Abstract
Decline in the productivity of Eucalyptus hybrid cutting production in the Guangdong Province of China is linked to cutting rot associated with several Calonectria spp. The aim of this study was to identify these fungi using morphological and DNA sequence comparisons. Two previously undescribed Calonectria spp., Ca. pseudoreteaudii sp. nov. and Ca. cerciana sp. nov. were identified together with Ca. pauciramosa. Calonectria pseudoreteaudii resides in the Ca. reteaudii complex and Ca. cerciana is closely related to Ca. morganii. Connected to the discovery of Ca. pseudoreteaudii, species in the Ca. reteaudii complex were re-considered and the group is shown to accommodate two cryptic species. These originate from Australia and are described as Ca. queenslandica sp. nov. and Ca. terrae-reginae sp. nov.
Keywords: Australia, Calonectria, China, Cylindrocladium, Eucalyptus, systematics
INTRODUCTION
Species of Calonectria (Ca.), and their Cylindrocladium (Cy.) anamorphs, are important plant pathogens worldwide (Crous 2002). Past taxonomic studies on these pathogens have focused on morphology and sexual compatibility to delimit new species (Peerally 1991, Crous & Wingfield 1994). More recently, DNA sequence comparisons have resulted in the recognition of several species complexes in Calonectria (Schoch et al. 1999, Crous et al. 2004, 2006a).
One of the newly recognised groups in Calonectria is the Ca. reteaudii complex (Crous & Kang 2001, Kang et al. 2001). The complex encompasses several Calonectria spp. with Cylindrocladium anamorphs morphologically similar to the Ca. reteaudii anamorph state, having clavate vesicles with multiseptate macroconidia. These include Cy. angustatum, Cy. hurae, Ca. leguminum and Ca. rumohrae (Crous 2002).
The Ca. morganii species complex (Crous et al. 1993, Schoch et al. 2001) includes Ca. insularis, Ca. morganii and Cy. hawksworthii (Schoch et al. 1999, 2001, Crous 2002). Species in this complex are characterised by ellipsoid to obpyriform to clavate vesicles, 1-septate conidia and orange to red perithecia producing 1-septate ascospores (Peerally 1991, Schoch et al. 1999, Crous 2002).
Species in both the Ca. reteaudii and the Ca. morganii complexes are responsible for a wide variety of disease symptoms on several plant hosts in subtropical and tropical regions of the world (Bolland et al. 1985, Peerally 1991, Sharma & Mohanan 1991, Booth et al. 2000, Crous 2002, Rodas et al. 2005). Disease symptoms include leaf blight (Sharma & Mohanan 1991, Booth et al. 2000, Rodas et al. 2005) and cutting rot (Sharma & Mohanan 1982, Sharma et al. 1984, Schoch et al. 1999, Crous 2002). Of these, leaf blight is most devastating in the tropical regions of South East Asia and South America and is particularly serious on Eucalyptus spp. (Booth et al. 2000, Crous & Kang 2001, Crous 2002, Rodas et al. 2005).
Decline in Eucalyptus hybrid cutting production due to cutting rot has recently been observed in a commercial forest nursery in the Guangdong Province of China. Initial investigations indicated that the causal agents were unknown Calonectria spp. that represented species in the Ca. reteaudii and Ca. morganii complexes (Zhou et al. 2008). The aim of this study was to identify these Calonectria spp. using morphological characteristics and phylogenetic inference. In addition, the taxonomic status and circumscription of species in the Ca. reteaudii species complex were re-considered.
MATERIALS AND METHODS
Isolates
Hybrid clonal Eucalyptus cuttings showing symptoms of cutting rot were collected in the nursery of the China Eucalypt Research Centre (CERC) in Guangdong Province, China. Diseased cuttings were placed in moist chambers and incubated for 48 h at room temperature to induce sporulation. Direct isolations were made onto malt extract agar (2 % w/v; MEA; Biolab, Midrand, South Africa) and cultures were incubated for 7 d at 25 °C under continuous near-ultraviolet light. For each isolate, single conidial cultures were prepared on MEA and representative strains are maintained in the culture collection (CMW) of the Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, Pretoria, South Africa, the Centraalbureau voor Schimmelcultures (CBS), Utrecht, The Netherlands, and the China Eucalypt Research Centre (CERC), ZhanJiang, China. Isolates of Ca. reteaudii were obtained from the culture collection of CBS, including representative isolates used in the study of Kang et al. (2001).
DNA sequence comparisons
Calonectria isolates were grown on MEA for 7 d. Mycelium was then scraped from the surfaces of Petri dishes, freeze-dried, and ground to a powder in liquid nitrogen, using a mortar and pestle. DNA was extracted from the powdered mycelium as described by Lombard et al. (2008). Three loci were amplified and sequenced. These included a fragment of the β-tubulin (BT) gene region using primers T1 (O’Donnell & Cigelnik 1997) and CYLTUB1R (Crous et al. 2004), a fragment of the histone H3 (HIS3) gene region using primers CYLH3F and CYLH3R (Crous et al. 2004) and a fragment of the Translation Elongation Factor-1α (TEF-1α) gene region using primers EF1-728F (Carbone & Kohn 1999) and EF2 (O’Donnell et al. 1998). The PCR reaction mixture used to amplify the different loci consisted of 2.5 units FastStart Taq polymerase (Roche Applied Science, USA), 10× PCR buffer, 1–1.5 mM MgCl2, 0.25 mM of each dNTP, 0.5 μm of each primer and ± 30 ng of fungal genomic DNA, made up to a total reaction volume of 25 μL with sterile distilled water.
Amplified fragments were purified using High Pure PCR Product Purification Kit (Roche, USA) and sequenced in both directions. For this purpose, the BigDye terminator sequencing kit (v3.1, Applied Biosystems, USA) and an ABI PRISMTM 3100 DNA sequencer (Applied Biosystems) were used. All PCRs and sequencing reactions were performed on an Eppendorf Mastercycler Personal PCR (Eppendorf AG, Germany) with cycling conditions as described in Crous et al. (2004, 2006a) for each locus.
Generated sequences were added to other sequences obtained from GenBank (http://www.ncbi.nlm.nih.gov) and these were assembled and aligned using Sequence Navigator (v1.0.1, Applied Biosystems) and MAFFT (v5.11, Katoh et al. 2005), respectively. The aligned sequences were then manually corrected where needed.
To determine whether the sequence datasets for the separate loci are congruent, tree topologies of 70 % reciprocal Neighbour-Joining bootstrap with Maximum Likelihood distances were compared visually to identify conflicts between partitions (Mason-Gamer & Kellogg 1996, Gueidan et al. 2007). Molecular evolution models for the separate partitions were determined in Modeltest v3.7 (Posada & Crandall 1998) and bootstrap analyses were run for 10k replicates.
PAUP (Phylogenetic Analysis Using Parsimony, v4.0b10, Swofford 2002) was used to analyse the DNA sequence datasets. Phylogenetic relationships were estimated by a heuristic searches with 1 000 random addition sequences and tree bisection-reconnection was used, with the branch swapping option set on ‘best trees’ only.
All characters were weighted equally and alignment gaps were treated as missing data. Measures calculated for parsimony included tree length (TL), consistency index (CI), retention index (RI) and rescaled consistence index (RC). Bootstrap analysis (Hillis & Bull 1993) was based on 1 000 replications. All sequences for the isolates studied were analysed using the Basic Local Alignment Search Tool for Nucleotide sequences (BLASTN, Altschul et al. 1990). The phylogenetic analysis was done on two separate sequence datasets.
