusitaniae I. Agronomia Lusitana 24: 87 – 131 . [Google Scholar]
- Mori Y, Sato Y, Takamatsu S. 2000. Evolutionary analysis of the powdery mildew fungi using nucleotide sequences of the nuclear ribosomal DNA. Mycologia 92: 74 – 93 . [Google Scholar]
- Nixon KC. 1999. The Parsimony Ratchet, a new method for rapid parsimony analysis. Cladistics 15: 407 – 414 . [DOI] [PubMed] [Google Scholar]
- Nomura Y. 1997. Taxonomical study of Erysiphaceae of Japan Yokendo Ltd., Tokyo, Japan: . [Google Scholar]
- Nylander JAA. 2004. MrModeltest v2 Evolutionary Biology Centre, Uppsala University; . Program distributed by the author . [Google Scholar]
- Saenz GS, Taylor JW. 1999. Phylogeny of the Erysiphales (powdery mildews) inferred from internal transcribed spacer ribosomal DNA sequences. Canadian Journal of Botany 77: 150 – 168 . [Google Scholar]
- Sandu-Ville C. 1967. Ciupercile Erysiphaceae din România Editura Academiei Republicii Socialiste România, Bucuresti, Romania: . [Google Scholar]
- Sequeira MPS de . 1969. Erysiphaceae Lusitaniae III. Agronomia Lusitana 30: 5 – 21 . [Google Scholar]
- Sequeira MPS de . 1975. Erysiphaceae Lusitaniae V. Agronomia Lusitana 36: 281 – 306 . [Google Scholar]
- Sequeira MPS de . 1978. Erysiphaceae Lusitaniae VI. Agronomia Lusitana 38: 297 – 320 . [Google Scholar]
- Sequeira MPS de . 1981. Erysiphaceae Lusitaniae VII. Agronomia Lusitana 41: 93 – 112 . [Google Scholar]
- Sequeira MPS de, Mendonça AA de . 1965. Erysiphaceae Lusitaniae II. Agronomia Lusitana 26: 21 – 43 . [Google Scholar]
- Sikes DS, Lewis PO. 2001. Beta software, version 1. PAUPRat: PAUP* implementation of the parsimony ratchet Distributed by the authors. Department of Ecology and Evolutionary Biology, University of Connecticut, Storrs, USA: . [Google Scholar]
- Swofford DL. 2001. PAUP: Phylogenetic analysis using parsimony (and other methods) 4.0b8 Sinauer, Sunderland, MA: . [Google Scholar]
- Takamatsu S, Havrylenko M, Wolcan SM, Matsuda S, Niinomi S. 2008. Molecular phylogeny and evolution of the genus Neoerysiphe (Erysiphaceae, Ascomycota). Mycological Research 112: 639 – 649 . [DOI] [PubMed] [Google Scholar]
- Takamatsu S, Matsuda S, Niinomi S, Havrylenko M. 2006. Molecular phylogeny supports a northern hemisphere origin of Golovinomyces (Ascomycota: Erysiphales). Mycological Research 110: 1093 – 1101 . [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 24: 4876 – 4882 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytyuk S. [as Voityuk], Heluta V, Nevo E. 2004. Neoerysiphe cumminsiana (Erysiphales, Eumycota), a new powdery mildew fungus in Israel. Flora Mediterranea 14: 267 – 273 . [Google Scholar]
- Voytyuk SO, Heluta VP, Wasser SP, Nevo E. 2006. Genus Neoerysiphe in Israel: species composition, host range and distribution. Mycotaxon 97: 247 – 256 . [Google Scholar]
- Walsh PS, Metzger DA, Higuchi R. 1991. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques 10: 506 – 513 . [PubMed] [Google Scholar]