Datasets were separated based on morphological characteristics to allow combinations of gene regions. The first dataset included 40 partial gene sequences per gene, representing 13 Calonectria spp. that form part of the Ca. reteaudii species complex and other morphologically similar species (Table 1). These included isolates used by Kang et al. (2001), and are Calonectria species with anamorph states producing large, multiseptate macroconidia and stipe extensions terminating in clavate vesicles. The second dataset consisted of 20 partial gene sequences per gene, representing 12 Calonectria spp. of the Ca. morganii species complex, and other morphologically similar species. This group of Calonectria species is characterised by smaller, 1–3-septate macrocondia, and stipe extensions terminating in ellipsoidal to obpyriform vesicles. Calonectria colombiensis (CBS 112221) and Cy. chinense (CBS 112744) were used as outgroup taxa in both analyses (Lombard et al. 2009). All sequences were deposited in GenBank and the alignments in TreeBASE (http://www.treebase.org).
Table 1.
Strains of Calonectria and Cylindrocladium used in the phylogenetic study.
| Species | Isolate number1 | GenBank accession no. |
Host | Origin | Collector | ||
|---|---|---|---|---|---|---|---|
| β-tubulin | Histone H3 | Translation elongation factor -1α | |||||
| Cy. angustatum | CBS 109169 | DQ190593 | DQ190695 | FJ918552 | Tillandsia capitata | USA | R.M. Leahy |
| CBS 109065T | AF207543 | DQ190656 | FJ918551 | T. capitata | USA | R.M. Leahy | |
| Ca. cerciana sp. nov. | CMW 25309 (= CBS 123693)T | FJ918510 | FJ918528 | FJ918559 | Eucalyptus urophylla × E. grandis cutting | China | M.J. Wingfield & X.D. Zhou |
| CMW 25290 (= CBS 123695) | FJ918511 | FJ918529 | FJ918560 | E. urophylla × E. grandis cutting | China | M.J. Wingfield & X.D. Zhou | |
| CMW 25288 | GQ25288 | GQ267243 | GQ267288 | E. urophylla × E. grandis cutting | China | M.J. Wingfield & X.D. Zhou | |
| Ca. colombiensis | CBS112221 | AY725620 | AY725663 | AY725712 | Soil | Colombia | M.J. Wingfield |
| Cy. chinense | CBS112744 | AY725618 | AY725660 | AY725709 | Soil | China | E.C.Y. Liew |
| Ca. brassicae | CBS 111869T | AF232857 | DQ190720 | FJ918567 | Argyreia sp. | South East Asia | |
| CBS 111478 | DQ190611 | DQ190719 | FJ918567 | Soil | Brazil | A.C. Alfenas | |
| Cy. hawksworthii | CBS 111870T | AF333407 | DQ190649 | FJ918558 | Nelumbo nucifera | Mauritius | A. Peerally |
| Cy. hurae | CBS 114551 | AF333408 | DQ190728 | FJ918548 | Rumohra adiantiformis | Brazil | A.C. Alfenas |
| Ca. insularis (= Cy. insulare) | CBS 114558 | AF210861 | FJ918526 | FJ918556 | Soil | Madagascar | P.W. Crous |
| CBS 114559 | AF210862 | FJ918525 | FJ918555 | Soil | Madagascar | C. L. Schoch | |
| Ca. leguminum (= Cy. leguminum) | CBS 728.68T | AF389837 | DQ190654 | FJ918547 | Annona squamosa | Brazil | M.B. Figueiredo |
| Cy. leucothoës | CBS 109166T | FJ918508 | FJ918523 | FJ918553 | Leucothoë axillaris | USA | N.E. El-Gholl |
| Cy. multiseptatum | CBS 112682T | DQ190573 | DQ190659 | FJ918535 | Eucalyptus sp. | Indonesia | M.J. Wingfield |
| Ca. pauciramosa (= Cy. pauciramosum) | CMW 5683 | FJ918514 | FJ918531 | FJ918565 | Eucalyptus sp. | Brazil | A.C. Alfenas |
| CMW 30823 | FJ918515 | FJ918532 | FJ918566 | E. grandis | South Africa | P.W. Crous | |
| CMW 25311 | FJ918516 | FJ918533 | FJ918569 | E. urophylla × E. grandis cutting | China | M.J. Wingfield & X.D. Zhou | |
| CMW 25283 | FJ918517 | FJ918534 | FJ918570 | E. urophylla × E. grandis cutting | China | M.J. Wingfield & X.D. Zhou | |
| Ca. pseudoreteaudii sp. nov. | CMW 25310 (= CBS 123694)T | FJ918504 | FJ918519 | FJ918541 | E. urophylla × E. grandis cutting | China | M.J. Wingfield & X.D. Zhou |
| CMW 25292 (= CBS 123696) | FJ918505 | FJ918520 | FJ918542 | E. urophylla × E. grandis cutting | China | M.J. Wingfield & X.D. Zhou | |
| CMW 25284 | GQ267205 | GQ267244 | GQ267289 | E. urophylla × E. grandis cutting | China | M.J. Wingfield & X.D. Zhou | |
| CMW 25285 | GQ267206 | GQ267245 | GQ267290 | E. urophylla × E. grandis cutting | China | M.J. Wingfield & X.D. Zhou | |
| Ca. pseudospathiphylli | CBS 109165 | FJ918513 | AF348241 | FJ918562 | Soil | Ecuador | M.J. Wingfield |
| Ca. pteridis | CBS 111793 | DQ190578 | DQ190679 | FJ918563 | Arachnoides adiantiformis | USA | |
| CBS 111871 | DQ190579 | DQ190680 | FJ918564 | Pinus sp. | Spain | T.L. Krugner | |
| Ca. queenslandica sp. nov. | CMW 30604 (= CBS 112146 = | ||||||
| CPC 3213 = DRFI000147)T | AF389835 | FJ918521 | FJ918543 | E. urophylla | Australia | B. Brown | |
| CMW 30603 (= CBS 112155 = | |||||||
| CPC 3210 = DFRI00172) | AF389834 | DQ190667 | FJ918544 | E. pellita | Australia | K.M. Old | |
| Ca. reteaudii | CBS 582.50 | AF389836 | DQ190673 | FJ918540 | Hibiscus sabdariffa | Indonesia | — |
| CBS 112143 | GQ240642 | DQ190660 | FJ918536 | E. camaldulensis | Vietnam | M.J. Dudzinski | |
| CBS 112144T | AF389833 | DQ190661 | FJ918537 | E. camaldulensis | Vietnam | M.J. Dudzinski | |
| CBS 112147 | AF389830 | DQ190663 | FJ918539 | E. camaldulensis | Vietnam | M.J. Dudzinski | |
| CBS 112153 | AF389831 | FJ918518 | FJ918538 | E. camaldulensis | Vietnam | M.J. Dudzinski | |
| CBS 113582 | GQ240643 | GQ240659 | GQ240675 | Eucalyptus sp. | Thailand | — | |
| CBS 113583 | GQ240644 | GQ240660 | GQ240676 | Eucalyptus sp. | Madagascar | P.W. Crous | |
| CMW 18446 | GQ240649 | GQ240665 | GQ240681 | E. urophylla | Indonesia | M.J. Wingfield | |
| CMW 18448 | GQ240650 | GQ240666 | GQ240682 | E. urophylla | Indonesia | M.J. Wingfield | |
| CMW 18450 | GQ240651 | GQ240667 | GQ240683 | E. grandis | Thailand | M.J. Wingfield | |
| CMW 18458 | GQ240654 | GQ240670 | GQ240686 | E. urophylla | Indonesia | M.J. Wingfield | |
| CMW 18462 | GQ240653 | GQ240669 | GQ240685 | E. urophylla | Indonesia | M.J. Wingfield | |
| CMW 18463 | GQ240646 | GQ240662 | GQ240678 | E. urophylla | Indonesia | M.J. Wingfield | |
| CMW 20597 | GQ240645 | GQ240661 | GQ240677 | E. grandis | Thailand | M.J. Wingfield | |
| CMW 31177 | GQ240657 | GQ240673 | GQ240689 | Eucalyptus sp. | Indonesia | M.J. Wingfield | |
| CMW 31178 | GQ240656 | GQ240672 | GQ240688 | Eucalyptus sp. | Indonesia | M.J. Wingfield | |
| CMW 31179 | GQ240655 | GQ240671 | GQ240687 | Eucalyptus sp. | Indonesia | M.J. Wingfield | |
| CMW 31188 | GQ240658 | GQ240674 | GQ240690 | Eucalyptus sp. | Indonesia | M.J. Wingfield | |
| CMW 31189 | GQ240647 | GQ240663 | GQ240679 | Eucalyptus sp. | Indonesia | M.J. Wingfield | |
| CMW 31190 | GQ240648 | GQ240664 | GQ240680 | Eucalyptus sp. | Indonesia | M.J. Wingfield | |
| CMW 31191 | GQ240652 | GQ240668 | GQ240684 | Eucalyptus sp. | Thailand | M.J. Wingfield | |
| Ca. rumohrae | CBS 111431T | AF232871 | DQ190675 | FJ918549 | Rumohra adiantiformis | Panama | J.W. Miller |
| CBS 109062 | AF232873 | DQ190676 | FJ918550 | Adiantum sp. | The Netherlands | R. Pieters | |
| Ca. morganii | CBS 110666 | FJ918509 | FJ918527 | FJ918557 | Rosa sp. | USA | N.E. Ell-Gholl |
| Ca. spathiphylli | CBS 116168T | FJ918512 | FJ918530 | FJ918561 | Spathiphyllum sp. | USA | C.L. Schoulties |
| Ca. spathulata | CBS 112689 | AF308463 | FJ918524 | FJ918554 | E. viminalis | Brazil | N.E. Ell-Gholl |
| Ca. terrae-reginae sp. nov. | CMW 30601 (= CBS 112151 = | ||||||
| CPC 3202 = DFRI00150 = Lynfield 417)T | FJ918506 | FJ918522 | FJ918545 | E. urophylla | Australia | C. Hanwood | |
| CMW 30602 (= CBS 112634) | FJ918507 | DQ190668 | FJ918546 | Xanthorrhoea australis | Australia | T. Baigent | |
1 CBS: Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands; CMW: culture collection of the Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria Pretoria, South Africa; T All ex-type cultures.
A Markov Chain Monte Carlo (MCMC) algorithm was used to generate phylogenetic trees with Bayesian probabilities using MrBayes v3.1.1 (Ronquist & Huelsenbeck 2003) for the combined sequence datasets. Models of nucleotide substitution for each gene were determined using MrModeltest v2.3 (Nylander 2004) and included for each gene partition. Three analyses of four MCMC chains were run from random trees for 1 000 000 generations and sampled every 100 generations. All runs converged on the same likelihood score and tree topology and therefore, the first 7 600 trees for the Ca. reteaudii complex and 2 000 trees for the Ca. morganii complex were discarded as the burn-in phase of each analysis and posterior probabilities determined from the remaining trees.
Sexual compatibility
A total of 29 single conidial Ca. reteaudii-like isolates (Table 1), originating from various geographical regions were crossed in all possible combinations including mating tester isolates CBS 112144 (+) and CBS 112147 (–) (Kang et al. 2001). Crosses were made as described in Schoch et al. (1999) on carnation leaf agar (CLA; Fisher et al. 1982, Crous et al. 1993). Control inoculations consisted of isolates crossed with themselves to determine whether they had a heterothallic or a homothallic mating system. The plates were stacked in plastic containers and incubated at 22 °C for 6 wk. Crosses were regarded as successful when isolate combinations produced perithecia extruding viable ascospores.
Taxonomy
For morphological identification of Calonectria isolates, single conidial cultures were prepared on MEA and synthetic nutrient-poor agar (SNA; Nirenburg 1981, Lombard et al. 2009). Inoculated plates were incubated at room temperature and examined after 7 d. Gross morphological characteristics of the anamorph state were determined by mounting fungal structures in lactic acid and 30 measurements at × 1 000 magnification were made for each isolate. The 95 % confidence levels were determined and extremes of conidial measurements are given in parentheses. For other structures, only extremes are presented. Optimal growth conditions for cultures were determined in the dark on MEA for each isolate, at temperatures ranging from 5–35 °C at 5 °C intervals. Colony colours were determined after 7 d on MEA at 25 °C in the dark, using the colour charts of Rayner (1970).
RESULTS
DNA phylogeny
Amplicons of ± 500 bases (BT and TEF-1α) and 450 bases (HIS3) were obtained for all isolates. The pooled sequence datasets for all three loci showed conflict in tree topology for the 70 % reciprocal bootstrap trees, with Ca. spathulata (CBS 112689) and Cy. multiseptatum (CBS 112682) grouping within different clusters for all three gene regions considered (results not shown). This conflict was resolved by separating the sequence datasets for those representing morphological and phylogenetically closely related species to Ca. reteaudii and Ca. morganii. Sequence datasets for BT, HIS3 and TEF-1α were then combined for the two separate datasets. Sequence alignments were deposited in TreeBASE as SN4542.
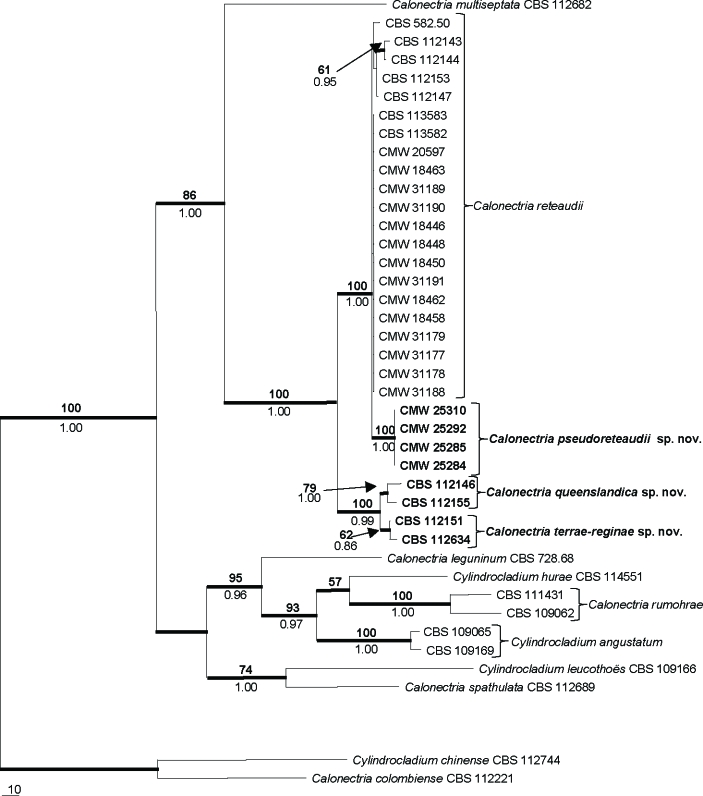
The combined sequence dataset for isolates representing Ca. reteaudii and other closely related species consisted of 1 540 characters, of which 926 were constant, 178 were parsimony uninformative and 436 were parsimony informative. Parsimony analysis of the alignment yielded six most parsimonious trees (TL = 1 316 steps; CI = 0.703; RI = 0.782; RC = 0.549), one of which is presented in Fig. 1. For Bayesian analyses, a HKY+G model was selected for BT, GTR+I+G for HIS3 and GTR+G for TEF-1α, and incorporated in the analyses. The consensus tree obtained from Bayesian analyses confirmed the tree topology obtained with parsimony as well as bootstrap support (Fig. 1).
Fig. 1.
One of six most parsimonious trees obtained from a heuristic search with 1 000 random additions of the combined sequences of β-tubulin, histone H3 and translation elongation factor-1α sequence alignments of the Ca. reteaudii complex and other closely related species. Scale bar shows 10 changes and bootstrap support values from 1 000 replicates are shown above the nodes in bold. Bayesian posterior probability values are indicated below the nodes. Thickened lines indicate branches in the strict consensus tree and the consensus tree of the Bayesian analyses. The tree was rooted to Ca. colombiensis (CBS 112221) and Cy. chinense (CBS 112744).
The isolates obtained from the Eucalyptus cuttings grouped in the Ca. reteaudii cluster, which formed a monophyletic group with a bootstrap value (BP) of 100 and a Bayesian posterior probability (PP) value of 1.00 (Fig. 1). This cluster segregated into two separate clades. The first of these, with a BP of 100 and PP value of 1.00 included the isolates from the Eucalyptus cuttings in a clade (BP = 100, PP = 1.00) separate from Ca. reteaudii, possibly representing a distinct species. The second clade (BP = 100, PP = 0.99) was comprised of two subclades (BP = 79, PP = 1.00 and BP = 62, PP = 0.86, respectively) representing isolates from the study of Kang et al. (2001), and also suggested the existence of distinct species.
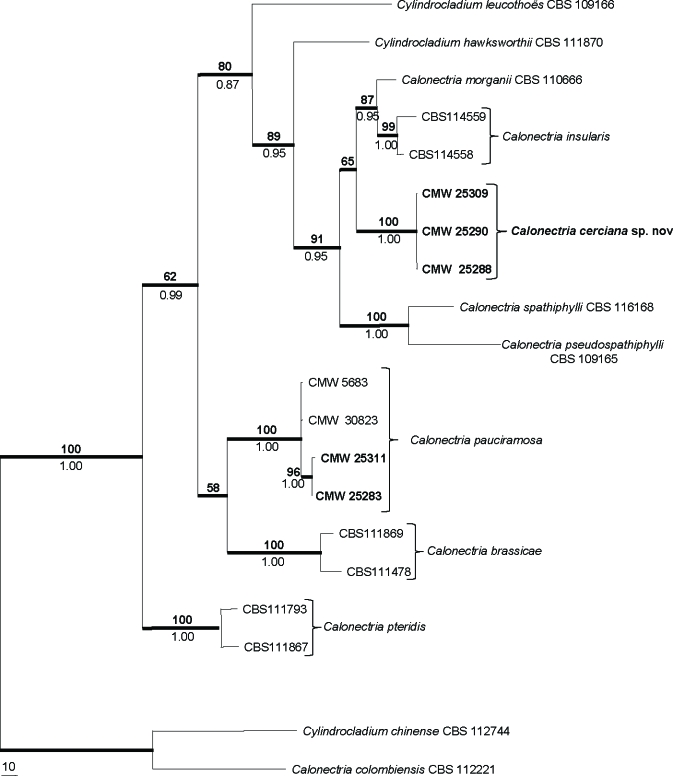
For the isolates that are closely related to Ca. morganii, the combined dataset consisted of 1 518 characters. Of these characters, 969 were constant, 161 were parsimony uninformative and 388 were parsimony informative. Parsimony analysis of the alignment yielded three most parsimonious trees (TL = 1 065 steps; CI = 0.736; RI = 0.783; RC = 0.577), one of which is represented in Fig. 2. For Bayesian analyses, a HKY+G model was selected for BT, GTR+I+G for HIS3, and GTR+G for TEF-1α, and incorporated in the analyses. The consensus tree obtained from Bayesian analyses confirmed the tree topology obtained with parsimony as well as bootstrap support (Fig. 2).
Fig. 2.
One of three most parsimonious trees obtained from a heuristic search with 1 000 random addition sequences of the combined sequences of β-tubulin, histone H3 and translation elongation factor-1α sequence alignments of the Ca. morganii complex and other closely related species. Scale bar shows 10 changes and bootstrap support values from 1 000 replicates are above the nodes in bold. Bayesian posterior probability values are indicated below the nodes. Thickened lines indicate branches in the strict consensus tree and the consensus tree of the Bayesian analyses. The tree was rooted to Ca. colombiensis (CBS 112221) and Cy. chinense (CBS 112744).
In the tree (Fig. 2), isolates from the Eucalyptus cuttings grouped in the cluster representing species in the Ca. morganii complex (BP = 89, PP = 0.95), and some in the cluster representing Ca. pauciramosa (BP = 100, PP = 1.00). However, those isolates grouping in the Ca. morganii cluster formed a separate clade (BP = 100, PP = 1.00), suggesting that they might represent a distinct species.
Sexual compatibility
Protoperithecia formed within 3 wk and mating tests produced viable perithecia within 6 wk on CLA. Crossing isolates CBS 112146, CBS 112151, CBS 112155 and CBS 112634 with isolates of Ca. reteaudii (represented in this phylogenetic study), failed to produce perithecia in any combination tested. Likewise, when isolates CMW 25284, CMW 25285, CMW 25292 and CMW 25310 were crossed with the isolates of Ca. reteaudii in this study, perithecia were also not found. However, the crossed isolates of Ca. reteaudii in this study, previously shown to represent different mating types by Kang et al. (2001), produced perithecia with viable ascospores. Isolate CBS 582.50, representing Ca. reteaudii, did not cross with any of the other Ca. reteaudii isolates, possibly due to loss of fertility.
Taxonomy
Morphological observations and DNA sequence comparisons (Fig. 2) showed that isolates CMW 25311 and CMW 25283 clearly represent the anamorph state of Ca. pauciramosa. Isolates CMW 25309, CMW 25290 and CMW 25288 represent an undescribed species closely related to other species in the Ca. morganii species complex, and it is consequently described as new. Isolates CMW 25310, CMW 25292, CMW 25285 and CMW 25284 obtained from the Eucalyptus cuttings are morphologically very similar to the anamorph state of Ca. reteaudii. However, some morphological differences were found and this fungus is treated as a new species. Based on DNA sequence analysis, mating strategy and morphological observations, isolates CBS 112146 (= CPC 3213) and CBS 112155 (= CPC 3210), which were previously regarded as Ca. reteaudii (Kang et al. 2001), are shown to represent a distinct species. A similar situation was found for isolates CBS 112151 (= CPC 3202) and CBS 112634 (= CPC 4233) also previously believed to represent Ca. reteaudii, and they are treated as a new species.
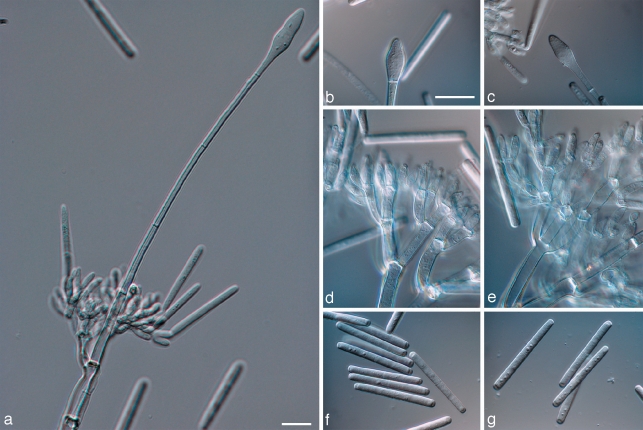
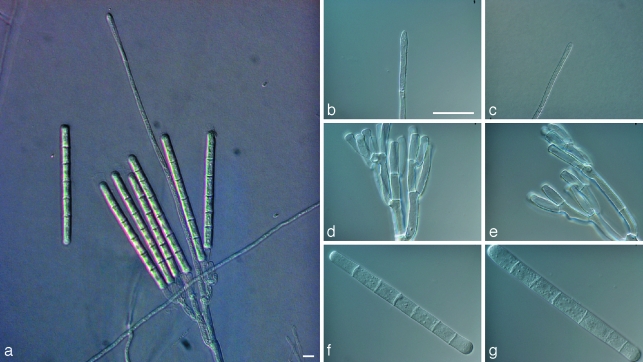
Calonectria cerciana L. Lombard, M.J. Wingf. & Crous, sp. nov. — MycoBank MB513263; Fig. 3
Fig. 3.
Calonectria cerciana. a. Macroconidiophore; b, c. fusiform to ellipsoidal vesicles; d, e. fertile branches with reniiform to doliiform phialides; f, g. macroconidia. — Scale bars = 10 μm.
Stipa extensiones septatae, rectae vel flexuosae, 148–222 μm longae, ad septum apicale 5–6 μm latae, vesiculo fusiforme vel obpyriforme 8–13 μm diametro terminantes. Macroconidia cylindrica, in extremitatibus ambabus rotundata, recta (37–)41–46(–49) × 5–6 μm, 1-septata.
Etymology. Name refers to the China Eucalypt Research Centre (CERC), a research institution that is pioneering the study of Eucalyptus diseases in China.
Teleomorph unknown. Conidiophores with a stipe bearing a suite of penicillate, fertile branches, a stipe extension, and a terminal vesicle; stipe septate, hyaline, smooth, 48–90 × 6–10 μm; stipe extensions septate, straight to flexuous, 148–222 μm long, 5–6 μm wide at the apical septum, terminating in fusiform to obpyriform vesicles, 8–13 μm diam. Conidiogenous apparatus 70–98 μm long, and 62–113 μm wide; primary branches aseptate or 1-septate, 21–31 × 5–7 μm; secondary branches aseptate, 15–22 × 4–5 μm; tertiary and additional branches (–4), aseptate, 10–20 × 4–5 μm, each terminal branch producing 2–6 phialides; phialides doliiform to reniform, hyaline, aseptate, 9–12 × 3 μm; apex with minute periclinal thickening and inconspicuous collarette. Macroconidia cylindrical, rounded at both ends, straight, (37–)41–46(–49) × 5–6 μm (av. = 44 × 5 μm), 1-septate, lacking a visible abscission scar, held in parallel cylindrical clusters by colourless slime. Megaconidia and microconidia not seen.
Culture characteristics — Colonies fast growing with optimal growth at 25 °C (growth at 15–30 °C) on MEA, reverse sepia-brown after 7 d; abundant white aerial mycelium and sporulation; chlamydospores abundant throughout the medium, forming microsclerotia.
Specimens examined. China, Guangdong Province, CERC nursery, on stems of E. urophylla × grandis hybrid cutting, Nov. 2007, M.J. Wingfield & X.D. Zhou, holotype PREM 60241, culture ex-type CMW 25309 = CBS 123693; CERC nursery, on stems of E. urophylla × grandis hybrid cutting, Nov. 2007, M.J. Wingfield & X.D. Zhou, PREM 60242, culture CMW 25290 = CBS 123695; CERC nursery, on stems of E. urophylla × grandis hybrid cutting, Nov. 2007, M.J. Wingfield & X.D. Zhou, culture CMW 25288.
Notes — Calonectria cerciana can be distinguished from Cy. hawksworthii (conidia av. 56 × 4 μm), Ca. morganii (conidia av. 45 × 4 μm) and Ca. insularis (conidia av. 45 × 4 μm) based on its fusoid to ellipsoidal vesicles. Macroconidia of Ca. cerciana are also slightly smaller (av. 44 × 5 μm) than those of the above-mentioned species.
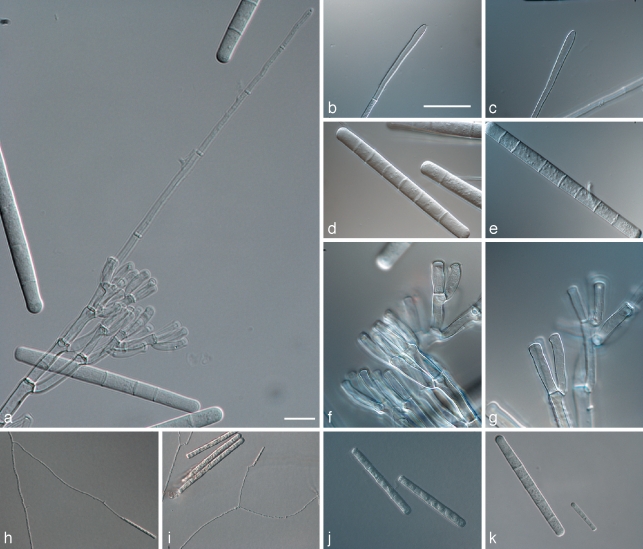
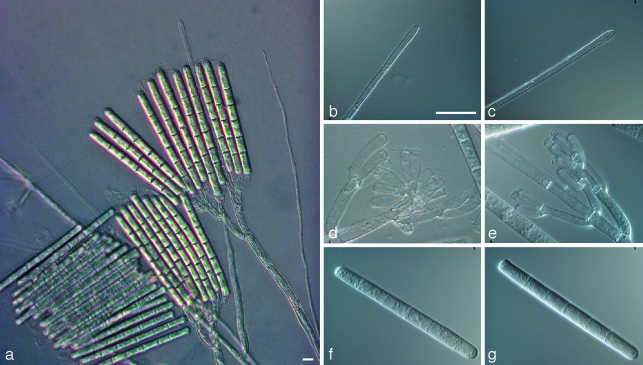
Calonectria pseudoreteaudii L. Lombard, M.J. Wingf. & Crous, sp. nov. — MycoBank MB513264; Fig. 4
Fig. 4.
Calonectria pseudoreteaudii. a. Macroconidiophore; b, c. clavate vesicle; d, e. macrocondia; f, g. fertile branches with cylindrical to allantoid phialides; h, i. microconidiophores; j. microconidia; k. comparison of macroconidia and microconidia. — Scale bars = 10 μm.
Stipa extensiones septatae, rectae vel flexuosae, 193–313 μm longae, ad septum apicale 5–6 μm latae, vesiculo anguste clavato, 3–5 μm diametro terminantes. Macroconidia cylindrica, apice rotundata, basi complanata, recta (88–)96–112(–119) × 7–9(–10) μm, 5–8-septata. Microconidia cylindrica, apice rotundata, basi complanata, (30–)34–54(–68) × 3–5(–6) μm, 1–3-septata, cum muco in glomerulis.
Etymology. Name reflects the fact that the species resembles the anamorph state of Ca. reteaudii.
Teleomorph unknown. Macroconidiophores with a stipe bearing a suite of penicillate fertile branches, a stipe extension, and a terminal vesicle; stipe septate, hyaline, smooth, 43–111 × 5–9 μm; stipe extensions septate, straight to flexuous, 193–313 μm long, 5–6 μm wide at the apical septum, terminating in a narrowly clavate vesicle, 3–5 μm diam. Conidiogenous apparatus 45–103 μm long, and 26–82 μm wide; primary branches aseptate or 1-septate, 29–42 × 5–6 μm; secondary branches aseptate, 20–36 × 3–6 μm; tertiary branches aseptate, 15–24 × 4–5 μm, each terminal branch producing 1–3 phialides; phialides cylindrical to allantoid, obpyriform when carried singly, hyaline, aseptate, 16–25 × 3–5 μm; apex with minute periclinal thickening and inconspicuous collarette. Macroconidia cylindrical, rounded at the apex, flattened at the base, straight, (88–)96–112(–119) × 7–9(–10) μm (av. = 104 × 8 μm), 5–8-septate, lacking a visible abscission scar, held in parallel cylindrical clusters by colourless slime. Microconidiophores simple with some lateral branching, comprising a stipe and a penicillate or subverticillate arrangement of fertile branches. Stipe septate, hyaline, thin-walled, 27–70 × 2–3 μm; primary branches aseptate, subcylindrical, straight to curved, 14–19 × 3–4 μm, terminating in 1–3 phialides that are straight to slightly curved, 11–18 × 3–4 μm; apex with minute periclinical thickening and collarette. Microconidia cylindrical, straight, rounded at the apex, flattened at the base, (30–)34–54(–68) × 3–5(–6) μm (av. 44 × 4 μm), 1–3-septate, held in fascicles by colourless slime. Megaconidia not seen.
Culture characteristics — Colonies fast growing with optimal growth at 25 °C (growth at 15–30 °C) on MEA, reverse amber to sepia-brown after 7 d; abundant white aerial mycelium with moderate sporulation; chlamydospores abundant throughout the medium, forming microsclerotia.
Specimens examined. China, Guangdong Province, CERC nursery, on stems of E. urophylla × grandis hybrid cutting, Nov. 2007, M.J. Wingfield & X.D. Zhou, holotype PREM 60290, culture ex-type CMW 25310 = CBS 123694; CERC nursery, on stems of E. urophylla × grandis hybrid cutting, Nov. 2007, M.J. Wingfield & X.D. Zhou, PREM 60291, culture CMW 25292 = CBS 123696; CERC nursery, on stems of E. urophylla × grandis hybrid cutting, Nov. 2007, M.J. Wingfield & X.D. Zhou, cultures CMW 25284 and CMW 25285.
Notes — The anamorph state of Ca. pseudoreteaudii can be distinguished from that of Ca. reteaudii based on its larger macro-conidia (av. = 104 × 8 μm vs 84 × 6.5 μm), as well as larger microconidia (av. = 44 × 4 μm vs 30 × 3 μm). The microconidiophores of Ca. pseudoreteaudii do not produce stipe extensions, a feature which is common in Ca. reteaudii.
Calonectria queenslandica L. Lombard, M.J. Wingf. & Crous, sp. nov. — MycoBank MB513265; Fig. 5
Fig. 5.
Calonectria queenslandica. a. Macroconidiophore; b, c. clavate vesicles; d, e. fertile branches with cylindrical to allantoid phialides; f, g. macroconidia — Scale bars = 10 μm.
Stipa extensiones septatae, rectae vel flexuosae, 105–156 μm longae, ad septum apicale 4–5 μm latae, vesiculo anguste clavato 3–4 μm diametro terminantes. Macroconidia cylindrica, in extremitatibus ambabus rotundata, recta (61–)65–73(–78) × (4–)5–6(–7) μm, 4–6-septata.
Etymology. Name refers to Queensland, Australia where the fungus was collected.
Teleomorph unknown. Conidiophores consisting of a stipe bearing a suite of penicillate fertile branches, a stipe extension, and a terminal vesicle; stipe septate, hyaline, smooth, 38–58 × 6–7 μm; stipe extensions septate, straight to flexuous, 105–156 μm long, 4–5 μm wide at the apical septum, terminating in narrowly clavate vesicle, 3–4 μm diam. Conidiogenous apparatus 39–64 μm long, and 27–68 μm wide; primary branches aseptate or 1-septate, 14–26 × 4–6 μm; secondary branches aseptate, 11–22 × 3–5 μm; tertiary branches aseptate, 13–17 × 3–5 μm, each terminal branch producing 1–3 phialides; cylindrical to allantoid, obpyriform when carried singly, hyaline, aseptate, 10–16 × 3–5 μm; apex with minute periclinal thickening and inconspicuous collarette. Macroconidia cylindrical, rounded at both ends, straight, (61–)65–73(–78) × (4–)5–6(–7) μm (av. = 69 × 6 μm), 4–6-septate, lacking a visible abscission scar, held in parallel cylindrical clusters by colourless slime. Megaconidia and microconidia not seen.
Culture characteristics — Colonies fast growing, with optimal growth at 25 °C (growth at 15–30 °C) on MEA; reverse sepia-brown after 7 d; abundant white aerial mycelium and sporulation; chlamydospores abundant throughout the medium, forming microsclerotia.
Specimens examined. Australia, Queensland, Lannercost, on leaves of E. urophylla, 15 Apr. 1991, B. Brown, holotype PREM 60243, culture ex-type CMW 30604 = CBS 112146 = CPC 3213 = DFRI00147; Queensland, Lannercost, on leaves of E. pellita, 10 Mar. 1999, P.Q Thu & K.M. Old, PREM 60244, culture CMW 30603 = CBS 112155 = CPC 3210 = DFRI00172.
Notes — Calonectria queenslandica can be distinguished from Ca. reteaudii and Ca. pseudoreteaudii based on its smaller macrocondia (av. = 69 × 6 μm) and shorter stipe extensions of the anamorph state. No microconidiophores were observed in Ca. queenslandica, although Ca. reteaudii and Ca. pseudoreteaudii readily produce these structures on SNA.
Calonectria terrae-reginae L. Lombard, M.J. Wingf. & Crous, sp. nov. — MycoBank 513266; Fig. 6
Fig. 6.
Calonectria terrae-reginae. a. Macroconidiophore; b, c. clavate vesicles; d, e. fertile branches with cylindrical to allantoid phialides; f, g. macroconidia. — Scale bars = 10 μm.
Stipa extensiones septatae, rectae vel flexuosae, 127–235 μm longae, ad septum apicale 4–6 μm latae, vesiculo anguste clavato 3–5 μm diametro terminantes. Macroconidia cylindrica, in extremitatibus ambabus rotundata, recta, 60–83(–87) × (4–)5–7(–8) μm, 4–6-septata.
Etymology. Name refers to Queensland, Australia, from where this fungus was isolated.
Teleomorph unknown. Conidiophores consisting of a stipe bearing a suite of penicillate fertile branches, a stipe extension, and a terminal vesicle; stipe septate, hyaline, smooth, 58–86 × 4–8 μm; stipe extensions septate, straight to flexuous, 127–235 μm long, 4–6 μm wide at the apical septum, terminating in narrowly clavate vesicle, 3–5 μm diam. Conidiogenous apparatus 35–54 μm long, and 33–48 μm wide; primary branches aseptate, 16–25 × 4–6 μm; secondary branches aseptate, 13–18 × 3–6 μm; tertiary branches aseptate, 10–14 × 3–5 μm, each terminal branch producing 1–3 phialides; cylindrical to allantiod, obpyriform when carried singly, hyaline, aseptate, 10–17 × 2–4 μm; apex with minute periclinal thickening and inconspicuous collarette. Macroconidia cylindrical, rounded at both ends, straight, 60–83(–87) × (4–)5–7(–8) μm (av. = 76 × 6 μm), 4–6-septate, lacking a visible abscission scar, held in parallel cylindrical clusters by colourless slime. Megaconidia and microconidia not seen.
Culture characteristics — Colonies fast growing with optimal growth temperature at 25 °C (growth at 15–30 °C) on MEA; reverse sepia-brown after 7 d; abundant white aerial mycelium and sporulation; chlamydospores abundant throughout the media, forming microsclerotia.
Specimens examined. Australia, Queensland, Cardwell, Meunga, on leaves of E. urophylla, 11 Apr. 1997, C. Hanwood, holotype PREM 60239, culture ex-type CMW 30601 = CBS 112151 = CPC 3202 = DFRI00150; Victoria, on Xanthorrhoea australis, T. Baigent, PREM 60240, culture CMW 30602 = CBS 112634 = CPC 4233 = Lynfield 417.
Notes — Calonectria terrae-reginae is distinct from Ca. queenslandica by having larger macroconidia (av. = 76 × 6 μm), although they are smaller than those of Ca. reteaudii and Ca. pseudoreteaudii.
DISCUSSION
This study emerged from the collection of isolates of Calonectria spp. from infected Eucalyptus cuttings in the Guangdong Province of China. The isolates were shown to represent three species including Ca. pauciramosa, and two new species that have been described as Ca. cerciana and Ca. pseudoreteaudii. The former species is related to taxa in the Ca. morganii complex, while the latter species resides in the Ca. reteaudii complex.
Taxonomic placement of Ca. pseudoreteaudii required a re-evaluation of the Ca. reteaudii complex. It was consequently found that the group has been poorly defined, and that it encompasses a number of cryptic species. This resulted in the description of Ca. pseudoreteaudii, Ca. queenslandica and Ca. terrae-reginae, three new sibling species in the Ca. reteaudii complex and distinguished based on phylogenetic inference and morphological comparisons with the ex-type culture of Ca. reteaudii (CBS 112144). Species in the Ca. reteaudii complex are important pathogens of Eucalyptus spp. causing Cylindrocladium leaf blight and cutting rot in Australia, South East Asia and South America (Pikethley 1976, Bolland et al. 1985, Sharma & Mohanan 1991, 1992, Booth et al. 2000, Crous & Kang 2001, Crous 2002, Rodas et al. 2005) and their refined taxonomy presented here will contribute to efforts to manage diseases caused by them.
Discovery of Ca. queenslandica and Ca. terrae-reginae was serendipitous as isolates used in the phylogenetic component of the study were largely the same as those used in a previous study by Kang et al. (2001). Although the latter study focused on the taxonomic position of Cy. quinqueseptatum (= Cy. reteaudii) and Ca. quinqueseptata (= Ca. leguminum), it employed a single gene region, and could thus not adequately define the variation in Ca. reteaudii, which was later recognised in multi-gene analyses (Crous et al. 2006a). Mating tests undertaken by Kang et al. (2001) indicated that 15 of the 20 Ca. reteaudii isolates used were capable of producing viable progeny. In the present study, it was not possible to successfully cross strains of Ca. queenslandica, Ca. terrae-reginae, and Ca. pseudoreteaudii, and only isolates of Ca. reteaudii could be induced to produce perithecia with viable ascospores.
Calonectria pauciramosa (anamorph: Cy. pauciramosum) is a well-known pathogen in Eucalyptus cutting nurseries (Schoch et al. 1999, Crous 2002). This pathogen resides in the Ca. scoparia species complex and is regarded as the dominant nursery pathogen of various plants in countries such as Australia, Italy, South Africa and the USA (Koike et al. 1999, Polizzi & Crous 1999, Schoch et al. 1999, 2001, Koike & Crous 2001). The present study represents the first report of this pathogen in China, but pathogenicity tests and diseases surveys will be required to determine its relevance in that country.
The description of Ca. cerciana from Eucalyptus cuttings adds a new species to the Ca. morganii species complex. Calonectria cerciana can be distinguished from the other species in the complex based on its smaller macroconidia and the formation of a fusiform to ellipsoidal vesicles. Crous (2002) and Schoch et al. (1999) found that there was a low level of fertility among species in this complex, and Schoch et al. (2001) used BT to show that they were closely related.
It is unknown whether Ca. cerciana and Ca. pseudoreteaudii are pathogens of Eucalyptus. Other species in the Ca. reteaudii (Sharma & Mohanan 1982, 1991, 1992) and Ca. morganii (Mohanan & Sharma 1985, Crous 2002) species complexes are known to be Eucalyptus pathogens, and this is probably also true for Ca. cerciana and Ca. pseudoreteaudii. However, the pathogenicity of these two Calonectria spp. must be tested and these studies would logically also consider the susceptibility of different Eucalyptus clones and hybrids being deployed in plantations.
Although no teleomorph states for the four newly described Calonectria spp. could be induced in this study, they have all been placed in Calonectria, and not in the anamorph genus Cylindrocladium. The decision to use the oldest generic name for a well-defined clade of fungi (Calonectria) is consistent with the approach taken previously by Lombard et al. (2009), and has also been followed in other groups of fungi such as Botryosphaeriaceae (Crous et al. 2006b, 2008, Phillips et al. 2008), Mycosphaerellaceae and Teratosphaeriaceae (Crous et al. 2009a, b), to name but a few. Although it might be considered a broad interpretation, it is allowed by the International Code of Botanical Nomenclature (Hawksworth 2005, McNeill et al. 2005). Based on these regulations and the fact that the anamorph genus Cylindrocladium (1892) is linked to the single teleomorph genus Calonectria (1867) (Rossman et al. 1999), all new species have been accommodated in Calonectria.
Acknowledgments
We thank members of the Tree Protection Cooperative Programme (TPCP), the Centraalbureau voor Schimmelcultures (CBS), and the University of Pretoria for financial and technical support to undertake the study. This study was initiated through the bilateral agreement between South Africa and China, and partially funded through the projects of 2007DFA31190, 2006BAD08A11 and CAFYBB2007024. We also thank Dr H. Glen, South African National Botanical Institute (SANBI), for the Latin descriptions and for valuable suggestions in naming the new species. The first author further acknowledges Drs J.Z. Groenewald, G.C. Hunter and C. Gueidan for advice regarding DNA sequence analyses.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic Local Alignment Search Tool. Journal of Molecular Biology 215: 403 – 410 . [DOI] [PubMed] [Google Scholar]
- Bolland L, Tierney JW, Tierney BJ. 1985. Studies on leaf spot and shoot blight of Eucalyptus caused by Cylindrocladium quinqueseptatum. European Journal of Forest Pathology 15: 385 – 397 . [Google Scholar]
- Booth TH, Jovanovic T, Old KM, Dudzinski MJ. 2000. Climatic mapping to identify high-risk areas for Cylindrocladium quinqueseptatum leaf blight on eucalypts in main land South East Asia and around the world. Environmental Pollution 108: 365 – 372 . [DOI] [PubMed] [Google Scholar]
- Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553 – 556 . [Google Scholar]
- Crous PW. 2002. Taxonomy and pathology of Cylindrocladium (Calonectria) and allied genera APS Press, St. Paul, Minnesota, USA: . [Google Scholar]
- Crous PW, Alfenas AC, Wingfield MJ. 1993. Calonectria scoparia and Calonectria morganii sp. nov., and variations among isolates of their Cylindrocladium anamorphs. Mycological Research 97: 701 – 708 . [Google Scholar]
- Crous PW, Groenewald JZ, Risède J-M, Simoneau P, Hyde KD. 2006a. Calonectria species and their Cylindrocladium anamorphs: species with clavate vesicles. Studies in Mycology 55: 213 – 226 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ, Risède J-M, Simoneau P, Hywel-Jones N. 2004. Calonectria species and their Cylindrocladium anamorphs: species with sphaeropedunculate vesicles. Studies in Mycology 50: 415 – 430 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Kang J-C . 2001. Phylogenetic confirmation of Calonectria spathulata and Cylindrocladium leucothoes based on morphology, and sequence data of the β-tubulin and ITS rRNA genes. Mycoscience 42: 51 – 57 . [Google Scholar]
- Crous PW, Slippers B, Wingfield MJ, Rheeder J, Marasas WFO, Phillips AJL, Alves A, Burgess T, Barber P, Groenewald JZ . 2006b. Phylogenetic lineages in the Botryosphaeriaceae. Studies in Mycology 55: 235 – 253 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Carnegie AJ, Wingfield MJ, Groenewald JZ . 2009a. Novel species of Mycosphaerellaceae and Teratosphaeriaceae. Persoonia 23: 119 – 146 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Carnegie AJ, Wingfield MJ, Hunter GC, Burgess TI, Andjic V, Barber PA, Groenewald JZ. 2009b. Unravelling Mycosphaerella: do you believe in genera? Persoonia 23: 99 – 118 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ. 1994. A monograph of Cylindrocladium, including anamorphs of Calonectria. Mycotaxon 51: 341 – 435 . [Google Scholar]
- Crous PW, Wood AR, Okada G, Groenewald JZ. 2008. Foliicolous microfungi occurring on Encephalartos. Persoonia 21: 135 – 146 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher NL, Burgess LW, Toussoun TA, Nelson PE. 1982. Carnation leaves as a substrate and for preserving cultures of Fusarium species. Phytopathology 72: 151 – 153 . [Google Scholar]
- Gueidan C, Roux C, Lutzoni F. 2007. Using multigene phylogeny analysis to assess generic delineation and character evolution in Verrucariaceae (Verrucariales, Ascomycota). Mycological Research 111: 1145 – 1168 . [DOI] [PubMed] [Google Scholar]
- Hawksworth DL. 2005. Two major changes in fungal nomenclature enacted in Vienna. Mycological Research 109: 1061 – 1062 . [Google Scholar]
- Hillis DM, Bull JJ. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42: 182 – 192 . [Google Scholar]
- Kang J-C, Crous PW, Old KM, Dudzinski MJ. 2001. Non-conspecificity of Cylindrocladium quinqueseptatum and Calonectria quinqueseptata based on a β-tubulin gene phylogeny and morphology. Canadian Journal of Botany 79: 1241 – 1247 . [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acid Research 33: 511 – 518 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike ST, Crous PW. 2001. First report of a root and crown rot disease of myrtle in California caused by Cylindrocladium pauciramosum. Plant Disease 85: 448 . [DOI] [PubMed] [Google Scholar]
- Koike ST, Henderson DM, Crous PW, Schoch CL, Tjosvold SA. 1999. A new root and crown rot disease of heath in California caused by Cylindrocladium pauciramosum. Plant Disease 83: 589 . [DOI] [PubMed] [Google Scholar]
- Lombard L, Bogale M, Montenegro F, Wingfield BD, Wingfield MJ. 2008. A new bark canker disease of the tropical hardwood tree Cedrelinga cateniformis in Ecuador. Fungal Diversity 31: 73 – 81 . [Google Scholar]
- Lombard L, Rodas CA, Crous PW, Wingfield BD, Wingfield MJ. 2009. Cylindrocladium species associated with dying Pinus cuttings. Persoonia 23: 41 – 47 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason-Gamer R, Kellogg E. 1996. Testing for phylogenetic conflict among molecular datasets in the tribe Tiriceae (Graminae). Systematic Biology 45: 524 – 545 . [Google Scholar]
- McNeill J, Stuessy TF, Turland NJ, Hörandl E. 2005. XVII International Botanical Congress: preliminary mail vote and report of Congress action on nomenclature proposals. Taxon 54: 1057 – 1064 . [Google Scholar]
- Mohanan C, Sharma JK. 1985. Cylindrocladium causing seedling diseases of Eucalyptus in Kerala, India. Transactions of the British Mycological Society 84: 538 – 539 . [Google Scholar]
- Nirenburg HI. 1981. A simplified method for identifying Fusarium spp. occurring on wheat. Canadian Journal of Botany 59: 1599 – 1609 . [Google Scholar]
- Nylander JAA. 2004. MrModeltest v.2. Programme distributed by the author. Evolutionary Biology Centre, Uppsala University; . [Google Scholar]
- O’Donnell K, Cigelnik E. 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103 – 116 . [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Kistler HC, Cigelnik E, Ploetz RC. 1998. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences of the United States of America 95: 2044 – 2049 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peerally A. 1991. The classification and phytopathology of Cylindrocladium species. Mycotaxon 40: 323 – 366 . [Google Scholar]
- Phillips AJL, Alves A, Pennycook SR, Johnston PR, Ramaley A, Akulov A, Crous PW. 2008. Resolving the phylogenetic and taxonomic status of dark-spored teleomorph genera in the Botryosphaeriaceae. Persoonia 21: 29 – 55 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikethley RN. 1976. Cylindrocladium quinqueseptatum on myrtaceous tree seedlings. Australian Plant Pathology Newsletter 5: 57 . [Google Scholar]
- Polizzi G, Crous PW. 1999. Root and collar rot of milkwort caused by Cylindrocladium pauciramosum, a new record for Europe. European Journal of Plant Pathology 105: 407 – 411 . [Google Scholar]
- Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817 – 818 . [DOI] [PubMed] [Google Scholar]
- Rayner RW. 1970. A mycological colour chart. Commonwealth Mycological Institute and British Mycological Society Kew, Surrey, UK: . [Google Scholar]
- Rodas CA, Lombard L, Gryzenhout M, Slippers B, Wingfield MJ. 2005. Cylindrocladium blight of Eucalyptus in Colombia. Australasian Plant Pathology 34: 143 – 149 . [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572 – 1574 . [DOI] [PubMed] [Google Scholar]
- Rossman AY, Samuels GJ, Rogerson CT, Lowen R. 1999. Genera of Bionectriaceae, Hypocreaceae and Nectriaceae (Hypocreales, Ascomycetes). Studies in Mycology 42: 1 – 248 . [Google Scholar]
- Schoch CL, Crous PW, Wingfield BD, Wingfield MJ. 1999. The Cylindrocladium candelabrum species complex includes four distinct mating populations. Mycologia 91: 286 – 298 . [Google Scholar]
- Schoch CL, Crous PW, Wingfield BD, Wingfield MJ. 2001. Phylogeny of Calonectria based on comparisons of β-tubulin DNA sequences. Mycological Research 105: 1045 – 1052 . [Google Scholar]
- Sharma JK, Mohanan C. 1982. Cylindrocladium spp. associated with various diseases of Eucalyptus in Kerala. European Journal of Forest Pathology 12: 129 – 136 . [Google Scholar]
- Sharma JK, Mohanan C. 1991. Pathogenic variation in Cylindrocladium quinqueseptatum causing leaf blight of Eucalyptus. European Journal of Forest Pathology 21: 210 – 217 . [Google Scholar]
- Sharma JK, Mohanan C. 1992. Relative susceptibility of Eucalyptus provenances to Cylindrocladium leaf blight in Kerala, India. European Journal of Forest Pathology 22: 257 – 265 . [Google Scholar]
- Sharma JK, Mohanan C, Florence EJM. 1984. Nursery diseases of Eucalyptus in Kerala. European Journal of Forest Pathology 14: 77 – 89 . [Google Scholar]
- Swofford DL. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods) v4.0b10. Computer programme Sinauer Associates, Sunderland, Massachusetts, USA: . [Google Scholar]
- Zhou XD, Xie YJ, Chen SF, Wingfield MJ. 2008. Diseases of eucalypt plantations in China: challenges and opportunities. Fungal Diversity 32: 1 – 7 . [Google Scholar]